Abstract
Three new species of Collybia in China, Collybia clavipes, C. carnea and C. violea, are originally reported and described based on morphological characteristics and molecular data. This study provides detailed morphological descriptions of these three new species of Collybia, which can be accurately distinguished from other species within the genus Collybia. Phylogenetic relationships of Clitocybaceae were analyzed using a four-loci combined dataset (ITS-nrLSU-rpb2-tef1-α), and the results show that the three newly discovered species of Collybia form three distinct lineages, respectively. Based on the combination of morphological and molecular methods, these three newly collected species of Collybia are confirmed as new to science. A theoretical basis is provided for the species diversity of Collybia.
1. Introduction
Based on phylogenetic analyses over the past two decades, many new genera have been established to accommodate traditional Clitocybe species [1], such as Cleistocybe Ammirati, A.D. Parker & Matheny [2], Trichocybe Vizzini [3], Atractosporocybe P. Alvarado, G. Moreno & Vizzini, Leucocybe Vizzini, P. Alvarado, G. Moreno & Consiglio and Rhizocybe Vizzini, G. Moreno, P. Alvarado & Consiglio [4], Pulverulina Matheny & K.W. Hughes 2020 [5] and Spodocybe Z.M. He & Zhu L. Yang [6]. However, it is noteworthy that Clitocybe remains polyphyletic [1,4,7,8,9,10,11,12,13]. When He et al. [14], based on phylogenetic and phylogenomic analyses, reconstructed the phylogenetic framework of Clitocybaceae and recircumscribed the genera within the family, a large number of traditional Clitocybe and Lepista species were transferred to different clades of the genus Collybia, and the two genera became monophyletic groups.
Species of Collybia have a widespread distribution all over the world. However, new taxa of Collybia had rarely been reported and described in China in recent years until He et al. [14], who proposed fourteen new Collybia species in China. In this present study, several interesting specimens of Collybia were collected from southwestern and northeastern China. To identify whether these specimens of Collybia are new to science, detailed phylogenetic and morphological examinations were conducted, resulting in possibly novel additions to the genus Collybia.
2. Materials and Methods
2.1. Specimens and Morphological Studies
Specimens of Collybia were collected from southwestern and northeastern China from 2019 to 2024. Some basidiomes were dehydrated with silica gel and used for DNA extraction; the remaining basidiomes were dehydrated with an electric drying oven at 50 °C and stored in the Fungal Herbarium of Shenyang Agricultural University (SYAU-FUNGI). Detailed information is given in Table 1. Methods for morphological observation followed Qi et al. [15]. The color codes followed the Methuen Handbook of Color [16]. For observation of the spore surface, the gills of the dried specimens were coated with gold and examined with a ZEISS Ultra Plus Scanning Electron Microscope (SEM) (Oberkochen, Germany).

Table 1.
Information on newly generated sequences in this study.
2.2. DNA Extraction, PCR, and Sequencing
Genomic DNA was extracted from dried specimens using the cetyltrimethylammonium bromide (CTAB) method [17]. Polymerase chain reaction (PCR) was performed to amplify sequences of the ITS, nrLSU, rpb2, and tef1-α regions. Primers ITS1 and ITS4 [18] were used for the ITS region; primers LROR and LR5 [19] were used for the nrLSU region; primers bRPB2-6F and bRPB2-7.1R [20] were used for the rpb2 region; and primers EF1-983F and EF1-1953R [21] were used for the tef1-α region. The PCR procedure was performed following Qi et al. [15].
2.3. Phylogenetic Analyses
Based on the latest phylogenetic studies on Clitocybaceae [14], high-quality sequences of Clitocybaceae species were retrieved and aligned with the newly generated sequences in this study that were checked and edited using Bioedit v7.0.9 [22]. Alignments were generated for single ITS, nrLSU, rpb2, and tef1-α datasets using MAFFT v7.313 [23]. Then, a four-locus combined dataset for Clitocybaceae, containing sequences of ITS, nrLSU, rpb2, and tef1-α, was generated by the PhyloSuite v1.2.1 software platform [24]. ModelFinder [25] was used for the selection of the best-fitting evolution model for the combined dataset. Maximum Likelihood (ML) analysis was performed with RAxML-8.2.10-WIN [26] with 1000 bootstrap replicates using the TIM2 model for the combined dataset. Bayesian Inference (BI) analysis was conducted with MrBayes v.3.2.6 [27] using the GTR+I+G model for the combined dataset, and the combined dataset was run for 8,000,000 generations. The best tree was viewed in FIGTREE v1.4.4 [28] and enhanced in Adobe Illustrator CC 2022.
