Abstract
Ectomycorrhizal fungi of the genus Tuber (Ascomycota) produce hypogeous ascomata commonly known as truffles. Despite their high ecological and economic importance, a considerable gap of knowledge exists concerning the diversity of Tuber species in the eastern Mediterranean region. In the frame of this study, more than 200 Tuber collections, originating from various regions of Greece, were examined. A new species to science, i.e., Tuber leptodermum, is formally described. Tuber leptodermum is grouped in the Maculatum clade, as revealed by the ITS and LSU rDNA concatenated phylogenetic tree, and appears as sister to T. foetidum. In addition, T. leptodermum exhibits distinct morphoanatomic features: it produces medium-sized, dark-brown ascomata with a thin pseudoparenchymatous peridium, composed of globose-to-angular cells and forms one-to-four-spored asci containing reticulate–alveolate, ellipsoid ascospores with broad meshes. Thirty other phylogenetic species are identified: seven of them (i.e., T. anniae, T. buendiae, T. conchae, T. dryophilum, T. monosporum, T. regianum and T. zambonelliae) constitute new records for the Greek mycobiota, while the presence of five other species is molecularly confirmed for the first time. Moreover, the existence of ten undescribed phylogenetic species is revealed, six of which are reported for the first time in Greece. Several taxonomic and phylogenetic issues and discrepancies in the genus Tuber are discussed in relation to the new findings.
1. Introduction
The term “true truffles” is used to describe the subterranean spherical and enclosed ascomata produced by fungi of the genus Tuber P.Micheli ex F.H.Wigg. (Pezizales, Ascomycota), which includes several gastronomically valuable and highly prized species [1,2,3]. These fungi form obligate ectomycorrhizal (ECM) relationships with various plant genera, which link the presence of truffles to that of their plant symbionts, depending on species-specific and environmental factors [4]. In addition, since truffle spores are mainly dispersed through the consumption of ascocarps by mycophagous animals, their distribution range is spatially limited [1,5]. During the last glacial periods, survival refugia for various species were established in favorable areas of southern Europe (e.g., in the Balkan peninsula); these refugia became the cradle for species recolonization of the continent [6]. Truffles follow a similar pattern of distribution, which is also affected by their unique biology and past biogeographic incidents [4,7,8,9].
The genus Tuber includes approx. 200 species which are grouped into 13 major phylogenetic clades, i.e., Aestivum, Excavatum, Gennadii, Gibbosum, Latisporum, Macrosporum, Maculatum, Melanosporum, Multimaculatum, Puberulum, Regianum, Rufum and Tumericum-Japonicum [1,10,11]. Several studies have dealt with the diversity of Tuber species in Europe; however, some regions—like the southern Balkans—still remain relatively understudied [2,12,13,14].
Regarding Greece in particular, the earliest documented reference of a “truffle” species can be found in the monumental work Flora Graeca [15], where “Tuber album” is mentioned; this finding, however, corresponds to Terfezia arenaria (Moris) Trappe, according to Chatin [16]. Xavier Laderer [17], a pharmacist at the palace during the reign of King Otto, reported the existence of Tuber cibarium. In addition, Chatin [16] described Terfezia gennadii based on collections made by Panagiotis Gennadios in the Peloponnese; this species was subsequently transferred to the genus Tuber, as T. gennadii (Chatin) Pat. Later, the first record of Tuber aestivum Vittad. was included in the Fungus-host index for Greece [18].
Hence, until the end of the 20th century, the diversity of the genus Tuber in Greece remained largely unknown, as reflected in the first national checklist of larger Ascomycota [19], which included the aforementioned records only. However, in recent years, the collection of truffles has become popular, while pioneering members of citizen science associations (e.g., the Greek Mushroom Society, GMS) have started identifying specimens using traditional taxonomy. A significant change in the knowledge of the genus Tuber in Greece is apparent from 2007 onwards, as reflected by the publication of a species checklist [20] and mushroom field guides [21,22,23,24,25]. The first study using DNA sequencing dealt with truffle specimens identified as T. borchii Vittad., T. gennadii, T. oligospermum (Tul. & C. Tul.) Trappe and T. puberulum Berk. & Broome [26], and was followed by reports of the presence of T. brumale Vittad. haplogroup II [8], T. magnatum Picco [9], T. mesentericum Vittad., T. bituminatum Berk. & Broome, T. suave Pacioni & M. Leonardi [27] and T. magentipunctatum Z. Merényi, I. Nagy, Stielow & Bratek [28]. In addition to these findings, two new species to science found in Greece have recently described, i.e., T. pulchrosporum Konstantinidis, Tsampazis, Slavova, Nakkas, Polemis, Fryssouli & Zervakis from material collected in spring to early summer in Bulgaria and continental Greece [29], and T. aereum Polemis, Daskalopoulos and Zervakis from specimens found in autumn and winter in Naxos Island [30]. At present, 26 species (including 4 phylogenetic species yet to be formally described) of this genus are known to exist in Greece, while the presence of only 15 of them is supported by molecular approaches.
The objective of the present work was to assess the diversity and distribution of the genus Tuber in Greece by examining numerous collections from a large range of geographic areas and a variety of habitats, through the combined use of morphoanatomical characters, ecological features and phylogenetics.
2. Materials and Methods
2.1. Specimens Studied
The biological material examined in the frame of this study was derived from the following sources: (a) from dried specimens previously deposited in the fungarium of the Laboratory of General and Agricultural Microbiology, Agricultural University of Athens (ACAM), or in personal collections of the GMS’s members [G. Konstantinidis (GK), V. Kaounas (VK), N. Tsilis (NT) and M. Gkilas (MG)]; and (b) from fresh ascomata sampled either by the authors or by collaborating truffle collectors. All the fresh samples received were subsequently dried and stored in the fungarium ACAM. In total, 242 collections (several of them including more than one specimen) were identified through the combined use of morphoanatomic criteria and DNA sequencing, while 63 of them were further subjected to phylogenetic analysis (Table 1; Supplementary Material, Table S1).
2.2. Morphoanatomic Features
Each sample, fresh or dried, was macroscopically examined and photographed using a Nikon SMZ18 stereoscope (Nikon Corporation, Tokyo, Japan) with the OCULAR vers. 2.0 software (Teledyne Photometrics, Tucson, AZ, USA). Color description was based on the “Flora of British Fungi: Colour Identification Chart” [31], and the respective codes are provided in braces. Microscopic observations were performed in water and in 3–5% (w/v) potassium hydroxide (KOH). Microanatomical features of the ascomata were examined using an Olympus BX53F2 (Olympus Corporation, Tokyo, Japan) or a Zeiss AxioImager A2 (Carl Zeiss Microscopy, Oberkochen, Germany) microscope, and were photographed with either an Olympus DP74 camera using the cellSens Entry software (Olympus Life Science, Waltham, MA, USA), or with an Axiocam 305 color camera using the ZEN vers. 2.3 lite software (Carl Zeiss Microscopy, Oberkochen, Germany), respectively. Photographs were taken under bright field, phase contrast and/or differential interference contrast (DIC) microscopy.
Initial evaluation of Tuber collections was based on their morphoanatomic and ecological features, in accordance with taxonomic keys included in various publications [27,28,32], and with a monograph on European Tuber species [12]. For selected specimens (e.g., those representing new species to science, and those corresponding to the first national records) the following microscopic features were thoroughly examined: the peridium width (total, and external if present), asci and stalk size, size of parenchymatic cells (if present), ascospore number per ascus, ascospore dimensions (per ascus category and in total), height of meshes or spines, and (in reticulate ascospores) number of meshes across the ascospore length. As a general rule, a minimum of 30 elements were measured for each feature by using Piximètre vers. 5.10 software (http://www.piximetre.fr/; France; accessed on 1 February 2025). Dimensions of various features are provided as follows: (minimum), range of length, (maximum), while the quotient (Q) value for ascospores is also provided, i.e., length divided by width. Average values (Av) were also calculated in the case of asci and ascospores; in addition, percentages are provided for the number of ascospores per ascus, and ranges are provided for the number of meshes along the spore length. Especially in the case of T. regianum Montecchi & Lazzari, the ascospore volume is also provided in accordance with Merenyi et al. [33] and Cseh et al. [28]: 0.523 × (width)2 × length.
2.3. DNA Extraction, Amplification and Sequencing of Phylogenetically Informative Markers
The total genomic DNA was isolated from dried or fresh material as follows: samples were ground to fine powder in the presence of liquid nitrogen, mixed with warm (60 °C) DNA lysis buffer (1% CTAB, 10 mM Na2EDTA, 0.7 M NaCl, and 50 mM Tris-HCl pH 8), and then extracted with phenol:chloroform:isoamyl alcohol (25:24:1), and precipitated with ethanol. The two regions of the nuclear ribosomal repeat unit, i.e., the internal transcribed spacer (ITS) and a fragment of the ribosomal large subunit gene (LSU), were amplified by the polymerase chain reaction (PCR) in a MiniAmp Plus Thermal Cycler (Applied Biosystems, CA, USA), using 0.8 μL of each primer (0.32 μM), 12.5 μL of FastGene® Optima Polymerase hotstart ready mix (NIPPON Genetics Europe, Düren, Germany), 11.5 μL of UltraPure™ DNase/RNase-Free Distilled Water (Thermo Fisher Scientific, Waltham, MA, USA) and 1 μL of DNA template (ca. 10 ng μL−1). The ITS was amplified through the use of ITS1 and the ITS1f/ITS4 primer pair [34,35] under the following conditions: initial denaturation at 95 °C for 4 min, followed by 35 cycles at 95 °C for 30 sec, annealing at 55 °C for 30 sec, extension at 72 °C for 1 min and final extension at 72 °C for 7 min. In addition, partial sequences of the LSU were amplified by the primer pair LROR/LR5 [36] under the following conditions: initial denaturation at 95 °C for 4 min, followed by 35 cycles at 94 °C for 1 min, annealing at 52 °C for 50 sec, extension at 72 °C for 1 min and final extension at 72 °C for 10 min. Electrophoresis was performed with 1% (w/v) agarose gel and purification of the PCR product was performed with the PureLink Quick PCR kit (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA), and the purified products were quantified using the MULTISKAN SkyHigh Microplate Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Sequences were obtained by using the same forward and reverse primers (as before in the amplification procedure) in an automated ABI sequencer (Life Technologies, USA) at CeMIA SA (Greece). FinchTV vers. 1.4.0 software (Geospiza, Inc., Denver, CO, USA) was used for chromatogram visualization and inspection of sequencing quality. Consensus sequences were generated using the SeqMan module of Lasergene vers. 7.1 software (DNAStar, Madison, WI, USA), manually edited to remove or replace all ambiguous characters, and cross-checked against public databases, including the international nucleotide sequence database collaboration (INSDC) [37] and UNITE [38]. Validated sequences were submitted to GenBank, and the pertinent accession numbers appear in Table 1 and in Supplementary Table S1.


Table 1.
Details of biological material from the genus Tuber used in the phylogenetic analyses performed in the frame of this study: the phylogenetic clade and taxon, collection code, geographic origin, GenBank accession no. for ITS and LSU sequences, and respective references. The material from which sequences were generated for the first time is presented in bold typeface, while material corresponding to type specimens is underlined.
Table 1.
Details of biological material from the genus Tuber used in the phylogenetic analyses performed in the frame of this study: the phylogenetic clade and taxon, collection code, geographic origin, GenBank accession no. for ITS and LSU sequences, and respective references. The material from which sequences were generated for the first time is presented in bold typeface, while material corresponding to type specimens is underlined.
| CLADE/Taxon | Collection Code | Origin | ITS | LSU | Reference | |
|---|---|---|---|---|---|---|
| AESTIVUM | ||||||
| 1 | T. aestivum | AQUI 10150 | Italy | MZ423173 | [32] | |
| 2 | T. aestivum | S8 | Slovakia | HQ285310 | [39] | |
| 3 | T. aestivum | 2PL-M(u) | Poland | KX028767 | [40] | |
| 4 | T. aestivum | ACAMTub361 | Greece | PP725740 | [41] | |
| 5 | T. aestivum | ACAMTub271 | Greece | PP918676 | This study | |
| 6 | T. aestivum | ACAMTub376 | Greece | PP918694 | This study | |
| 7 | T. bituminatum | AQUI 9833 | Italy | OL711611 | [27] | |
| 8 | T. bituminatum | AQUI 8579 | Greece | OL711613 | [27] | |
| 9 | T. bituminatum | ACAMTub310 | Greece | PP918764 | This study | |
| 10 | T. bituminatum | ACAMTub421 | Greece | PP918781 | This study | |
| 11 | T. magnatum | TO HG3458 | Italy | MZ423175 | [32] | |
| 12 | T. magnatum | HHD19491 | Turkey | PP239641 | Unpublished | |
| 13 | T. magnatum | B1 | Italy | AJ586268 | [42] | |
| 14 | T. magnatum | ACAMTub055 | Greece | PP918868 | This study | |
| 15 | T. magnatum | ACAMTub156 | Greece | PP918870 | This study | |
| 16 | T. mesentericum | AQUI 9717 | Italy | OL711593 | [27] | |
| 17 | T. mesentericum | AQUI 10238 | France | OL711600 | [27] | |
| 18 | T. mesentericum | AQUI 10230 | Greece | OL711607 | [27] | |
| 19 | T. mesentericum | ACAMTub111 | Greece | PP918880 | This study | |
| 20 | T. mesentericum | ACAMTub250 | Greece | PP918885 | This study | |
| 21 | T. panniferum | JT12835 | Spain | HM485380 | [43] | |
| 22 | T. panniferum | MUB:Fung-0973 | Spain | MN962725 | [44] | |
| 23 | T. panniferum | ACAMTub269 | Greece | PP918896 | This study | |
| 24 | T. panniferum | ACAMTub385 | Greece | PP918897 | This study | |
| 25 | T. pulchrosporum | VN091 | Greece | MK113975 | [29] | |
| 26 | T. pulchrosporum | 1961 F0388 | Bulgaria | MK113982 | [29] | |
| 27 | T. suave | AQUI 7131 | Italy | OL711623 | [27] | |
| 28 | T. suave | AQUI 10229 | Greece | OL711627 | [27] | |
| EXCAVATUM | ||||||
| 29 | T. excavatum | AP-T63a | Slovenia | FM205555 | Unpublished | |
| 30 | T. excavatum | Tub10 | Italy | HM152012 | Unpublished | |
| 31 | T. aff. excavatum 1 | ACAMTub151 | Greece | PP918825 | This study | |
| 32 | T. aff. excavatum 1 | ACAMTub162 | Greece | PP918826 | This study | |
| 33 | T. excavatum | SFI:TUBEXC/251008B | Slovenia | FN433141 | Unpublished | |
| 34 | T. fulgens | AP-T60 | Slovenia | FM205568 | Unpublished | |
| 35 | T. excavatum | zb3377 | Hungary | HM152023 | Unpublished | |
| 36 | T. excavatum | AP-T11 | Slovenia | FM205554 | Unpublished | |
| 37 | T. aff. excavatum 2 | ACAMTub028 | Greece | PP918828 | This study | |
| 38 | T. aff. excavatum 2 | ACAMTub316 | Greece | PP918829 | This study | |
| 39 | T. excavatum | SFI:TUBEXC/121008 | Slovenia | FN433142 | Unpublished | |
| 40 | T. excavatum | SA1TE | Poland | KJ524533 | Unpublished | |
| 41 | T. aff. excavatum 3 | ACAMTub168 | Greece | PP918841 | This study | |
| 42 | T. aff. excavatum 3 | ACAMTub320a | Greece | PP918851 | This study | |
| 43 | T. excavatum | BM100 | Spain | FJ748899 | [45] | |
| 44 | T. excavatum | MA:Fungi:54695 | Spain | FM205561 | Unpublished | |
| 45 | T. fulgens | zb3335_205 | Hungary | HM152021 | Unpublished | |
| 46 | T. fulgens | CMI-Unibo_5034 | Iran | MW884551 | [46] | |
| 47 | T. fulgens | HMT44 | Italy | HM151979 | Unpublished | |
| 48 | T. fulgens | ACAMTub170 | Greece | PP918853 | This study | |
| 49 | T. fulgens | ACAMTub312 | Greece | PP918854 | This study | |
| 50 | T. iranicum | CMI-Unibo 4939 | Iran | MN854634 | [47] | |
| 51 | T. iranicum | CMI-Unibo 4965 | Iran | MN854635 | [47] | |
| GENNADII | ||||||
| 52 | T. gennadii | B M1904 | Italy | HM485361 | [43] | |
| 53 | T. gennadii | AH39251 | Spain | JN392211 | [26] | |
| 54 | T. aff. gennadii | ACAMTub050 | Greece | PP918857 | This study | |
| 55 | T. aff. gennadii | ACAMTub131 | Greece | PP918858 | This study | |
| 56 | T. gennadii | AH38957 | Spain | JN392204 | [26] | |
| 57 | T. gennadii | VK2141 | Greece | JN392205 | [26] | |
| 58 | T. gennadii | AH38955 | Spain | JN392207 | [26] | |
| 59 | T. gennadii | MRG496 | Spain | OQ565278 | [48] | |
| 60 | T. conchae | MRG486 | Spain | OQ565276 | [48] | |
| 61 | T. conchae | ACAMTub046 | Greece | PP918816 | This study | |
| 62 | T. lacunosum | AH38912 | Morocco | JN392214 | [26] | |
| 63 | T. lacunosum | MRG692 | Spain | OQ565280 | [48] | |
| 64 | T. lucentum | j957 | Spain | MN437527 | [49] | |
| 65 | T. lucentum | MUB:Fung-j825 | Spain | NR184909 | [49] | |
| MACROSPORUM | ||||||
| 66 | T. calosporum | HKAS:88790 | China | KT444598 | [50] | |
| 67 | T. calosporum | HKAS:88751 | China | KT444600 | [50] | |
| 68 | T. glabrum | BJTCFan228 | China | KF002731 | [51] | |
| 69 | T. glabrum | BJTCFan232 | China | KF002727 | [51] | |
| 70 | T. macrosporum | CMI-Unibo_4937 | Iran | MW884553 | [46] | |
| 71 | T. macrosporum | CMI-Unibo_4938 | Iran | MW884554 | [46] | |
| 72 | T. macrosporum | FHS-449 | Serbia | FM205664 | [14] | |
| 73 | T. macrosporum | IC28092233 | Spain | PP948901 | Unpublished | |
| 74 | T. macrosporum | ITA_014s | Italy | KP738351 | [52] | |
| 75 | T. macrosporum | ACAMTub052 | Greece | PP918859 | This study | |
| 76 | T. macrosporum | ACAMTub110 | Greece | PP918860 | This study | |
| 77 | T. macrosporum | ACAMTub252 | Greece | PP918862 | This study | |
| 78 | T. macrosporum | ITA_003V | Italy | KP738380 | [52] | |
| 79 | T. macrosporum | ITA_008 | Italy | KP738388 | [52] | |
| 80 | T. macrosporum | FRA_002 | France | KP738361 | [52] | |
| 81 | T. monosporum | ACAMTub455 | Greece | PP918907 | PP892729 | This study |
| 82 | T. sinomonosporum | BJTCFan150 | China | KF002729 | [51] | |
| MACULATUM | ||||||
| 83 | T. arnoldianum | RH1619 | USA | KU186913 | [53] | |
| 84 | T. arnoldianum | FLAS:F-71902 | USA | OR762689 | PP337424 | Unpublished |
| 85 | T. arnoldianum | FLAS:F-71880 | USA | OR762672 | PP337406 | Unpublished |
| 86 | T. arnoldianum | AMC105 | USA | OP413000 | Unpublished | |
| 87 | T. aztecorum | ITCV 993 ITCV | Mexico | NR159044 | [54] | |
| 88 | T. aztecorum | GG993 FLAS | Mexico | KY271791 | [54] | |
| 89 | T. beyerlei | JT32597 | USA | HM485408 | [43] | |
| 90 | T. beyerlei | Berch 0042 | Canada | KP972075 | [55] | |
| 91 | T. brennemanii | MES653 | USA | MF611779 | KY565252 | [56] |
| 92 | T. brennemanii | RH1746 | USA | MF611791 | KY565258 | [56] |
| 93 | T. excelsum-reticulatum | BJTC FAN863 | China | OM265272 | OM366224 | [11] |
| 94 | T. excelsum-reticulatum | BJTC FAN758 | China | OM265264 | OM366218 | [11] |
| 95 | T. foetidum | ZB2489 | Hungary | JQ288906 | JF261362 | Unpublished |
| 96 | T. foetidum | ZB2452 | Hungary | JQ288905 | JF261361 | Unpublished |
| 97 | T. foetidum | JCAS0140001000096 | Spain | MH703905 | [57] | |
| 98 | T. foetidum | ZB3454 | Finland | FN568055 | [58] | |
| 99 | T. foetidum | B-2489 | Hungary | AJ557544 | [13] | |
| 100 | T. foetidum | B-2452 | Hungary | AJ557543 | [13] | |
| 101 | T. leptodermum | ACAMTub475 | Greece | PQ877422 | PQ881954 | This study |
| 102 | T. leptodermum | ACAMTub359 | Greece | PP918908 | PP905688 | This study |
| 103 | T. leptodermum | ACAMTub379 | Greece | PP918909 | PP905689 | This study |
| 104 | T. leptodermum | ACAMTub380 | Greece | PP918910 | PP905690 | This study |
| 105 | T. leptodermum | ACAMTub382 | Greece | PP918911 | PP905691 | This study |
| 106 | T. hubeiense | HMAS 60233 | China | KT067688 | KT067694 | [11] |
| 107 | T. lusitanicum | MUB:Fung-0986 | Spain | MT621651 | MT705332 | [59] |
| 108 | T. lusitanicum | MUB:Fung-0988 | Spain | MT621653 | [59] | |
| 109 | T. lusitanicum | MUB:Fung-0990 | Spain | MT621655 | [59] | |
| 110 | T. maculatum | Italy | AF003919 | Unpublished | ||
| 111 | T. maculatum | herbarium 1967 | Italy | EU753269 | Unpublished | |
| 112 | T. maculatum | Imatra_3 | Finland | MZ389968 | [60] | |
| 113 | T. maculatum | FLAB22 | Uruguay | PP297668 | [61] | |
| 114 | T. maculatum | BJTC FAN876 | China | OM265278 | OM366228 | [11] |
| 115 | T. maculatum | BJTC FAN868 | China | OM265274 | OM366227 | [11] |
| 116 | T. miquihuanense | ITCV885 | Mexico | HM485414 | JF419292 | [43]; [62] |
| 117 | T. pseudomagnatum | BJTC FAN163 | China | JQ771192 | [63] | |
| 118 | T. pseudomagnatum | BJTC FAN299 | China | OM265244 | OM366195 | [11] |
| 119 | T. pseudomagnatum | BJTC FAN392 | China | KP276184 | KP276193 | [64] |
| 120 | T. pseudomagnatum | BJTC FAN390 | China | OM265243 | [11] | |
| 121 | T. pseudomagnatum | BJTC FAN389 | China | OM265242 | [11] | |
| 122 | T. rapaeodorum | BG Kew K(M)128884 | England | EU784429 | [65] | |
| 123 | T. rapaeodorum | CMI-UNIBO 2483 | Armenia | DQ011849 | Unpublished | |
| 124 | T. walkeri | RH5211 | USA | JF419260 | [62] | |
| 125 | T. walkeri | FLAS:MES-274 | USA | MT156441 | Unpublished | |
| 126 | T. walkeri | RH794 | USA | JF419258 | JF419308 | [62] |
| 127 | T. whetstonense | JT25783 | USA | HM485392 | JF419304 | [43]; [62] |
| 128 | T. whetstonense | SOC 756 OSC 111412 | USA | AY830855 | [66] | |
| 129 | T. whetstonense | FLAS:MES-210 | USA | MT156439 | Unpublished | |
| 130 | T. wumengense | BJTC FAN218A | China | KT067682 | KT067707 | [64] |
| 131 | T. wumengense | HMAS 60229 | China | KT067687 | [64] | |
| 132 | T. wumengense | BJTC FAN586 | China | OM265254 | [11] | |
| MELANOSPORUM | ||||||
| 133 | T. brumale | G1126 | Greece | KF550998 | [8] | |
| 134 | T. brumale | BTb2p2 | Turkey | KF551063 | [8] | |
| 135 | T. brumale | FHS-460 | Serbia | FM205660 | [14] | |
| 136 | T. brumale | CMI-Unibo_5001 | Iran | MW829419 | [46] | |
| 137 | T. brumale | MA:Fungi:28373 | Spain | FM205550 | [14] | |
| 138 | T. brumale | TB59k1 | France | KF550974 | [8] | |
| 139 | T. brumale | ACAMTub159 | Greece | OP850807 | This study | |
| 140 | T. brumale | ACAMTub233 | Greece | PP918804 | This study | |
| 141 | T. brumale | ACAMTub456 | Greece | PP918814 | This study | |
| 142 | T. cryptobrumale | HNHM<HUN>:BP107922 | Hungary | KU203777 | [33] | |
| 143 | T. cryptobrumale | HNHM<HUN>:BP107923 | Hungary | KU203778 | [33] | |
| 144 | T. indicum | Tind-gs01 | China | DQ375490 | [67] | |
| 145 | T. indicum | Tind-gs05 | China | DQ375491 | [67] | |