3. Results
3.1. Phylogenetic Analyses
In the preliminary molecular analysis, C. clavipes sequences showed 96.4% similarity to C. hunanensis (NR_198326) with 98% query coverage for ITS, 99.4% similarity to C. tibetica (NG_243147) with 100% query coverage for nrLSU, 97.2% similarity to Clitocybe dealbata (DQ825407) with 100% query coverage for rpb2, and 80.5% similarity to C. bisterigmata (OP558049) with 97% query coverage for tef1-α. Collybia carnea sequences showed 97.8% similarity to C. pannosa (NR_198323) with 100% query coverage for ITS, 99.5% similarity to C. phyllophila (MK277723) with 99% query coverage for nrLSU, 96.2% similarity to C. xylogena (OP956582) with 92% query coverage for rpb2, and 94.6% similarity to C. xylogena (OP672233) with 60% query coverage for tef1-α. Collybia violea sequences showed 95.5% similarity to C. nuda (OP626948) with 100% query coverage for ITS, 98.7% similarity to C. sordida (OP646393) with 100% query coverage for nrLSU, 85.4% similarity to C. nuda (KJ136110) with 100% query coverage for rpb2, and 82.8% similarity to C. nuda (MG702630) with 100% query coverage for tef1-α.
In the phylogenetic analysis, this study analyzed 411 sequences (109 ITS, 108 nrLSU, 98 rpb2, 96 tef1-α) from 110 samples representing 44 species of Clitocybaceae. The combined dataset ITS-nrLSU-rpb2-tef1-α of Clitocybaceae comprised a total of 2792 positions. ML and BI resulted in almost identical tree topologies, and the BI tree was selected for display (Figure 1). The phylogenetic relationships of Clitocybaceae (Figure 1) were consistent with previous work [14].
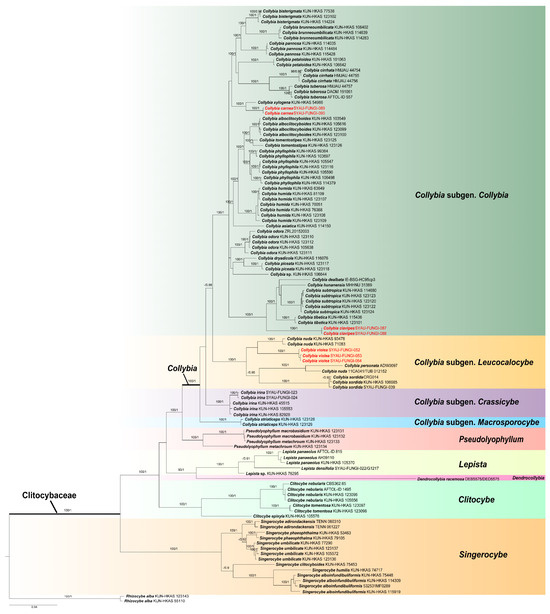
Figure 1.
Phylogram of Clitocybaceae generated based on the combined ITS-nrLSU-rpb2-tef1-α dataset, with Rhizocybe alba as an outgroup. Bootstrap values (BP) ≥ 90% and Bayesian posterior probabilities (PP) ≥ 0.90 are shown around the branches. Newly generated sequences are shown in red.
The optimal topology indicates that Clitocybaceae represents a monophyletic lineage with high support (100% BP, 1.00 PP). As shown in the phylogenetic analyses of Clitocybaceae (Figure 1), a total of six clades can be recognized within the family as the following genera: Collybia, Pseudolyophyllum, Lepista, Dendrocollybia, Singerocybe and Clitocybe. This is in line with He et al. [14]. Sequences of Collybia species are divided into four clades, which is in line with He et al. [14]. In this present study, three new species, C. clavipes, C. carnea and C. violea, form a single clade with high support (100% BP, 1.00 PP). In detail, C. clavipes and C. carnea are located within the Collybia subgen. Collybia, and C. violea is located within the Collybia subgen. Leucocalocybe.