| 146 | T. melanosporum | AQUI 10152 | Italy | MZ423176 | [32] | |
| 147 | T. melanosporum | MEL178 | France | EU555385 | [68] | |
| 148 | T. melanosporum | ACAMTub256 | Greece | PP918878 | This study | |
| 149 | T. petrophilum | LJU-GIS-TUBsp_xx1010A | Serbia | HG810883 | [69] | |
| 150 | T. petrophilum | LJU-GIS-TUBsp_xx1010D | Serbia | HG810886 | [69] | |
| 151 | T. thracicum | SOMF:30882 | Bulgaria | OR666020 | [70] | |
| 152 | T. thracicum | SOMF:30883 | Bulgaria | OR666027 | [70] | |
| PUBERULUM | ||||||
| 153 | T. oligospermum | CMI-UNIBO 4230 | Morocco | KF021624 | [71] | |
| 154 | T. oligospermum | CMI-UNIBO 4234 | Morocco | KF021622 | [71] | |
| 155 | T. oligospermum | AH39002 | Italy | JN392248 | [26] | |
| 156 | T. oligospermum | AH37861 | Italy | JN392251 | [26] | |
| 157 | T. oligospermum | GK3060 | Greece | JN392249 | [26] | |
| 158 | T. aff. oligospermum 1 | ACAMTub075 | Greece | PP918887 | This study | |
| 159 | T. oligospermum | MUB:Fung-1049 | Spain | PQ281432 | Unpublished | |
| 160 | T. oligospermum | MUB:Fung-1050 | Israel | PQ281433 | Unpublished | |
| 161 | T. oligospermum | GK5388 | Greece | JN392263 | [26] | |
| 162 | T. aff. oligospermum 2 | ACAMTub062 | Greece | PP918889 | This study | |
| 163 | T. oligospermum | HAI-D-102 | Israel | JN392271 | [26] | |
| 164 | T. oligospermum | VK2042 | Greece | JN392254 | [26] | |
| 165 | Soil sample | S052 | Iran | UDB0765470 | Unpublished | |
| 166 | T. aff. oligospermum 3 | ACAMTub133 | Greece | PP918893 | This study | |
| 167 | T. anniae | JT13209 | USA | HM485338 | [43] | |
| 168 | T. anniae | Juva_1 | Finland | MZ389970 | [60] | |
| 169 | T. anniae | ACAMTub024 | Greece | PP918746 | This study | |
| 170 | T. borchii | AP-T30 | Slovenia | FM205497 | [14] | |
| 171 | T. borchii | CMI-Unibo_5047 | Iran | MW829422 | [46] | |
| 172 | T. borchii | 17Bo | Italy | DQ679802 | [72] | |
| 173 | T. borchii | ACAMTub135 | Greece | PP918787 | This study | |
| 174 | T. borchii | ACAMTub334 | Greece | PP918792 | This study | |
| 175 | T. borchii | ACAMTub371A | Greece | PP918796 | This study | |
| 176 | T. borchii | AQUI 10151 | Italy | MZ423174 | [32] | |
| 177 | T. borchii | GK5597 | Greece | JN392228 | [26] | |
| 178 | T. borchii | CMI-UNIBO 3026 | Italy | FJ554493 | [72] | |
| 179 | T. borchii | MUB:Fung-1036 | Spain | PQ273773 | Unpublished | |
| 180 | T. borchii | ACAMTub120A | Greece | PP918786 | This study | |
| 181 | T. borchii | ACAMTub207 | Greece | PP918802 | This study | |
| 182 | T. borchii | ACAMTub377 | Greece | PP918803 | This study | |
| 183 | T. dryophilum | Italy | AF003917 | Unpublished | ||
| 184 | T. puberulum | GK4352 | Greece | JN392223 | [26] | |
| 185 | T. dryophilum | GB69 | Italy | HM485353 | [10] | |
| 186 | T. puberulum | GK5601 | Greece | JN392224 | [26] | |
| 187 | T. dryophillum | ACAMTub043 | Greece | PP918819 | This study | |
| 188 | T. dryophillum | ACAMTub134 | Greece | PP918821 | This study | |
| 189 | T. dryophillum | ACAMTub182 | Greece | PP918822 | This study | |
| 190 | T. dryophillum | ACAMTub204 | Greece | PP918824 | This study | |
| 191 | T. lijiangense | BJTC FAN307 | China | KP276188 | [64] | |
| 192 | T. cf. lijiangense | BJTC FAN148 | China | OM286798 | [11] | |
| 193 | T. puberulum | Berk. & Broome TL11885 | Denmark | AJ969626 | [73] | |
| 194 | T. puberulum | Berk. & Broome TL3857 | Denmark | AJ969625 | [73] | |
| REGIANUM | ||||||
| 195 | T. magentipunctatum | ZB4293 | Hungary | JQ288909 | [74] | |
| 196 | T. magentipunctatum | MO793 | Italy | KY420089 | [28] | |
| 197 | T. magentipunctatum | GK 4552 | Greece | KY420088 | [28] | |
| 198 | T. magenctipunctatum | ACAMTub126 | Greece | PP918865 | This study | |
| 199 | T. magenctipunctatum | ACAMTub381 | Greece | PP918866 | This study | |
| 200 | T. regianum | M46 | Spain | KY420100 | [28] | |
| 201 | T. regianum | ZB3081 | Slovakia | KY420098 | [74] | |
| 202 | T. regianum | IC13071106 | Spain | KY420103 | [74] | |
| 203 | T. regianum | ACAMTub226 | Greece | PP918898 | This study | |
| RUFUM | ||||||
| 204 | T. aereum | ACAMTub245 | Greece | PP425915 | [30] | |
| 205 | T. aereum | ACAMTub467 | Greece | PQ336045 | This study | |
| 206 | T. nitidum | AH39101 | Morocco | JX402092 | [75] | |
| 207 | T. nitidum | MUB:Fung-0934 | Spain | MN962721 | [76] | |
| 208 | T. rufum | SFI:TUBRUF/111208 | N. Macedonia | FN433159 | Unpublished | |
| 209 | T. nitidum | ACAMTub039 | Greece | PP918707 | This study | |
| 210 | T. nitidum | ACAMTub375 | Greece | PP918708 | This study | |
| 211 | T. rufum | 1798 | Italy | EF362473 | [77] | |
| 212 | T. rufum | 17114 | Italy | JF908745 | [78] | |
| 213 | T. rufum f. ferrugineum | FHS-393 | Serbia | FM205676 | [14] | |
| 214 | T. aff. rufum 1 | ACAMTub251 | Greece | PP918918 | This study | |
| 215 | T. rufum | 1785 | Italy | EF362475 | [77] | |
| 216 | T. rufum | FHS-XX12 | Serbia | FM205690 | [14] | |
| 217 | Tuber sp. 57 | JT13224 | USA | JQ925650 | [10] | |
| 218 | T. aff. rufum 2 | ACAMTub315 | Greece | PP918919 | This study | |
| 219 | T. buendiae | MUB:Fung-0974 | Spain | MT006095 | [76] | |
| 220 | T. buendiae | ACAMTub350 | Greece | PP918815 | This study | |
| 221 | T. melosporum | AH31737 | Spain | JX402095 | [75] | |
| 222 | T. melosporum | MA-33360 | Spain | OQ866602 | Unpublished | |
| 223 | T. pustulatum | AQUI 9725 | Spain | MK211278 | [79] | |
| 224 | T. rufum | MA:Fungi:28397 | Spain | FM205615 | [14] | |
| 225 | Tuber sp. | TUF113700 | Estonia | UDB027742 | Unpublished | |
| 226 | T. rufum f. lucidum | CMI-Unibo_4944 | Iran | MW829417 | [46] | |
| 227 | T. rufum f. lucidum | AZ2097 | Italy | FJ809888 | [80] | |
| 228 | T. rufum | GK4422 | Greece | JX402094 | [75] | |
| 229 | T. aff. rufum 3 | ACAMTub425 | Greece | PP918727 | This study | |
| 230 | T. aff. rufum 3 | ACAMTub067 | Greece | PP918709 | This study | |
| 231 | T. aff. rufum 3 | ACAMTub230 | Greece | PP918710 | This study | |
| 232 | T. aff. rufum 3 | ACAMTub317 | Greece | PP918714 | This study | |
| 233 | T. zambonelliae | MUB:Fung-0995 | Spain | MW632951 | [81] | |
| 234 | T. zambonelliae | MUB:Fung-1007 | Spain | MW632955 | [81] | |
| 235 | T. zambonelliae | ACAMTub040 | Greece | PP918921 | This study | |
| 236 | T. zambonelliae | ACAMTub079 | Greece | PP918922 | This study | |
| OUTGROUP | ||||||
| 237 | Choiromyces helanshanensis | HKAS 80634 | China | NR153899 | [82] |
ITS: internal transcribed spacer; LSU: large subunit 25-28S RNA.
2.4. Phylogenetic Analysis
For the phylogenetic analyses, sequences generated in this study, as well as reference sequences of Tuber species derived from the GenBank (https://www.ncbi.nlm.nih.gov/genbank/; accessed on 10 March 2025) and UNITE (https://unite.ut.ee/; accessed on 10 March 2025), were used (Table 1). Sequence alignments were implemented using the ClustalW algorithm of MEGA11 software [83]. To exclude poorly aligned or ambiguous regions of the sequences, trimming was performed manually. The phylogenetic trees were constructed with the maximum likelihood (ML) method via the IQ-TREE vers. 2.4.0 software by using the GTR + gamma substitution model, estimated as the best-fit model by the same software [84,85]. ML support values in the branches were obtained using ultrafast bootstrap (1000 replicates), implemented in the IQ-TREE software [86]. Bayesian inference (BI) analysis was performed utilizing the BEAST vers. 10.5.0 software [87], employing the same nucleotide substitution model as in the ML method. The Effective Sample Sizes (ESS > 200) were evaluated by the Tracer vers. 1.7.2 software [88]. A single chain of 10 million generations was run, logging every 100 generations. A maximum clade credibility (MCC) tree was generated using TreeAnnotator vers. 10.5.0 [87] with 10% burn-in. Visualization of the phylogenetic tree was performed with the iTOL vers. 6 web-based software [89]. Bootstrap support (≥70%) and BPP (≥0.95) values are shown in the tree nodes, while Choiromyces helanshanensis (sequence from the holotype) was used as an outgroup in all cases. The tree scale indicates the expected number of changes per site. The resulting phylogenetic trees are deposited in TreeBASE (http://www.treebase.org; accessed on 20 March 2025) with accession ID 32081.
Each taxonomic annotation appearing on the phylogenetic trees corresponds to a well-defined species or to a non-described phylospecies, which is provisionally named by using the epithet of the most related taxon and the term “aff.” (affinity). Although we acknowledge the limitations mentioned by earlier studies regarding the generalized use of the ITS region as a universal DNA barcode marker for fungi, we followed the widely adopted threshold for separating species of the genus Tuber, i.e., <96% for ITS sequence identity in pairwise comparisons [10,11,27]. The respective values were estimated with the aid of the BLASTn web-based tool [90], accessed through the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi; accessed on 10 March 2025).
3. Results
3.1. Phylogeny of the Genus Tuber
A total of 242 ITS and six LSU sequences were generated for the first time (Supplementary Material, Table S1); among them, 63 ITS and five LSU sequences were selected for inclusion in the phylogenetic trees of this study (Figure 1, Figure 2 and Figure 3, Supplementary Figure S1). In addition to the Maculatum clade, which appears compressed in the ML ITS tree (Figure 1), the following Tuber clades (as these were defined by Bonito et al. [10], and Bonito and Smith [1]) are included: Aestivum, Excavatum, Gennadii, Macrosporum, Melanosporum, Puberulum, Regianum and Rufum. They consist mostly of species of European origin and/or distribution, and contain 187 sequences (other than those of the Maculatum clade), 58 of which derive from this work and 129 of which are sequences deposited in NCBI and UNITE databases (16 of them correspond to specimens from Greece generated in the frame of previous studies) (Table 1).
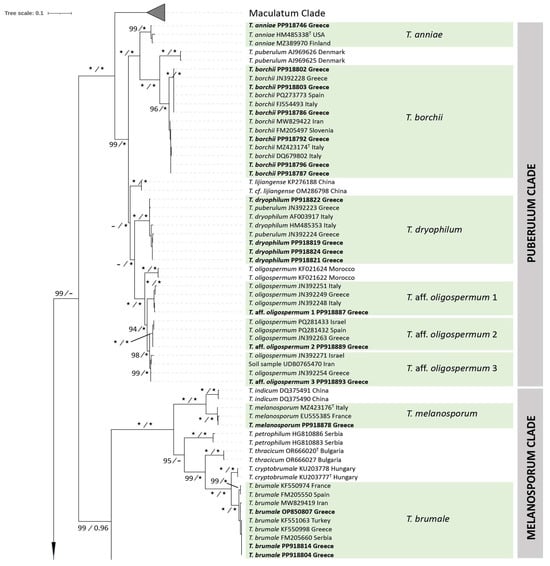
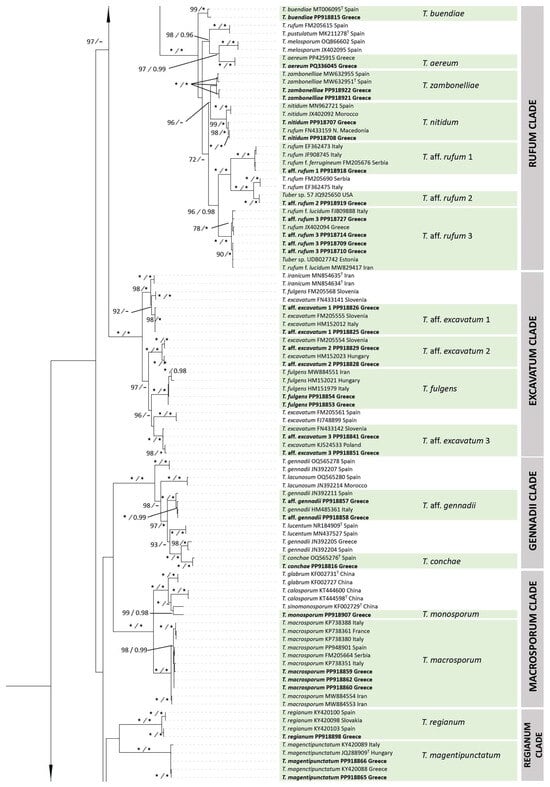
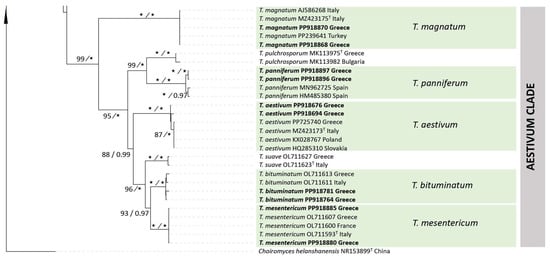
Figure 1.