3.2. Taxonomy
Collybia clavipes A.G. Xu & Y. Qi, sp. nov. (Figure 2)
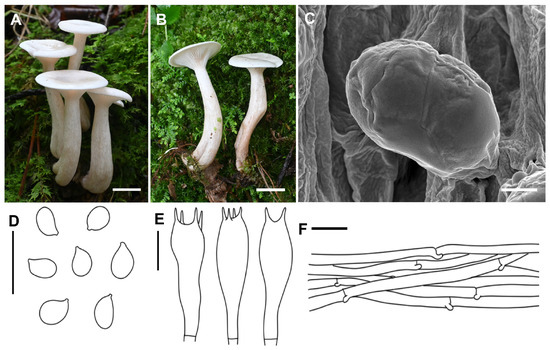
Figure 2.
Morphological features of Collybia clavipes (SYAU-FUNGI-087, holotype). (A,B) Habitat and ba basidiomes; (C) SEM images of basidiospores. Line drawings of (D) basidiospores; (E) basidia; (F) pileipellis. Scale bar: (A,B) 1 cm; (C) 1 μm; (D–F) 10 μm.
MycoBank: MB858330
Etymology: “clavipes” refers to the clavate stipe.
Diagnosis: Differed from other known species of the subgenus by having small and white basidiomata, a clavate stipe and ellipsoid to sublacrymoid basidiospores.
Holotype: China, Xizang, Jilong County, in a mixed forest, 3104 m alt., 2 August 2023, A.G. Xu & Y. Qi (SYAU-FUNGI-087, holotype).
Description: Basidiomata small. Pileus 1.0–3.0 cm in width, applanate to slightly depressed at center; surface glabrous, white (1A1), hygrophanous; margin not striate, slightly inrolled. Lamellae decurrent, moderately crowded, 2–3 mm wide, white (1A1), with numerous tiers of lamellulae; edge entire or slightly eroded. Stipe 2.5–4.5 cm long × 0.5–1.2 cm wide, central, subclavate, hollow, gradually becoming enlarged downwards, sometimes slightly curved, surface white (1A1), brownish (6D4–8), longitudinally striate. Context thin, white.
Basidiospores (4.0) 5.0–6.0 (6.5) × 3.0–4.0 (4.5) μm, Q = 1.32–1.87, Qav = 1.64, ellipsoid to sublacrymoid, inamyloid, surface smooth under both the light microscope and electron microscope, not adherent, almost always single. Basidia 25.6–32.0 × 6.0–9.5 μm, clavate, four-spored, sterigmata up to 3.0–6.0 μm in length. Cystidia absent. Hymenophoral trama regular, hyphae cylindrical, hyphae up to 3.5–9.6 µm wide. Pileipellis hyphae cylindrical, hyphae up to 2.0–10.5 µm wide, with many branches. Stipitipellis hyphae cylindrical, hyphae up to 3.2–9.5 µm wide. Clamp connections present.
Habitat: Generally gregarious, more rarely solitary; saprotrophic on needle and broad leaf litter, in a mixed forest; summer.
Distribution: Only known from Xizang, China.
Additional material studied: China, Xizang, Jilong County, in a mixed forest, 3014 m alt., 20 August 2024, A.G. Xu & Y. Qi (SYAU-FUNGI-088).
Notes: Within Collybia subgen. Collybia, several species also have small and white basidiomes, such as C. asiatica, C. bisterigmata and C. pannosa [14]. However, the clavate stipe and ellipsoid to sublacrymoid basidiospores of C. clavipes make it easily distinguished from these species, which generally have a cylindrical stipe and non-sublacrymoid basidiospores. Furthermore, C. asiatica differs from C. clavipes by its infundibuliform basidiome and having basidiospores always adhering in tetrads [14]; C. bisterigmata differs by having two-spored basidia and larger basidiospores (6.0–7.5 (8.0) × 4.0–6.0 μm) [14]; C. pannosa differs by having felty pileus, basidiospores sometimes adhering in tetrads and irregularly arranged pileipellis [14].
Collybia carnea A.G. Xu & Y. Qi, sp. nov. (Figure 3)
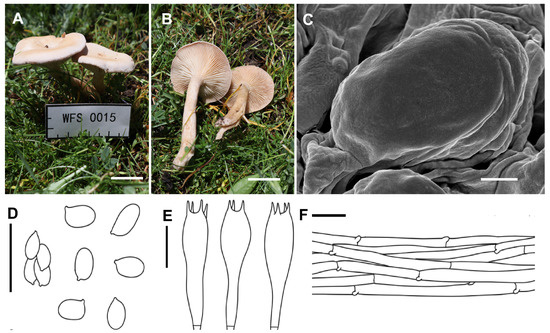
Figure 3.
Morphological features of Collybia carnea (SYAU-FUNGI-089, holotype). (A,B) Habitat and ba basidiomes; (C) SEM images of basidiospores. Line drawings of (D) basidiospores; (E) basidia; (F) pileipellis. Scale bar: (A,B) 1 cm; (C) 1 μm; (D–F) 10 μm.
MycoBank: MB858331
Etymology: “carnea” referring to a carneous basidiome.
Diagnosis: Differed from C. dealbata by having a carneous basidiome.
Holotype: China, Xizang, Dangxiong County, Ara wetland, in a mixed forest, 3104 m alt., 2 August 2024, A.G. Xu & Y. Qi (SYAU-FUNGI-089, holotype).
Description: Basidiomata small. Pileus 2.0–3.0 cm in width, plano-convex to nearly applanate, somewhat depressed at center; surface pinkish brown (5A2–3 to 6A2), becoming light brown (6B2–3) when old, white–pruinose, sometimes slightly lobate, not hygrophanous; margin not striate, inrolled, sometimes irregular, undulating. Lamellae adnate to subdecurrent, moderately crowded, thin, 1.0–2.0 mm wide, concolorous with pileus, with numerous tiers of lamellulae; edge entire. Stipe 4.0–5.0 cm long × 0.7–1.0 cm wide, central, cylindrical, hollow, sometimes slightly curved, concolorous with pileus, longitudinally striate, base not inflated, sometimes covered with whitish mycelium. Context about 1 cm thick, fleshy.
Basidiospores 4.5–6.0 × 2.5–4.0 (4.5) μm, Q = 1.30–2.02, Qav = 1.67, ellipsoid to elongate, inamyloid, surface smooth under both the light microscope and electron microscope, often single, rarely adherent. Basidia 25.5–31.0 × 6.2–8.6 μm, clavate, four-spored, hyaline, with sterigmata up to 2.0–6.5 μm in length. Cystidia absent. Hymenophoral trama regular, hyphae cylindrical, hyphae up to 3.5–9.6 µm wide. Pileipellis hyphae cylindrical, hyphae up to 3.0–9.5 µm wide, with many branches. Stipitipellis hyphae cylindrical, hyphae up to 3.0–8.5 µm wide. Clamp connections present.
Habitat: Scattered on rich soil among grasses, at the edge of a mixed forest, summer.
Distribution: Only known from Xizang, China.
Additional material studied: China, Xizang, Dangxiong County, on rich soil among grasses at the edge of a mixed forest, 3214 m alt., 20 August 2024, A.G. Xu & Y. Qi (SYAU-FUNGI-090).
Notes: Phylogenetically, Collybia carnea represents an independent clade and is a sister taxon to C. xylogena (Figure 1). Even so, C. xylogena can be easily distinguished by its brown to grayish brown pileus, relatively larger basidiospores (5.0–8.0 × 3–4.5 μm), and growing on wood [14]. Morphologically, C. carnea is closely related to C. dealbata, which also produces a small basidiome, a white–pruinose pileus with inrolled margin and medium-sized basidiospores, which are always mostly single [29,30]). However, Collybia carnea have a carneous basidiome, while C. dealbata have a white basidiome.
Collybia violea X.D. Yu & H.B. Guo, sp. nov. (Figure 4)
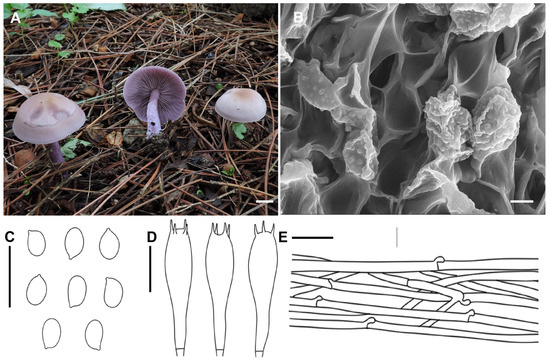
Figure 4.
Morphological features of Collybia violea (SYAU-FUNGI-052, holotype). (A) Habitat and basidiomes; (B) SEM images of basidiospores. Line drawings of (C) basidiospores; (D) basidia; (E) pileipellis. Scale bar: (A) 1 cm; (B) 2 μm; (C–E) 10 μm.