A maximum likelihood (ML) tree, based on ITS sequences, depicting the phylogeny of Tuber spp. in Greece. Choiromyces helanshanensis is used as an outgroup. Bootstrap support (>70%) for ML and Bayesian posterior probabilities (>0.95) are indicated at the tree nodes; values of 100% and 1.00, respectively, are marked by asterisks (*). Sequences generated in this study appear in bold typeface, while the Maculatum clade appears collapsed, and is presented in Figure 2, Figure 3 and Figure 4. Species including sequences generated by this study are marked in green. Clades are presented in the vertical line on the right side of the figure. The use of sequences from type specimens is indicated by “T” placed as superscript after the respective GenBank accession number.
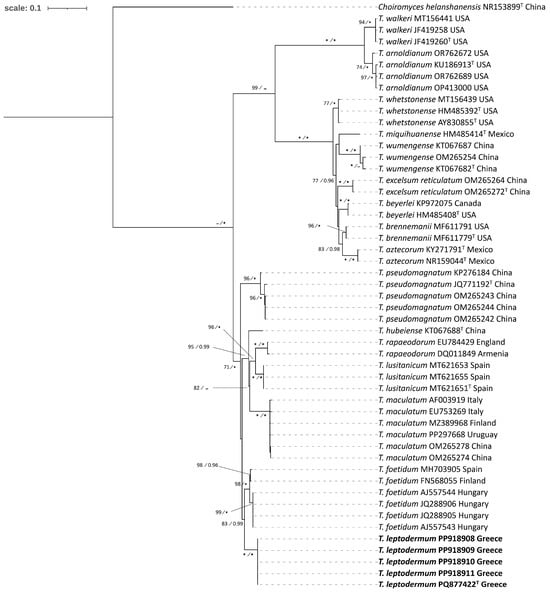
Figure 2.
A maximum likelihood (ML) tree, based on ITS sequences, depicting the phylogeny of the Maculatum clade by including T. leptodermum sp. nov., which appears in bold typeface. Choiromyces helanshanensis is used as an outgroup. Bootstrap support (>70%) for ML and Bayesian posterior probabilities (>0.95) are indicated at the tree nodes; values of 100% and 1.00, respectively, are marked by asterisks (*). The use of sequences from type specimens is indicated by “T” placed as superscript after the respective GenBank accession number.
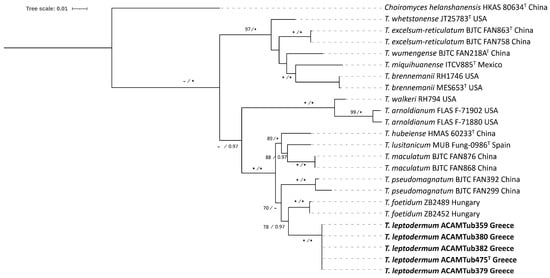
Figure 3.
A maximum likelihood (ML) concatenated tree, based on ITS + LSU sequences, depicting the phylogeny of the Maculatum clade by including T. leptodermum sp. nov., which appears in bold typeface. Choiromyces helanshanensis is used as an outgroup. Bootstrap support (>70%) for ML and Bayesian posterior probabilities (>0.95) are indicated at the tree nodes; values of 100% and 1.00, respectively, are marked by asterisks (*). The use of sequences from type specimens is indicated by “T” placed as superscript after the respective GenBank accession number.
The length of the aligned ITS dataset was 566 nucleotides, and the resulting phylogenetic trees inferred from ML and BI analyses demonstrated consistent topologies (with no supported conflicts) showing high ML-BS and BPP support values.
In the Aestivum clade, specimens from this study are included in five highly supported groups (ML-BS 100%, BPP 1.00) which correspond to well-defined species, i.e., T. aestivum, T. bituminatum, T. magnatum Picco, T. mesentericum and T. panniferum Tul. & C. Tul. All of them, with the exception of T. panniferum, demonstrated high sequence identity (97.2% to 100%) with the reference material examined. In contrast, T. panniferum collections from the islands of Crete and Naxos growing in association with Quercus spp. (ACAMTub243, ACAMTub269, and ACAMTub385) showed some variation (97.3–97.5% sequence identity) when compared to three sequences available in international databases.
As regards the Excavatum clade, our analysis presents six to seven phylospecies; Greek specimens are distributed in four of them, with high support (98–100%, 1.00), i.e., T. fulgens, and another three are hereby provisionally named as T. aff. excavatum 1 (ACAMTub151, ACAMTub162), T. aff. excavatum 2 (ACAMTub028, ACAMTub316) and T. aff. excavatum 3 (ACAMTub168); each of them also includes collections of European origin.
In the Gennadii clade, the existence of six phylogenetic species was detected, with high support (98-100%, 1.00). Sequences from specimens studied in this work are grouped within two of them, one being the recently described T. conchae M. Romero & P. Alvarado. The other is one of the three phylospecies consisting of specimens identified as T. gennadii; hence, our two collections from the Attica area (ACAMTub050 and ACAMTub131) are grouped within a phylospecies provisionally labelled as T. aff. gennadii, which also includes specimens from Italy and Spain.
The Macrosporum clade is divided in two main well-supported groups (100%, 1.00). One of them corresponds to the Tuber macrosporum Vittad. species complex, and is further divided into three subgroups (98–100%, 0.99–1.00); all Greek T. macrosporum specimens originate from Central and Northern Greece and grow in association with a large range of angiosperms, and are grouped into one of these subgroups. In addition, one of the collections (ACAMTub455 from Paggaio Mt. in Fagus sylvatica) studied in this work corresponds to the European T. monosporum (Mattir.) Vizzini, on the basis of morphoanatomical features alone; no reference sequence for this species exists in the international databases.
As regards the Melanosporum clade, the majority of the respective specimens from Greece are grouped within the T. brumale species complex, while others (e.g., ACAMTub256) are identified as T. melanosporum (99.6–100% sequence identity with the species epitype, MZ423176), and originate from areas in the vicinity of truffle orchards.
Greek specimens in the Puberulum clade are grouped into at least six phylospecies with high support (96–100%, 1.00): T. anniae Vittad. (98.4% sequence identity with T. anniae holotype), T. borchii Vittad. s.l. (97.5–100% sequence identity with species lectotype MZ423174), T. dryophilum Tul. & C. Tul. (98.5–99.6%, when compared to reference sequence AF003917) and the T. oligospermum complex, including three phylogenetic species provisionally named T. aff. oligospermum 1, T. aff. oligospermum 2 and T. aff. oligospermum 3. The first and second phylospecies each include one collection studied in this work (ACAMTub075—Attica area and ACAMTub062—Fthiotida area, respectively), while the third includes five collections, all from Attica (Supplementary Table S1); this material is associated with P. halepensis, P. pinea, Quercus coccifera, Q. ilex, Quercus sp. and Cistus monspeliensis.
The presence of two species of the Regianum clade was detected in Greece, i.e., T. magenctipunctatum and T. regianum. Specimens of the former demonstrated high sequence identity (99.8–100%) with the species holotype, whereas the latter is represented by a single collection (ACAMTub226, Prespes area in F. sylvatica) which showed 98.0–100% sequence identity with the six T. regianum sequences deposited in GenBank.
Regarding the Rufum clade, Greek collections were grouped into seven phylospecies. Four of them correspond to known species: T. aereum (100% sequence identity with the holotype PP425915), T. buendiae Ant. Rodr. & Morte (97.8% sequence identity with the holotype MT006095), T. nitidum and T. zambonelliae (97.6–98.1% sequence identity with the holotype, MW632951). The other three form part of phylogenetically distinct groups (100%, 1.00) within the T. rufum species complex, which are provisionally named T. aff. rufum 1, T. aff. rufum 2 and T. aff. rufum 3. The first includes one collection from this study (ACAMTub251 from Olympos Mt.), and specimens identified as T. rufum from Italy and as T. rufum f. ferrugineum from Serbia; the second consists of one Greek collection (ACAMTub315 from the Prespes area) and one other specimen, labeled as Tuber sp. 57, from the USA. The third includes 23 collections from various regions of continental Greece (Supplementary Table S1), as well as specimens from Iran and Italy, identified as T. rufum f. lucidum, and from Estonia, identified as Tuber sp.
In particular, as concerns the Maculatum clade, a total of 51 ITS and 24 LSU sequences were analyzed; among these, 5 ITS and 5 LSU sequences were generated for the first time (Table 1). For the analysis of the two loci, the final alignment included 24 specimens and spanned 1406 nucleotides, comprising 544 nucleotides from the ITS and 862 from the LSU. The tree topologies obtained from the ML and BI analysis were similar, and the former was selected for the purposes of this study.
Taxa within the Maculatum clade present a worldwide distribution, by including collections originating from Europe, Asia, and North, Central and South America. European species are represented by T. maculatum Vittad., T. foetidum Vittad., T. rapaeodorum Tul. & C. Tul. and T. lusitanicum Ant. Rodr. & Muñoz-Mohedano. Most importantly, the phylogenetic analysis of the Maculatum clade revealed the presence of a new species to science, i.e., Tuber leptodermum, which forms a monophyletic group with 100% bootstrap support and a posterior probability of 1 (100%, 1.00) in individual ITS and LSU phylogenies and in concatenated ITS+LSU phylogenies (Figure 2 and Figure 3, Supplementary Figure S1).
In the ITS and in the concatenated ITS+LSU phylogenies, T. leptodermum was resolved as a sister to T. foetidum (83%, 0.99 and 78%, 0.97, respectively), while other phylogenetically related taxa were T. hubeiense L. Fan, T. lusitanicum, T. maculatum, T. pseudomagnatum L. Fan and T. rapaeodorum. Based on a megablast search of the INSDC (GenBank) nucleotide database, the closest hits using the ITS sequence of the T. leptodermum type material were collections identified as T. foetidum from Spain (MH703905, MT621657, MT621658) and Serbia (FM205704), presenting sequence identity values of 94.6–95.7%.
3.2. Taxonomy
Tuber leptodermum V. Daskalopoulos, G. Konstantinidis, N. Tsilis, E. Polemis & G.I. Zervakis, sp. nov. (Figure 4 and Figure 5).

Figure 4.
Tuber leptodermum sp. nov.: (a) representative biotope of occurrence (Grevena, collection code ACAMTub379); (b) mature ascomata in situ (holotype; ACAMTub475); (c) mature ascomata in situ (ACAMTub382).
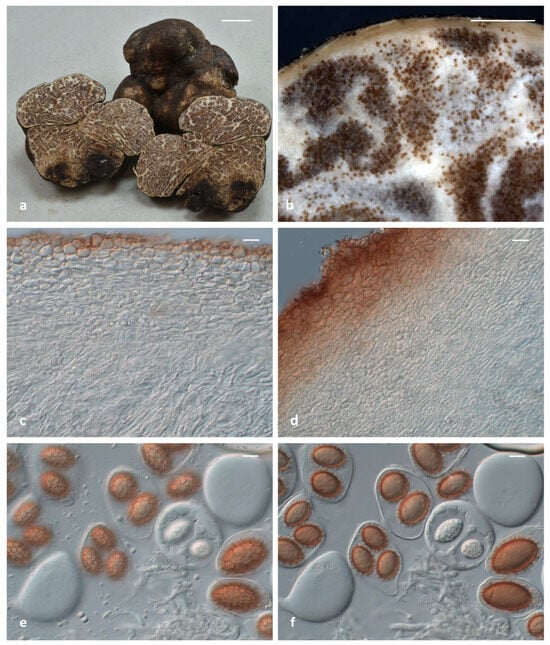
Figure 5.
Tuber leptodermum sp. nov.: (a) mature ascomata ex situ (holotype; collection code ACAMTub475); (b) macroscopic details of the gleba and the peridium (holotype; ACAMTub475); (c) a peridium structure resembling a stonewall, colored only at the outer margin (ACAMTub380); (d) a peridium structure also colored towards the internal layer (ACAMTub379); (e) ascospore details featuring ornamentation of the polygonal meshes (ACAMTub380); (f) ascospore details with clear depiction of their outline and the size of the alveoli (ACAMTub380). Bars: (a) ascomata 1 cm; (b) gleba 1 mm; (c–f) microscopic features 20 μm.
MycoBank: MB858411
Etymology: The epithet leptodermum derives from the Greek words “λεπτό” (“lepto”, meaning thin) and “δέρμα” (“derma”, meaning skin), and refers to the thin peridium of the new species.
Diagnosis: Ascoma medium sized, dark-brown, peridum thin, up to 150 (170) μm, pseudoparenchymatous, composed of globose-to-angular cells, exoperidium easily scratched off to reveal a light-colored endoperidium; asci 1–4-spored (rather equally varied); ascospores ellipsoid, reticulate–alveolate, with broad meshes. It is found in riparian habitats dominated by Populus and/or Salix spp. T. leptodermum is a phylogenetically distinct species in the Maculatum clade, and represents a sister taxon to T. foetidum, as revealed by the ITS and the concatenated ITS+LSU phylogeny.
Holotype: GREECE. KOZANI: Municipality of Voio, Tsotyli greater area, elevation approx. 700 m, found in soil under Populus alba, 12 October 2024, coll. N. Tsilis (holotype ACAMTub475, designated here). GenBank: ITS = PQ877422; LSU = PQ881954.