MycoBank: MB858332
Etymology: “violea” refers to violet basidiome.
Diagnosis: Distinguished from C. sordida and C. nuda by its slightly darker pileus color, and slightly smaller basidiospores.
Holotype: China, Liaoning Province, Shenyang City, Dongling Park, on rich soil among grasses, 177 m alt., 17 July 2019, X.D. Yu & H.B. Guo (SYAU-FUNGI-052, holotype).
Description: Basidiomata medium. Pileus 4.0–7.0 cm in width, at first convex, then gradually applanate, sometimes slightly depressed; surface lilac to grayish lilac (15 B2–3), becoming light brown (6C2–3) to ochraceous (6D5–8) from the center outwards; margin slightly inflexed when young, sometimes uplifted, irregular, undulating when mature. Lamellae adnexed to emarginate to subdecurrent, moderately crowded, thin, 2.0–3.0 mm wide, grayish violet to violet (17 B2–4), with numerous tiers of lamellulae; edge entire or slightly eroded. Stipe 2.5–5.0 cm long × 0.4–0.8 cm wide, central, cylindrical, solid or hollow, concolorous with lamellae, longitudinally striate, base slightly inflated. Context about moderately thick, fleshy.
Basidiospores 4.5–7.0 × 3.2–4.0 μm, Q = 1.30–1.82, Qav = 1.65, ellipsoid, inamyloid, surface smooth under the light microscope, finely verruculose under the electron microscope, not adherent, almost always single. Basidia 25.5–30.5 × 7.0–9.6 μm, clavate, four-spored, with sterigmata up to 3.0–5.5 μm in length. Cystidia absent. Hymenophoral trama regular, hyphae cylindrical, hyphae up to 3.0–9.2 µm wide. Pileipellis hyphae cylindrical, hyphae up to 2.5–9.5 µm wide, with many branches. Stipitipellis hyphae cylindrical, hyphae up to 2.0–8.5 µm wide. Clamp connections present.
Habitat: Saprophytic in small groups on rich soil among grasses, on roadsides.
Distribution: Known from northeastern China.
Additional material studied: China, Liaoning Province, Shenyang City, on the campus of Shenyang Agricultural University, on rich soil among grasses, 121 m alt., 31 August 2019, X.D. Yu & H.B. Guo (SYAU-FUNGI-053); China, Liaoning Province, Shenyang City, on the campus of Shenyang Agricultural University, on rich soil among grasses, 121 m alt., 31 August 2019, X.D. Yu & H.B. Guo (SYAU-FUNGI-054).
Notes: Collybia sordida and C. nuda could be easily mistaken for C. violea because they have lilac to grayish lilac basidiomata. Collybia sordida mainly differs by its hygrophanous and more dull-colored (pale bluish violet then pinkish beige with tinge olive at the center and in concentric zones on drying) pileus [31], and slightly larger basidiospores (7 × 4 μm [31]; 6–9 × 4–5 μm [32]). C. nuda can distinguish from C. violea by having much larger basidiomata (8–15 cm [31]; 2.5–10 cm [32]; 5–15 cm [33]) and slightly larger basidiospores (8 × 5 μm [31]; 5.5–9 × 4–5.5 μm [32]; 6.5–8.5 × 3.9–4.8 μm [33]).
4. Discussion
In the combined ITS-nrLSU-rpb2-tef1-α phylogenetic tree (Figure 1), Clitocybaceae forms a monophyletic clade, which is consistent with the previous study [14]. The three new species of Collybia described in this study occupy three independent positions, respectively. Collybia clavipes forms a distinct clade within the clade containing C. dealbata, C. hunanensis, C. subtropica and C. tibetica. Collybia carnea occupies a clade with the sister taxon, C. xylogena, but morphologically, C. carnea differs from C. xylogena by having a pinkish basidiome, smaller spores and growing on soil. Collybia violea forms a sister group with C. nuda, but morphologically, C. violea can be distinguished by having a smaller basidiome and smaller spores.
In this study, two new species belongs to the Collybia subgen. Collybia, which is a species-rich subgenus, with useful diagnostic features, such as basidiospore adhesion, two-spored or four-spored basidia, muscarine existence or non-existence, and growth substrates [14]. To some extent, this study further confirmed these diagnostic features’ usefulness to separate supraspecific taxa within the subgenus. For example, C. clavipes clusters with C. subtropica, C. hunanensis, C. dealbata and C. tibetica (Figure 1), all of which have four-spored basidia, basidiospores which are almost always single and grow on leaf litter or soil [14]. However, four-spored C. carnea has basidiospores which are always single and grow on soil, while two-spored C. xylogena has basidiospores which are often in tetrads and grow on rotten wood [14], although C. carnea clusters together with C. xylogena (Figure 1). This indicates that there may be still a number of new taxa remaining to be discovered within this clade.
Collybia violea belongs to Collybia subgen. Leucocalocybe. Due to interspecific similarities and phenotypic plasticity, C. violea has always been misidentified as C. sordida or C. nuda. However, our study finds there are distinct differences between C. violea and these species in both molecular data and morphological characteristics. Furthermore, previous phylogenetic analysis [14] based on worldwide ITS sequences of this subgenus suggested that there are five subclades of C. nuda. In the future, large amounts of related specimens need to be collected and detailed morphological studies need to be conducted to resolve the puzzles concerning which subclade could represent the true C. nuda.
Author Contributions
Conceptualization, Y.Q. and X.Y.; methodology, Y.G.; software, F.W.; validation, R.Y., Y.P. and X.Y.; formal analysis, Y.Q.; investigation, H.G.; resources, A.X.; data curation, L.Y.; writing—original draft preparation, Y.Q.; writing—review and editing, Y.Q. and X.Y.; visualization, A.X.; supervision, X.Y.; project administration, A.X.; funding acquisition, R.Y. and X.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China (No. 32370008), Investigation and Evaluation of Characteristic Biological Resources in Xizang (XZ202301YD0007C), Natural Science Foundation of Liaoning Province (2024-MSLH-344), Development Plan of Xizang (XZ202201ZY0010N) and Shenyang Institute of Technology Experimental Technology Fund Project (SYJS202407).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All of the data that support the findings of this study are available in the main text.
Conflicts of Interest
Author Ying Pei was employed by the company Shenyang Research Institute of Chemical Industry. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Matheny, P.B.; Hofstetter, V.; Aime, M.C.; Moncalvo, J.M.; Ge, Z.M.; Yang, Z.L.; Slot, J.C.; Ammirati, J.F.; Baroni, T.J.; Bougher, N.L.; et al. Major clades of Agaricales: A multilocus phylogenetic overview. Mycologia 2006, 98, 982–995. [Google Scholar] [CrossRef] [PubMed]
- Ammirati, J.F.; Parker, A.D.; Matheny, P.B. Cleistocybe, a new genus of Agaricales. Mycoscience 2007, 48, 282–289. [Google Scholar] [CrossRef]
- Vizzini, A.; Musumeci, E.; Murat, C. Trichocybe, a new genus for Clitocybe puberula (Agaricomycetes, Agaricales). Fungal Divers. 2010, 42, 97–105. [Google Scholar] [CrossRef]
- Alvarado, P.; Moreno, G.; Vizzini, A.; Consiglio, G.; Manjón, J.L.; Setti, L. Atractosporocybe, Leucocybe and Rhizocybe: Three new clitocyboid genera in the Tricholomatoid clade (Agaricales) with notes on Clitocybe and Lepista. Mycologia 2015, 107, 123–136. [Google Scholar] [CrossRef]
- Matheny, P.B.; Hughes, K.W.; Kalichman, J.; Lebeuf, R. Pulverulina, a new genus of Agaricales for Clitocybe ulmicola. Southeast. Nat. 2020, 19, 447–459. [Google Scholar] [CrossRef]
- He, Z.M.; Yang, Z.L. A new clitocyboid genus Spodocybe and a new subfamily Cuphophylloideae in the family Hygrophoraceae (Agaricales). MycoKeys 2021, 79, 129–148. [Google Scholar] [CrossRef] [PubMed]