Description: Ascomata hypogeous, 20–50 mm in diameter, generally irregular, tuberiform, knotty and lobed but also subglobose, with firm and solid texture, surface felty smooth to minutely verrucose to areolate in spots, color cigar-brown {16} to dark-umber {36} or darker-brown vinaceous {25} to purplish-chestnut {21} (Figure 5a). Peridium (50) 80–150 (170) µm thick, external layer (exoperidium) (15) 25–85 (130) µm thick, pseudoparenchymatous, of subglobose to irregularly angular cells (textura globulosa-angularis), dark-brick {20} in color, without any protruding cystidioid hyphae, cells (8.5) 11–18.5 (29) × (6.5) 7.5–13.5 (16.5) µm; internal layer (endoperidium) (50) 70–110 (125) µm thick, hyaline, with relatively smaller cells in comparison with the external layer and progressively more elongated towards the gleba (Figure 5b–d). Gleba firm, solid, and whitish at first, becoming fawn {29} to umber {18} or date-brown {24} at maturity, ornamented with numerous branching white veins (Figure 5a,b). Odor intense, penetrating, reminiscent of gas mixed with a somewhat spicy aromatic scent, like horseradish (Armoracia rusticana). Taste mild and sweetish, occasionally with a mustard-like aftertaste. Asci excluding stalk (70.5) 80–109 (122.5) × (47) 58–83.5 (945) µm, Av = 94.6 × 72.3 µm (n = 100), irregularly subglobose, broadly ellipsoid or pyriform, generally without, occasionally with a short stalk measuring 33–52 × 9–18.5 µm, with 1–4 ascospores per ascus (1-spored 24%, 2-spored 37%, 3-spored 27%, 4-spored 12%, n = 500), walls approx. 1 μm thick (Figure 5e,f). Ascospores (23.0) 28.9–53.6 (61.7) × (17.8) 22.7–36.9 (42.5) µm, Av: 40.4 × 28.8, Q = (1.01) 1.23–1.57 (1.93), Qav: 1.40 (n = 400), excluding ornamentation, cinnamon-to-rusty brown at maturity, generally broadly ellipsoid-to-ellipsoid, but also sometimes subglobose, ovoid or even oblong, and reticulate–alveolate, with broad meshes, (2.4) 3.6–5.9 (10.4) µm in height; meshes regular, closed and regularly polygonal (5-6 sides), 3-5 (6) lengthwise (3 = 15%, 4 = 43%, 5 = 33%, 6 = 9%, n = 100) (Figure 5e,f). Ascospores from one-spored asci (39.8) 44.7–58.7 (61.7) × (26.1) 31.6–39.8 (42.5) µm, Av = 52 × 35.7 µm; Q = (1.2) 1.3–1.6 (1.8), Qav = 1.5 (n = 100); two-spored asci (30.6) 36.5–47.6 (53.2) × (23.5) 25.5–33.2 (40.2) µm, Av = 42.6 × 29.5 µm; Q = (1) 1.3–1.6 (1.9), Qav = 1.5 (n = 100); three-spored asci (24.5) 28.5–40.4 (47.2) × (19.4) 22.3–29.4 (33.3) µm, Av = 34.6 × 26 µm; Q = (1.1) 1.2–1.5 (1.6), Qav = 1.3 (n = 100); four-spored asci (23) 27.5–38 (43.4) × (17.8) 21.7–27.6 (29.3) µm, Av = 32.5 × 24.1 µm; Q = (1) 1.2–1.5 (1.8), Qav = 1.4 (n = 100).
Distribution and ecology: Tuber leptodermum grows in relatively humid, deciduous forests or stands of Populus, Quercus, Carpinus, Corylus and/or Salix, predominantly near water streams and lakes, in habitats where T. macrosporum and (more often) T. magnatum also appear (Figure 4). The ascomata were found from October to November, at the Kozani and Grevena prefectures in NW Greece.
Additional specimens examined: GREECE. KOZANI: under Populus, Quercus, Carpinus and Corylus, 25 November 2021, coll. P. Kladopoulou (ACAMTub359; GenBank: ITS = PP918908; LSU = PP905688); GREECE. KOZANI: under trees of the genera Populus (P. alba and P. tremula) and Quercus, 28 November 2021, coll. N. Tsilis, GK13815 (ACAMTub382; GenBank: ITS = PP918911; LSU = PP905691); GREECE. GREVENA: under Populus (P. alba, P. nigra and P. tremula), Quercus, Carpinus, Corylus and Salix, 7 November 2021, coll. I. Kalampoukas, GK13481 (ACAMTub379; ITS = PP918909; LSU = PP905689); GREECE. GREVENA: under Quercus and Salix, 17 November 2021, coll. I. Kalampoukas GK13509 (ACAMTub380; ITS = PP918910; LSU = PP905690).
Notes: T. leptodermum sp. nov. is distinct from other related species of the Maculatum clade, i.e., T. maculatum, T. rapaeodorum, T. lusitanicum, T. hubeiense, T. pseudomagnatum and T. foetidum, as demonstrated in the ITS and LSU, and in the concatenated ITS+LSU phylogenies (Figure 2 and Figure 3, Supplementary Figure S1). As regards morphology, T. leptodermum produces medium-sized (20–50 mm diam), dark-brown ascomata with a thin (up to 150 (170) μm) pseudoparenchymatous peridium, a fawn-to-umber or date-brown gleba when mature, and cinnamon-to-rusty brown ascospores with broad meshes; mature ascomata have an intense and penetrating odor reminiscent of gas mixed with a somewhat spicy aromatic scent, like horseradish. It is mostly found in riparian habitats with predominantly Populus spp. Comparisons of such features with those of the aforementioned phylogenetically related taxa reveal several differences in the size and color of ascomata, in the thickness (mainly) and/or structure of the peridium, in the presence of hair-like hyphae on the peridium surface, in the gleba color and/or in the shape, color and ornamentation of the ascospores. Especially as regards T. foetidum, which is the closest phylogenetic taxon to T. leptodermum, it is morphologically very similar to T. leptodermum, but it is distinguished from the latter by the sparse presence of very short hair-like hyphae on the peridium surface, a thicker (200–350 µm) peridium and a fetid odor [12,13].
As regards the seven Tuber species which are reported for the first time in Greece, a summary of the key morphoanatomic features of the respective collections, together with details about their distribution and ecology, are included in Supplementary Table S2. In addition, representative photos of their ascomata and microscopic characters are provided in Figure 6.
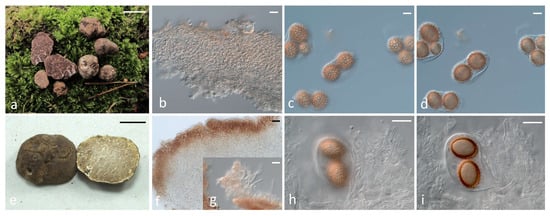

Figure 6.
Tuber species corresponding to first records for the Greek mycobiota: (a–d) T. anniae ascomata, peridium structure and ascospores; (e–i) T. buendiae ascomata, peridium structure, dermatocystidia and ascospores; (j–m) T. conchae ascomata, peridium structure and ascospores; (n–q) T. dryophilum ascomata, peridium structure, peridium with dermatocystidia and ascospores; (r–u) T. monosporum ascomata, peridium structure and ascospores; (v–y) T. regianum ascomata, peridium structure and ascospores, (z–ac); T. zambonelliae ascomata, peridium structure and ascospores. Bars: ascomata 1 cm; microscopic features 20 μm.
4. Discussion
The new truffle species, T. leptodermum, produces medium-sized (20–50 mm diam), dark-brown truffles which exhibit distinct morphological features, i.e. a felty smooth or sparsely warty, thin peridium (up to 150 (170) µm), pseudoparenchymatic, composed of globose-to-angular cells, and an exoperidium which is easily scratched off to reveal a light-colored endoperidium. In addition, the asci contain one to four reticulate–alveolate, ellipsoid ascospores with broad meshes. It was found to occur in riparian habitats, mainly under Populus and Salix spp., in autumn. T. leptodermum is a phylogenetically distinct species of the Maculatum clade, and is most closely related to T. foetidum, and then to T. hubeiense, T. maculatum, T. rapaeodorum, T. pseudomagnatum and T. lusitanicum, on the basis of the phylogenetic study performed.
When T. leptodermum was compared to the aforementioned species, the following differences were noted: T. hubeiense grows under Pinus armandii by forming small-sized (5–9 mm) whitish ascomata with a peridium of 180–250 µm, and yellow–brown-to-golden-brown ascospores [64]; T. lusitanicum also produces small-sized (5–20 mm) ascomata that are white at first, then pale-yellowish, sometimes with a reddish tinge, and are darker at maturity, with a thick peridium (300–500 µm) bearing sparse hair-like hyphae, an olive-brown-to-dark-brown gleba and yellowish-brown ascospores [59]; T. maculatum forms ascomata with a thick peridium (300–400 µm), prosenchymatous throughout, but often with scattered isodiametric cells or clusters of such cells, and with ellipsoid, light-golden-brown ascospores, ornamented with a regular reticulum 2–5 μm deep [91]; T. pseudomagnatum produces yellow–white ascomata with blackish gleba and small elliptic ascospores with a large meshed reticulum [63]; T. rapaeodorum produces small-sized (up to 25 mm), white-to-ochraceous or ochraceous-yellow (with dark-brown patches) ascomata with a thick peridium (250–600 µm), a snuff-brown-to-dark-purplish brown gleba and a horseradish- or radish-like odor [13].
In particular, as regards comparisons between T. foetidum and T. leptodermum, which appear to be the closest relatives (sequence identity values up to 95%), the two species demonstrate a high similarity in most of their morphoanatomical features; however, the former could be distinguished by the sparse presence of very short hair-like hyphae on the peridium surface, a thicker (200–350 µm) peridium and a fetid odor [12,13]. A phylogenetically distant species that could be confused with T. leptodermum is T. macrosporum; the latter also grows in similar (riparian) habitats, but it has is garlic-like odor and forms larger, darker colored ascospores with more meshes along the spore length.
Within the Aestivum clade, collections from our material were grouped into five species: T. aestivum, T. bituminatum, T. magnatum, T. mesentericum and T. panniferum; the presence of T. panniferum in Greece is molecularly confirmed for the first time. In addition, the occurrence of two other species of this clade, i.e., T. pulchrosporum and T. suave, has been previously recorded in various regions of the country and in the Attica area, respectively [27,29]. Especially as regards T. aestivum, a small number of seven-spored asci was noted in the majority of our collections, while a single eight-spored ascus was observed in one specimen only (Figure 7); hence, when seven collections of T. aestivum were examined, the presence of one-, two-, three-, four-, five-, six- and seven-spored asci was calculated at (percentages of total) 2.4%, 10.0%, 14.9%, 23.0%, 32.4%, 15.0% and 2.3%, respectively (n = 660). To the best of our knowledge, there is only one previous report on the presence of seven- and eight-spored asci in T. aestivum ascomata [92], despite the quite extensive pertinent literature. Noteworthy morphoanatomical variability was also detected in T. mesentericum; in measurements of ascospores in six collections, the quotient was Q = (1.01) 1.22 × 1.72 (2.17), with Qav = 1.47 (n = 613). These measurements differ substantially from the respective observations made by Leonardi et al. [27], suggesting the existence of more elongated ascospores in Greek T. mesentericum specimens. Furthermore, the latter is apparently limited to the northern part of the country, whereas T. bituminatum seems to be much more widespread and common (Supplementary Table S1).
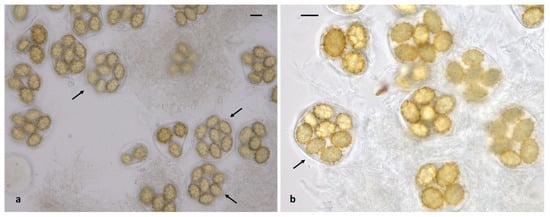
Figure 7.
Tuber aestivum asci and ascospores; arrows indicate the presence of (a) 7-spored asci and (b) an 8-spored ascus. Bars: 20 μm.
In the phylogenetic analysis of our study, the Excavatum clade consisted of six highly supported phylospecies which included collections initially determined either as T. excavatum Vittad. or as T. fulgens Quél. (the presence of the latter is molecularly confirmed for the first time); however, none of them could be assigned with certainty to either one of these species, due to a lack of sequences from the respective type material. Most of the Greek specimens were grouped in T. aff. excavatum 3, which corresponds to “T. excavatum Clade IV” sensu Puliga et al. [47]; this phylospecies exhibits a large distribution throughout mainland Greece in coniferous, deciduous or mixed forests. Two additional phylospecies, i.e., T. aff. excavatum 1 and T. aff. excavatum 2, corresponding to “T. excavatum Clade I” and “T. excavatum Clade III” sensu Puliga et al. [47], respectively, include collections from the northern part of the country growing in association with broadleaved trees only. It should be noted that the existence of species within the Excavatum clade in Greece is molecularly confirmed for the first time.
As regards the Gennadii clade, T. conchae is reported for the first time in Greece. According to the original description (Romero and Alvarado in Crous et al. [48]), this species differs from T. gennadii because the latter “… has smaller (0.5–2 cm diam) and globose or subglobose ascomata (rarely wrinkled or lobulated), develops long cracks or locules in the gleba with age, and is probably associated to a different host plant (Tuberaria guttata)”. Interestingly, our collection’s features do not support the distinction of the two species based on these differences, since it consists of a small ascoma of ca. 2 cm diam, its gleba possesses locules and cracks, and it was found in the vicinity of T. guttata (Figure 6, Supplementary Table S2); still, the ITS sequence of the Greek collection exhibits 99.6% identity with the species holotype from Spain, leaving no doubts about its identity. It should be noted that this same T. conchae collection was previously identified as T. gennadii by using an LSU sequence only [26]. T. gennadii was described by Chatin [16] from material deriving from the Peloponnese, Greece. Recent collections from other regions of the country were examined and were identified as such together with specimens from Italy, Spain and north Morocco [23,26]; these correspond to three distinct phylospecies in our ITS tree (Figure 1). Our collections were provisionally labeled as Tuber aff. gennadii, and are grouped within one of the three aforementioned phylospecies by also including a specimen from Italy (BM1904, Genebank HM485361) which is considered to represent T. gennadii [43]. Therefore, the true phylogenetic position of this species will remain ambiguous until the holotype is sequenced.
The presence of T. macrosporum is molecularly confirmed for the first time in Greece. The respective collections are included—together with other sequences deriving from various areas of the Mediterranean Basin—in of one of the three phylogenetic groups formed with high support in the ITS tree (Figure 1); this particular group corresponds to T. macrosporum Clade I (sensu Benucci et al. [52]), while another group represents T. macrosporum Clade II, comprising sequences from France and Italy. The identity values between the sequences of T. macrosporum Clade I and II are less than 96.5%. The third group seems to be further separated, and consists of material originating from Iran [46]. Another species of the Macrosporum clade is T. monosporum, as revealed by the identity of a single collection from Greece which possesses the typical (and unique) morphoanatomical features of the species, and it was identified as such, although no pertinent sequences have been available until now in the international databases. This finding constitutes a new record for Greek mycobiota. It is worth mentioning that T. monosporum forms ascospores that differ considerably from those of other Tuber spp., since their surface has polygonal craters (like inward meshes) resembling a golf ball (Figure 6, Supplementary Table S2); therefore, it was originally placed in a distinct genus, i.e., Paradoxa monospora Mattir. However, Vizzini [93] transferred it to the genus Tuber as T. monosporum (Mattir.) Vizzini, and our phylogenetic analysis supports this placement.
As regards the Melanosporum clade in our phylogeny, it consists of six phylogroups, three corresponding to well-known species, i.e., T. brumale (Europe), T. melanosporum Vittad. (Europe) and T. indicum Cooke & Massee (China), whereas the rest represent the recently described T. cryptobrumale Merényi, T. Varga & Bratek, T. petrophilum Milenković, P. Jovan., Grebenc, Ivančević & Marković and T. thracicum Slavova, M. Leonardi, A. Paz, Assyov & Pacioni, all originating from the Balkan peninsula. Specimens from Greece are grouped either within the T. brumale species complex, or identified as T. melanosporum. Especially as regards the former, recent results indicate that the Melanosporum clade may have undergone evolutionary diversification, separating populations from eastern and southeastern Europe from those present in central and western Europe [8,9,70]. In addition, Mereneyi et al. [8] demonstrated that the “T. brumale aggregate” comprises three phylospecies corresponding to haplogroups I and II in Clade A and Clade B. The latter coincides with T. cryptobrumale [33], while haplogroup II exhibits a distribution which is limited to the wider Balkan region, including Greece, as our findings confirm; however, haplogroups I and II have yet to be formally described as distinct species. Concerning T. melanosporum, its occurrence in Greece has not been verified in areas other than those in close proximity to truffle orchards where this species is cultivated. Hence, and in the absence of other pertinent records in the country (or in neighboring regions), we believe that T. melanosporum is not a native species.
In the Puberulum clade, T. dryophilum is a relatively small, whitish and hairless truffle (or with very short hairs), with ellipsoid ascospores and a spore reticulum with very wide meshes, i.e., two to three along the spore length, as well as a plectenchymatous peridium structure [12]. However, the results of Lancellotti et al. [94] and our own observations on the respective material indicate the existence of a pseudoparenchymatic peridium with large angular cells and more rounded ascospores with 4–7 (8) meshes lengthwise (Figure 6; Supplementary Table S2). Thus, it is quite possible that specimens of T. dryophilum studied by Montecchi and Sarasini [12] correspond to another taxon from the reference collection of T. dryophilum (AF003917), as the latter is defined by Lancelotti et al. [94], and our specimens were compared with this. It is worth noting that ambiguities in discriminating the small whitish truffles of the Puberulum group s.l. (which includes the Maculatum and Gibbosum clades) are well documented, particularly by Lancellotti et al. [94], who stated that only 4 out of 44 sequences initially labeled as T. dryophilum were correctly identified.
As regards the rest of the species within the Puberulum clade, T. anniae W. Colgan & Trappe was first described in North America, growing in association with conifers. The respective Greek specimen is the first ever recorded in Greece, and it is morphoanatomically very similar to the holotype of T. anniae and other pertinent descriptions [11,95,96,97], by sharing the same pseudoparenchymatic peridium structure and the coniferous habitat (Pinus sylvestris, N. Greece). Furthermore, in our ITS phylogeny, T. borchii appears as a sister group to T. anniae (100%, 1.00; Figure 1). According to previous studies [32,72] this species consists of two “Haplotypes” with no apparent morphoanatomical or ecological differences between them. The majority of the Greek collections identified as T. borchii correspond to “Haplotype 1”, while three were grouped in “Haplotype 2” (Supplementary Table S1). As concerns T. oligospermum, the results of our phylogenetic analysis are in agreement with those of Alvarado et al. [26] by demonstrating that it corresponds to a complex of (at least) four distinct phylogenetic species with a predominantly Mediterranean distribution, three of which comprise specimens from Greece. To better distinguishing the latter, we have provisionally named them as T. aff. oligospermum 1, T. aff. oligospermum 2 and T. aff. oligospermum 3. Still, the phylogenetic grouping of T. oligospermum s.str. remains unclear, and to elucidate this issue, sequencing of the type specimen is required. Interestingly enough, our phylogenetic analysis shows that specimens previously reported as “T. puberulum” [26], i.e., GK4352 (JN392223) from Xanthi and GK5601 (JN392224) from Kozani, are grouped in T. dryophilum; thus, the presence of the latter in Greece is reported for the first time. In contrast, the existence of T. puberulum Berk. & Broome in the country has yet to be confirmed. This species was originally described from a collection found in the United Kingdom, and according to Lancellotti et al. [94], it appears almost exclusively in central and northern Europe, while it has not been recorded in Italy and in the Balkan peninsula.
The Regianum clade is composed of only three European species, i.e., T. bernardini Gori, T. megentipuncatum and T. regianum, demonstrating similar/overlapping morphoanatomic features, which makes it difficult to distinguish them by classical taxonomy only. Cseh et al. [28] reported that they could be discriminated by their relative ascospore size and volume, as well as by the average size of their ascospore meshes. However, our T. regianum collection possesses the T. magenctipunctatum ascospore pattern, both as regards size and volume. The presence of T. regianum is reported for the first time in Greece.
The Rufum clade includes some of the most taxonomically challenging groups, since it consists of many closely related (and several still undescribed) species exhibiting high phenotypic similarity, particularly within the T. rufum complex and in Mediterranean habitats [76,81,98]. In the frame of our work, the existence of T. buendiae and T. zambonelliae is recorded for the first time in Greece, thus expanding the known distribution of these species, since their presence had been reported in Spain and Morocco only. Moreover, sequences from Greek collections show a relatively high variation (up to 97.5% sequence identity values) compared to the holotypes of the two species. In addition, the existence of T. nitidum in Greece is molecularly confirmed for the first time, while T. aereum is also reported on Andros island, very close to the type locality (Naxos island; [30]). Apart from the four aforementioned taxa, our collections are grouped into three other well-supported phylospecies which were provisionally named T. aff. rufum 1, T. aff. rufum 2 and T. aff. rufum 3; the first two are recorded for the first time in Greece, while the existence of the third was also reported by Alvarado et al. [75]. Still, none of them could be assigned with certainty to T. rufum, due to absence of sequences from the respective type material. In the past, the presence of T. rufum was recorded in Greece as T. rufum var. rufum Pollini [20,21,99], as T. rufum var. apiculatum E. Fisch. [20] and as T. rufum f. lucidum (H. Bonnet) Montecchi & Lazzari [21], but it is not known yet to which of the aforementioned phylospecies these findings correspond to.
The findings of this study substantially increase our knowledge of truffle diversity, the range of associated plants, and their geographic distribution in Greece (Table 2). As a result, a total of 40 species (including 10 undescribed phylogenetic species) is now known to exist, as compared to 26 species which were previously reported, while the number of molecularly confirmed species has increased from 15 to 20.


Table 2.
A summarized review of the known diversity of the genus Tuber in Greece, also including the findings of this study: taxa grouped per phylogenetic clade, associated plant(s), geographic distribution according to administrative regions of Greece, and pertinent references. Species whose presence has been confirmed by molecular approaches are presented in bold. Abbreviations for the administrative regions of Greece are as follows: East Macedonia and Thrace (EM & T), Western Macedonia (WM), Central Macedonia (CM), Epirus (Ep), Thessaly (Th), Western Greece (WG), Central Greece (CG), Attica (At), Peloponnese (P), Ionian Islands (I), North Aegean (NA), South Aegean (SA) and Crete (Cr). A map depicting the administrative regions of Greece is provided in the Supplementary Materials, Figure S2.
Table 2.
A summarized review of the known diversity of the genus Tuber in Greece, also including the findings of this study: taxa grouped per phylogenetic clade, associated plant(s), geographic distribution according to administrative regions of Greece, and pertinent references. Species whose presence has been confirmed by molecular approaches are presented in bold. Abbreviations for the administrative regions of Greece are as follows: East Macedonia and Thrace (EM & T), Western Macedonia (WM), Central Macedonia (CM), Epirus (Ep), Thessaly (Th), Western Greece (WG), Central Greece (CG), Attica (At), Peloponnese (P), Ionian Islands (I), North Aegean (NA), South Aegean (SA) and Crete (Cr). A map depicting the administrative regions of Greece is provided in the Supplementary Materials, Figure S2.
| A/A | CLADE/Taxon | Associated Plant(s) | Distribution | References * |
|---|---|---|---|---|
| AESTIVUM | ||||
| 1 | T. aestivum Vittad. 1 | Abies, Pinus, Quercus, Corylus, Cistus | EM & T, CM, WM, Ep, Th, WG, CG, At, P, SA | [18,20,21,41,100,101,102,103]; this study |
| 2 | T. bituminatum Berk. & Broome | Abies cephalonica, Abies borisii-regis, Pinus nigra, Quercus pubescens, Quercus coccifera, Quercus, Fagus sylvatica | WM, CM, Th, CG, P | [27]; this study |
| 3 | T. magnatum Picco | Populus, Carpinus, Quercus, Salix, Tilia, Corylus, Pinus | WM, CM, Th | [9,25,102]; this study |
| 4 | T. mesentericum Vittad. 1 | F. sylvatica, Fagus, Corylus avelana, Corylus, Carpinus betulus, Carpinus, Quercus, Populus, Salix, Tilia, Pinus nigra, Pinus | EM & T, WΜ, CM | [27,104]; this study |
| 5 | T. panniferum Tul. & C. Tul. | Q. pubescens, Q. coccifera, Quercus ilex, Quercus | EM & T, CM, SA, Cr | [20,21,102]; this study |
| 6 | T. pulchrosporum Konstantinidis, Tsampazis, Slavova, Nakkas, Polemis, Fryssouli & Zervakis | Quercus, Corylus, Carpinus, Pinus | EM & T, WM, Ep, Th, WG, At | [29] |
| 7 | T. suave Pacioni & M. Leonardi | Q. coccifera, Pinus halepensis | At | [27] |
| EXCAVATUM | ||||
| 8 | T. excavatum Vittad. | Abies, Pinus, Quercus, Corylus, Tilia, Salix | EM & T, CM, WM, Ep, P | [20,21,99] |
| 9 | T. aff. excavatum 1 | Corylus | WM | This study |
| 10 | T. aff. excavatum 2 | F. sylvatica | EM & T, WM | This study |
| 11 | T. aff. excavatum 3 | A. cephalonica, Q. coccifera, Q. ilex, Quercus, F. sylvatica, Carpinus, P. halepensis | EM & T, CM, WM, CG, At, P | This study |
| 12 | T. fulgens Quél. | Abies, F. sylvatica, Corylus, Carpinus, Quercus | EM & T, CM, WM, Th | [21]; this study |
| GENNADII | ||||
| 13 | T. asa Tul. & C. Tul. 2 | Cistus | At | [23] |
| 14 | T. conchae M. Romero & P. Alvarado 3 | Tuberaria guttata | EM & T | This study |
| 15 | T. gennadii (Chatin) Pat. 4 | Helianthemum, Cistaceae | EM & T, CG, At, P | [16,21,23,26,105] |
| 16 | T. aff. gennadii | T. guttata, P. halepensis, Pinus pinea, Q. coccifera | At | This study |
| MACROSPORUM | ||||
| 17 | T. macrosporum Vittad. | Corylus avelana, Corylus, Carpinus, Quercus, Salix, Tilia, Populus | EM & T, WM, CM, Th | [24,102]; this study |
| 18 | T. monosporum(Mattir.) Vizzini | F. sylvatica | WM | This study |
| MACULATUM | ||||
| 19 | T. leptodermum Daskalopoulos, Konstantinidis, Tsilis, Polemis & Zervakis | Populus alba, Populus tremula, Populus nigra, Quercus, Salix, Corylus, Carpinus | WM | This study |
| MELANOSPORUM | ||||
| 20 | T. brumale Vittad. 5 | Quercus, Carpinus, Fagus | EM & T, Ep, CM, WM, P | [8,20,21,33,104]; this study |
| 21 | T. melanosporum Vittad. 6 | Quercus, Carpinus | CM | [20,21,102,104]; this study |
| 22 | T. cibarium Corda | [17] | ||
| PUBERULUM | ||||
| 23 | T. anniae W. Colgan & Trappe | Pinus sylvestris | CM | This study |
| 24 | T. borchii Vittad. | Abies, Pinus, Quercus, Erica | EM & T, CM, WM, Ep, Th, WG, CG, At, P, NA, SA | [20,21,26,99,102,103,104]; this study |
| 25 | T. dryophilum Tul. & C. Tul. | Quercus, Carpinus, Pinus | EM & T, WM, Th, At, P | This study |
| 26 | T. aff. oligospermum 1 | P. halepensis, P. pinea, Q. coccifera | At | [26]; this study |
| 27 | T. aff. oligospermum 2 | Quercus | CG | [26]; this study |
| 28 | T. aff. oligospermum 3 | P. halepensis, Q. ilex, C. monspeliensis | At | [26]; this study |
| 29 | T. puberulum Berk. & Broome 7 | Quercus | EM & T, WM | [20,26,104] |
| REGIANUM | ||||
| 30 | T. magentipunctatum Merényi, I. Nagy, Stielow & Bratek | A. cephalonica, C. avelana | WM, P | [28] |
| 31 | T. regianum Montecchi & Lazzari | F. sylvatica | WM | This study |
| RUFUM | ||||
| 32 | T. aereum Polemis, Daskalopoulos & Zervakis | Q. coccifera, Q. pubescens, Q. macrolepis | SA | [30] |
| 33 | T. buendiae Ant. Rodr. & Morte | A. cephalonica | CG | This study |
| 34 | T. ferrugineum Vittad. | Abies, Corylus, Quercus | CM, WM, Th, CG, At, P, NA | [21,99,106] |
| 35 | T. nitidum Vittad. 8 | Quercus, Carpinus | EM & T, Ep, Th, At | [20]; this study |
| 36 | Tuber rufum Pollini 9 | Abies, Quercus, Carpinus, Cistus | EM & T, WM, CM, Th, At | [20,21,99] |
| 37 | T. aff. rufum 1 | Populus, Carpinus, Salix, Tilia | CM | This study |
| 38 | T. aff. rufum 2 | F. sylvatica | WM | This study |
| 39 | T. aff. rufum 3 10 | A. cephalonica, Pinus nigra, Pinus, Q. coccifera, Quercus, F. sylvatica | EM & T, WM, Ep, Th, CG, At, P | [75]; this study |
| 40 | T. zambonelliae Ant. Rodr. & Morte | P. halepensis, Q. ilex, Quercus, Carpinus | WM, At | This study |
1 Reported findings prior to 2021 could correspond to any one of the following species: T. aestivum, T. mesentericum, T. bituminatum and T. suave. 2 The collections initially identified as T. asa [23] were later assessed as T. oligospermum [26]. 3 The collection was initially identified as Loculotuber gennadii [21], and later as T. gennadii [26]. 4 The collections identified as T. gennadii by Alvarado et al. [26] correspond to more than one phylospecies, which also include T. aff. gennadii. 5 Reported as T. brumale or T. brumale f. moschatum in the pertinent literature. 6 The only molecularly confirmed record of this species that does not originate directly from a truffle orchard with cultivated T. melanosporum (but in the vicinity of one) derives from this study. 7 The collections examined by Alvarado et al. [26] were identified as T. dryophilum by this study. 8 Reported as Tuber rufum var. nitidum (Vittad.) Mont. & Lazzari by Diamandis and Perlerou [20]. 9 Reported either as T. rufum, T. rufum var. rufum, or T. rufum f. lucidum or T. rufum var. apiculatum in the pertinent literature. 10 Reported as T. rufum by Alvarado et al. [75]. * References [8,9,26,27,28,29,30,41,75] include molecularly identified specimens.