- Moncalvo, J.M.; Vilgalys, R.; Redhead, S.A.; Johnson, J.E.; James, T.Y.; Catherine Aime, M.; Hofstetter, V.; Verduin, S.J.; Larsson, E.; Baroni, T.J.; et al. One hundred and seventeen clades of euagarics. Mol. Phylogenetics Evol. 2002, 23, 357–400. [Google Scholar] [CrossRef]
- Walther, G.; Garnica, S.; Weiß, M. The systematic relevance of conidiogenesis modes in the gilled Agaricales. Mycol. Res. 2005, 109, 525–544. [Google Scholar] [CrossRef]
- Binder, M.; Larsson, K.H.; Matheny, P.B.; Hibbett, D.S. Amylocorticiales ord. nov. and Jaapiales ord. nov.: Early diverging clades of agaricomycetidae dominated by corticioid forms. Mycologia 2010, 102, 865–880. [Google Scholar] [CrossRef]
- Sánchez-García, M.; Matheny, P.B.; Palfner, G.; Lodge, D.J. Deconstructing the Tricholomataceae (Agaricales) and introduction of the new genera Albomagister, Corneriella, Pogonoloma and Pseudotricholoma. Taxonomy 2014, 63, 993–1007. [Google Scholar] [CrossRef]
- Alvarado, P.; Moreau, P.A.; Sesli, E.; Khodja, L.Y.; Contu, M.; Vizzini, A. Phylogenetic studies on Bonomyces (Tricholomatineae, Agaricales) and two new combinations from Clitocybe. Cryptogam. Mycol. 2018, 39, 149–168. [Google Scholar] [CrossRef]
- Alvarado, P.; Moreau, P.A.; Dima, B.; Vizzini, A.; Consiglio, G.; Moreno, G.; Setti, L.; Kekki, T.; Huhtinen, S.; Liimatainen, K.; et al. Pseudoclitocybaceae fam. nov. (Agaricales, Tricholomatineae), a new arrangement at family, genus and species level. Fungal Divers. 2018, 90, 109–133. [Google Scholar] [CrossRef]
- He, Z.M.; Yang, Z.L. The genera Bonomyces, Harmajaea and Notholepista from Northwestern China: Two new species and a new record. Mycol. Prog. 2022, 21, 26. [Google Scholar] [CrossRef]
- He, Z.M.; Chen, Z.H.; Bau, T.; Wang, G.S.; Yang, Z.L. Systematic arrangement within the family Clitocybaceae (Tricholomatineae, Agaricales): Phylogenetic and phylogenomic evidence, morphological data and muscarine-producing innovation. Fungal Divers. 2023, 123, 1–47. [Google Scholar] [CrossRef]
- Qi, Y.; Yu, X.D.; Hou, J.X.; Guo, H.B.; Yang, R.H.; Xu, A.G. Addition to the genus Harmajaea (Agaricales, Pseudoclitocybaceae): A new and a known species from China. Phytotaxa 2024, 665, 157–166. [Google Scholar] [CrossRef]
- Kornerup, A.; Wanscher, J.H. Methuen Handbook of Colour; Methuen and Co., Ltd.: London, UK, 1963; 242p. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes from phylogenetics. In PCR Protocols: Methods and Applications; Innes, M.A., Gelfand, D.H., Sninsky, J.S., White, T.J., Eds.; Academic Press: London, UK, 1990; pp. 315–322. [Google Scholar]
- Vilgalys, R.; Hestsr, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- Matheny, P.B. Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Mol. Phylogenetics Evol. 2005, 35, 1–20. [Google Scholar] [CrossRef]
- Matheny, P.B.; Wang, Z.; Binder, M.; Curtis, J.M.; Lim, Y.W.; Nilsson, H.; Hughes, K.W.; Hofstetter, V.; Ammirati, J.F.; Schoch, C.L.; et al. Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Mol. Phylogenetics Evol. 2007, 43, 430–451. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Haeseler, A.V.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.4.4: Tree Figure Drawing Tool. Available online: https://github.com/rambaut/figtree/releases (accessed on 18 September 2024).
- Harmaja, H. The genus Clitocybe (Agaricales) in Fennoscandia. Karstenia 1969, 10, 5–121. [Google Scholar] [CrossRef]
- Bigelow, H.E. North American species of Clitocybe; Part I; Strauss & Cramer GmbH: Vaduz, Liechtenstein, 1982. [Google Scholar]
- Bon, M. The Mushrooms and Toadstools of Britain and North-Western Europe; Hodder & Stoughton: London, UK, 1987. [Google Scholar]
- Breitenbach, J.; Kränzlin, F. Fungi of Switzerland, Vol. 3-Boletes and Agarics, 1st ed.; Mykologia Lucerne: Lucerne, Switzerland, 1991; 361p. [Google Scholar]
- Buczacki, S. Collins Fungi Guide: The Most Complete Field Guide to the Mushrooms and Toadstools of Britain & Europe; Harpercollins Pub Ltd.: New Delhi, India, 2014. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).