5. Conclusions
In the frame of the study, the presence of 31 phylospecies of the genus Tuber is reported in Greece. These correspond to one new species to science (i.e., Tuber leptodermum); to 20 well-described taxa, 7 of which (T. anniae, T. buendiae, T. conchae, T. dryophilum, T. monosporum, T. regianum, and T. zambonelliae) constitute first national records; and to 10 undescribed phylogenetic species within the T. excavatum, T. gennadii, T. oligospermum and T. rufum species complexes (the existence of 6 is revealed for the first time). Collections of the T. excavatum complex are grouped into three phylospecies, while those initially identified as T. oligospermum and T. rufum appear as members of three distinct clades in each one of the respective species complexes. Moreover, sequences from Greece labeled as T. gennadii correspond to two distinct phylospecies, while the presence of T. puberulum in Greece could not be confirmed, since all specimens previously identified as such were grouped within T. dryophilum. T. melanosporum is considered non-native to the country, and T. bituminatum seems to be much more common than the phylogenetically related and morphologically similar T. mesentericum. In addition, a sequence representing T. monosporum has been generated (and becomes available) for the first time, representing a collection deriving from Northern Greece. Our results, as well as the outcome of other pertinent studies, emphasize the need for epitypification (or neotypification) of several truffle species occurring in Europe. In addition, a combined effort of truffle taxonomists could focus on the description of cryptic species within the Rufum and Excavatum clades, and among the small whitish truffles (mainly in the Puberulum and Gennadii clades), where the corresponding collections demonstrate high levels of phenotypic similarity and considerable sequence variation.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/jof11050358/s1: Figure S1: A maximum likelihood (ML) tree, based on LSU sequences, depicting the phylogeny of the Maculatum clade by including Tuber leptodermum sp. nov., which appears in bold typeface. Choiromyces helanshanensis is used as an outgroup. Bootstrap support (>70%) for ML and Bayesian posterior probabilities (>0.95) are indicated at the tree nodes; values of 100% and 1.00, respectively, are marked by asterisks. The use of sequences from type specimens is indicated by “T” placed as superscript after the respective GenBank accession number. Figure S2: A map depicting the administrative regions of Greece. Abbreviations (and the respective colors) are as follows: East Macedonia and Thrace (EM & T—in red), Western Macedonia (WM—in purple), Central Macedonia (CM—in green), Epirus (Ep—in blue), Thessaly (Th—in orange), Western Greece (WG—in yellow), Central Greece (CG—in red), Attica (At—in purple), Peloponnese (P—in blue), Ionian Islands (I—in magenta), North Aegean (NA—in yellow), South Aegean (SA—in green) and Crete (Cr—in orange). Table S1. Biological material of the genus Tuber deriving from Greece and examined in the frame of this study: phylogenetic clade, taxon, fungarium (collection) code and collector’s code, geographic origin, associated plant(s) and GenBank accession no. for ITS and LSU sequences. Collections used in the phylogenetic analyses are presented in bold typeface. Table S2: Detailed descriptions of Tuber anniae, T. buendiae, T. conchae, T. dryophilum, T. monosporum, T. regianum and T. zambonelliae, which constitute first records for the Greek mycobiota. The number of samples, as well as the numbers of all measurements, are shown in brackets (n).
Author Contributions
Conceptualization, V.D., E.P. and G.I.Z.; methodology, V.D., E.P., V.F., V.N.K. and G.I.Z.; software, V.D., E.P., V.F., V.N.K. and G.I.Z.; validation, V.D., E.P., G.K., V.K., V.N.K. and G.I.Z.; formal analysis, V.D., E.P., V.N.K. and G.I.Z.; investigation, V.D., E.P., G.K., V.K. and N.T.; resources, E.P. and G.I.Z.; data curation, V.D., E.P., V.N.K. and G.I.Z.; writing—original draft preparation, V.D., E.P. and G.I.Z.; writing—review and editing, V.D., E.P., V.F., V.N.K. and G.I.Z.; visualization, V.D., E.P., G.K., V.K. and N.T.; supervision, E.P., V.N.K. and G.I.Z.; project administration, E.P. and G.I.Z.; funding acquisition, E.P. and G.I.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This project was partly supported by the Hellenic Foundation for Research and Innovation (H.F.R.I.) under the “2nd Call for H.F.R.I. Research Projects to support Post-Doctoral Researchers” (Project Number: 1057).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in the manuscript are available on request from the corresponding author. In addition, sequences generated by this study are deposited in GenBank, and phylogenetic trees and pertinent data are deposited in TreeBASE.
Acknowledgments
We would like to thank the following individuals for providing truffle specimens: P. Aidonopoulos, M. Alvanos, N. Arabatzis, N. Arapopoulou, G. Argyropoulos, Ch. Bogiatzis, Ch. Chrysopoulos, G. Donatiello, D. Drivas, D. Giannouhidis, K. Giatra, M. Gkillas, G. Harokopakis, I. Kalaboukas, P. Kladopoulou, V. Koutsoukis, S. Kyriacou, E. Lahouvaris, D. Lazaridis, P. Litsas, G. Mertzanis, E. Mouratidis, V. Mylonas, K. Nola, P. Ntonados, N. Pallas, P. Panagiotidis, V. Panagiotopoulos, D. Panteloglou, E. Papadopoulos, F. Paraskevaiadis, S. Patsahis, G. Setkos, N. Sgouridi, P. Siafarikas, D. Sofos, E. Stergiou, D. Tabouras, N. Theodorou, S. Tsirovasilis, E. Tziatziou, E. Vamvakas, P. Varypatis, E. Velissaris, Al. Ziogas, Ar. Ziogas and V. Ziogas. In addition, we acknowledge the support, in various stages of our research, of G. Bekiaris, A.C. Christinaki, S. Christodoulou, K. Geballa-Koukoulas, A.M. Kortsinoglou, E. Pavlou, N.S. Sotiropoulou, and C. Tsiouni.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bonito, G.M.; Smith, M.E. General Systematic Position of the Truffles: Evolutionary Theories. In True Truffle (Tuber spp.) in the World: Soil Ecology, Systematics and Biochemistry; Zambonelli, A., Iotti, M., Murat, C., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 3–18. [Google Scholar] [CrossRef]
- Zambonelli, A.; Iotti, M.; Murat, C. (Eds.) True Truffle (Tuber spp.) in the World; Soil Biology 47; Springer International Publishing: Cham, Switzerland, 2016; 436p. [Google Scholar] [CrossRef]
- Graziosi, S.; Hall, I.R.; Zambonelli, A. The Mysteries of the White Truffle: Its Biology, Ecology and Cultivation. Encyclopedia 2022, 2, 1959–1971. [Google Scholar] [CrossRef]
- Qin, J.; Feng, B. Life Cycle and Phylogeography of True Truffles. Genes 2022, 13, 145. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.W.; Thomas, H.W. Mycorrhizal Fungi and Invertebrates: Impacts on Tuber melanosporum Ascospore Dispersal and Lifecycle by Isopod Mycophagy. Food Webs 2022, 33, e00260. [Google Scholar] [CrossRef]
- Cheddadi, R.; Bennett, K.D. Past Climate Changes and the Role of Refugia in the Temperate Northern Hemisphere. Past Glob. Changes Mag. 2020, 28, 1. [Google Scholar] [CrossRef]
- García-Cunchillos, I.; Sánchez, S.; Barriuso, J.J.; Pérez-Collazos, E. Population Genetics of the Westernmost Distribution of the Glaciations-Surviving Black Truffle Tuber melanosporum. Mycorrhiza 2014, 24, 89–100. [Google Scholar] [CrossRef]
- Merényi, Z.; Varga, T.; Geml, J.; Orczán, Á.K.; Chevalier, G.; Bratek, Z. Phylogeny and Phylogeography of the Tuber brumale aggr. Mycorrhiza 2014, 24, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Belfiori, B.; D’Angelo, V.; Riccioni, C.; Leonardi, M.; Paolocci, F.; Pacioni, G.; Rubini, A. Genetic Structure and Phylogeography of Tuber magnatum Populations. Diversity 2020, 12, 44. [Google Scholar] [CrossRef]
- Bonito, G.; Smith, M.E.; Nowak, M.; Healy, R.A.; Guevara, G.; Cázares, E.; Vilgalys, R. Historical Biogeography and Diversification of Truffles in the Tuberaceae and Their Newly Identified Southern Hemisphere Sister Lineage. PLoS ONE 2013, 8, e52765. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Li, T.; Xu, Y.Y.; Yan, X.Y. Species Diversity, Phylogeny, Endemism and Geography of the Truffle Genus Tuber in China Based on Morphological and Molecular Data. Persoonia 2022, 48, 175–202. [Google Scholar] [CrossRef] [PubMed]
- Montecchi, A.; Sarasini, M. Funghi Ipogei d’Europa; AMB Fondazione Centro Studi Micologici: Trento, Italy, 2000; 714p. [Google Scholar]
- Halász, K.; Bratek, Z.; Szegő, D.; Rudnóy, S.; Rácz, I.; Lásztity, D.; Trappe, J.M. Tests of Species Concepts of the Small, White, European Group of Tuber spp. Based on Morphology and rDNA ITS Sequences with Special Reference to Tuber rapaeodorum. Mycol. Prog. 2005, 4, 281–290. [Google Scholar] [CrossRef]
- Marjanovic, Z.; Grebenc, T.; Markovic, M.; Glisic, A.; Milenkovic, M. Ecological Specificity and Molecular Diversity of Truffles (Tuber spp.) Originating from Mid-West of the Balkan Peninsula. Sydowia 2010, 62, 67–87. [Google Scholar]
- Sibthorp, J.; Smith, J.E. Florae Graecae prodromus; Sive Plantarum omnium Enumeratio; Typis Richardi Taylor: London, UK, 1806–1813; 438p. [Google Scholar]
- Chatin, A. Truffes (Terfaz) de Grèce, Terfezia gennadii. Bull. Soc. Bot. Fr. 1896, 43, 611–617. [Google Scholar] [CrossRef]
- Landerer, X. Über die in Griechenland Vorkommenden Kryptogamischen Pflanzen und Deren Bedeutung bei den Alten Griechen. Oesterr. Bot. Wochenschr. 1854, 38, 82–84. [Google Scholar] [CrossRef]
- Pantidou, M.E. Fungus-Host Index for Greece; Benaki Phytopathological Institute: Kifisia, Greece, 1973; 382p. [Google Scholar]
- Zervakis, G.; Lizoň, P.; Dimou, D.; Polemis, E. Annotated Check-List of the Greek Macrofungi. II. Ascomycotina. Mycotaxon 1999, 72, 487–506. [Google Scholar]
- Diamandis, S.; Perlerou, C. Recent Records of Hypogeous Fungi in Greece. Acta Mycol. 2008, 43, 2. [Google Scholar] [CrossRef][Green Version]
- Konstantinidis, G. Mushrooms (Mushroom-Collector’s Photographic Guide), 1st ed.; G. Konstantinidis: Grevena, Greece, 2009; 559p. (In Greek) [Google Scholar]
- Konstantinidis, G. Mushrooms of Greece, a Pocket Guide; G. Konstantinidis: Grevena, Greece, 2020; 383p. (In Greek) [Google Scholar]
- Agnello, C.; Kaounas, V. Tuber asa and Tuber genadii. A Close Morphological Study of Two Species Often Confused in the Past with a Brief Historical Bibliographic Summary. Ascomycete.org 2011, 3, 65–74. [Google Scholar]
- Gyosheva, M.; Assyov, B.; Konstantinidis, G.; Stoykov, D. Collections of Tuber macrosporum from the Balkan Peninsula (Bulgaria and Greece). Ascomycete.org 2012, 4, 75–78. [Google Scholar]
- Christopoulos, V.; Psoma, P.; Diamandis, S. Site Characteristics of Tuber magnatum in Greece. Acta Mycol. 2013, 48, 27–32. [Google Scholar] [CrossRef]
- Alvarado, P.; Moreno, G.; Manjón, J.L. Comparison between Tuber gennadii and T. oligospermum Lineages Reveals the Existence of the New Species T. cistophilum (Tuberaceae, Pezizales). Mycologia 2012, 104, 894–910. [Google Scholar] [CrossRef]
- Leonardi, M.; Salvi, D.; Iotti, M.; Rana, G.L.; Paz-Conde, A.; Pacioni, G. Multilocus Phylogeography of the Tuber mesentericum Complex Unearths Three Highly Divergent Cryptic Species. J. Fungi 2021, 7, 1090. [Google Scholar] [CrossRef]
- Cseh, P.; Merényi, Z.; Bóna, L.; Varga, T.; Bóka, K.; Nagy, I.; Bratek, Z. Taxonomic Characterisation of the Regianum Clade (Tuber) and the Trait Evolution of Spore Size Among True Truffles. Mycol. Prog. 2024, 23, 11. [Google Scholar] [CrossRef]
- Polemis, E.; Konstantinidis, G.; Fryssouli, V.; Slavova, M.; Tsampazis, T.; Nakkas, V.; Assyov, B.; Kaounas, V.; Zervakis, G.I. Tuber pulchrosporum sp. nov., a Black Truffle of the Aestivum Clade (Tuberaceae, Pezizales) from the Balkan Peninsula. MycoKeys 2019, 47, 35–51. [Google Scholar] [CrossRef]
- Crous, P.W.; Jurjević, Z.; Balashov, S.; De la Peña-Lastra, S.; Mateos, A.; Pinruan, U.; Osieck, E.R.; Altés, A.; Czachura, P.; Esteve-Raventós, F.; et al. Fungal Planet Description Sheets: 1614–1696. Fungal Syst. Evol. 2024, 13, 183–440. [Google Scholar] [CrossRef] [PubMed]
- Flora of British Fungi: Colour Identification Chart; Royal Botanic Garden Edinburgh: Edinburgh, UK, 1969; p. 6.
- Leonardi, M.; Iotti, M.; Mello, A.; Vizzini, A.; Paz-Conde, A.; Trappe, J.; Pacioni, G. Typification of the Four Most Investigated and Valuable Truffles: Tuber aestivum Vittad., T. borchii Vittad., T. magnatum Picco, and T. melanosporum Vittad. Cryptogam. Mycol. 2021, 42, 149–170. [Google Scholar] [CrossRef]
- Merényi, Z.; Varga, T.; Hubai, A.G.; Pitlik, P.; Erős, Á.; Trappe, J.M.; Bratek, Z. Challenges in the Delimitation of Morphologically Similar Species: A Case Study of Tuber brumale Agg. (Ascomycota, Pezizales). Mycol. Prog. 2017, 16, 613–624. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS Primers with Enhanced Specificity for Basidiomycetes—Application to the Identification of Mycorrhizae and Rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Vilgalys, R.; Hester, M. Rapid Genetic Identification and Mapping of Enzymatically Amplified Ribosomal DNA from Several Cryptococcus Species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- Arita, M.; Karsch-Mizrachi, I.; Cochrane, G. The International Nucleotide Sequence Database Collaboration. Nucleic Acids Res. 2021, 49, D121–D124. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Abarenkov, K. The UNITE Database for Molecular Identification of Fungi: Handling Dark Taxa and Parallel Taxonomic Classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Gryndler, M.; Hršelová, H.; Soukupová, L.; Streiblová, E.; Valda, S.; Borovička, J.; Miko, M. Detection of Summer Truffle (Tuber aestivum Vittad.) in Ectomycorrhizae and in Soil Using Specific Primers. FEMS Microbiol. Lett. 2011, 318, 84–91. [Google Scholar] [CrossRef]
- Hilszczańska, D.; Siebyła, M.; Horak, J.; Król, M.; Podsadni, P.; Steckiewicz, P.; Turło, J. Comparison of Chemical Composition in Tuber aestivum Vittad. of Different Geographical Origin. Chem. Biodivers. 2016, 13, 1617–1629. [Google Scholar] [CrossRef]
- Daskalopoulos, V.; Polemis, E.; Kioupidi, I.E.; Trigas, P.; Zervakis, G.I. Evaluation of the Colonization of Plants from Five Quercus Taxa Native to Greece by Tuber aestivum (Ascomycota, Pezizales). Life 2024, 14, 852. [Google Scholar] [CrossRef] [PubMed]
- Mello, A.; Murat, C.; Vizzini, A.; Gavazza, V.; Bonfante, P. Tuber magnatum Pico, a Species of Limited Geographical Distribution: Its Genetic Diversity Inside and Outside a Truffle Ground. Environ. Microbiol. 2005, 7, 55–65. [Google Scholar] [CrossRef]
- Bonito, G.M.; Gryganskyi, A.P.; Trappe, J.M.; Vilgalys, R. A Global Meta-Analysis of Tuber ITS rDNA Sequences: Species Diversity, Host Associations and Long-Distance Dispersal. Mol. Ecol. 2010, 19, 4994–5008. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Osieck, E.R.; Jurjević, Ž.; Boers, J.; Van Iperen, A.L.; Starink-Willemse, M.; Maggs-Kölling, G. Fungal Planet description sheets: 1284–1382. Persoonia 2021, 47, 178. [Google Scholar] [CrossRef] [PubMed]
- Bonito, G.; Trappe, J.M.; Donovan, S.; Vilgalys, R. The Asian Black Truffle Tuber indicum Can Form Ectomycorrhizas with North American Host Plants and Complete Its Life Cycle in Non-Native Soils. Fungal Ecol. 2011, 4, 83–93. [Google Scholar] [CrossRef]
- Puliga, F.; Illice, M.; Iotti, M.; Leonardi, P.; Baghdadi, A.; Mozafari, A.A.; Zambonelli, A. True Truffle Diversity in Iran. Ital. J. Mycol. 2021, 50, 52–62. [Google Scholar] [CrossRef]
- Puliga, F.; Illice, M.; Iotti, M.; Baldo, D.; Zambonelli, A. Tuber iranicum sp. nov., a Truffle Species Belonging to the Excavatum Clade. Mycologia 2020, 112, 932–940. [Google Scholar] [CrossRef]
- Crous, P.W.; Osieck, E.R.; Shivas, R.G.; Tan, Y.P.; Bishop-Hurley, S.L.; Esteve-Raventós, F.; Larsson, E.; Luangsa-Ard, J.J.; Pancorbo, F.; Balashov, S.; et al. Fungal Planet Description Sheets: 1478–1549. Persoonia 2023, 50, 158–310. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Lombard, L.; Roets, F.; Swart, W.J.; Alvarado, P.; Popov, E.S. Fungal Planet Description Sheets: 951–1041. Persoonia 2019, 43, 223. [Google Scholar] [CrossRef]
- Wan, S.P.; Wang, X.H.; Zheng, Y.; Yu, F.Q. Tuber shidianense and Tuber calosporum, Two New Truffle Species from Southwest China. Mycoscience 2016, 57, 393–399. [Google Scholar] [CrossRef]
- Fan, L.; Feng, S.; Cao, J.Z. The Phylogenetic Position of Tuber glabrum sp. nov. and T. sinomonosporum nom. nov., Two Paradoxa-like Truffle Species from China. Mycol. Prog. 2014, 13, 241–246. [Google Scholar] [CrossRef]
- Benucci, G.M.N.; Csorbai, A.G.; Falini, L.B.; Marozzi, G.; Suriano, E.; Sitta, N.; Donnini, D. Taxonomy, Biology and Ecology of Tuber macrosporum Vittad. and Tuber mesentericum Vittad. In True Truffle (Tuber spp.) in the World; Zambonelli, A., Iotti, M., Murat, C., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 69–86. [Google Scholar] [CrossRef]
- Healy, R.A.; Zurier, H.; Bonito, G.; Smith, M.E.; Pfister, D.H. Mycorrhizal Detection of Native and Non-Native Truffles in a Historic Arboretum and the Discovery of a New North American Species, Tuber arnoldianum sp. nov. Mycorrhiza 2016, 26, 781–792. [Google Scholar] [CrossRef] [PubMed]
- Guevara-Guerrero, G.; Bonito, G.; Smith, M.E.; Healy, R.; Grupe II, A.C.; Cázares, E.; Trappe, J.M. Tuber aztecorum sp. nov., a Truffle Species from Mexico Belonging to the Maculatum Clade (Tuberaceae, Pezizales). MycoKeys 2018, 30, 61. [Google Scholar] [CrossRef] [PubMed]
- Berch, S.M.; Bonito, G. Truffle Diversity (Tuber, Tuberaceae) in British Columbia. Mycorrhiza 2016, 26, 587–594. [Google Scholar] [CrossRef]
- Grupe, A.C.; Sulzbacher, M.A.; Grebenc, T.; Healy, R.; Bonito, G.; Smith, M.E. Tuber brennemanii and Tuber floridanum: Two New Tuber Species Are among the Most Commonly Detected Ectomycorrhizal Taxa within Commercial Pecan (Carya illinoinensis) Orchards. Mycologia 2018, 110, 780–790. [Google Scholar] [CrossRef]
- Castro, J.; Alonso-Díaz, J.; Fraga, A. Hongos Hipogeos de la Provincia de Lugo: Tuber foetidum. Bol. Soc. Micol. Lugo 2018, 5, 31–37. [Google Scholar]
- Orczán, K.A.; Turunen, O.; Merényi, Z.; Rudnóy, S.; Bratek, Z.; Shamekh, S. Tuber foetidum Found in Finland. Mycotaxon 2010, 114, 127–133. [Google Scholar] [CrossRef]
- Crous, P.W.; Cowan, D.A.; Maggs-Kölling, G.; Yilmaz, N.; Larsson, E.; Angelini, C.; Mostowfizadeh-Ghalamfarsa, R. Fungal Planet Description Sheets: 1112–1181. Persoonia 2020, 45, 251. [Google Scholar] [CrossRef]
- Marathe, S.J.; Marozzi, G.; Lorenzi, L.; Donnini, D.; Baciarelli Falini, L.; Bashein, A.M.; Shamekh, S. Whitish Truffles Found in Finland: Soil Characteristics, and Identification Based on Morphological and Molecular Properties. Arch. Microbiol. 2023, 205, 111. [Google Scholar] [CrossRef]
- Kuhar, F.; Tejedor-Calvo, E.; Sequeira, A.; Pelissero, D.; Cosse, M.; Donnini, D.; Nouhra, E. Comprehensive Characterization of Tuber maculatum, New in Uruguay: Morphological, Molecular, and Aromatic Analyses. J. Fungi 2024, 10, 421. [Google Scholar] [CrossRef]
- Guevara, G.; Bonito, G.; Trappe, J.M.; Cázares, E.; Williams, G.; Healy, R.A.; Vilgalys, R. New North American Truffles (Tuber spp.) and Their Ectomycorrhizal Associations. Mycologia 2013, 105, 194–209. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Cao, J.Z. Two new species of white truffle from China. Mycotaxon 2013, 121, 297–304. [Google Scholar] [CrossRef]
- Fan, L.; Zhang, P.R.; Yan, X.Y.; Li, Y. Phylogenetic Analyses of Chinese Tuber Species That Resemble T. borchii Reveal the Existence of the New Species T. hubeiense and T. wumengense. Mycologia 2016, 108, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Brock, P.M.; Döring, H.; Bidartondo, M.I. How to Know Unknown Fungi: The Role of a Herbarium. New Phytol. 2009, 181, 719–724. [Google Scholar] [CrossRef]
- Frank, J.L.; Barry, S.; Southworth, D. Mammal Mycophagy and Dispersal of Mycorrhizal Inoculum in Oregon White Oak Woodlands. Northwest Sci. 2006, 80, 264. [Google Scholar]
- Wang, Y.J.; Tan, Z.M.; Zhang, D.C.; Murat, C.; Jeandroz, S.; Tacon, F.L. Phylogenetic and Populational Study of the Tuber indicum Complex. Mycol. Res. 2006, 110, 1034–1045. [Google Scholar] [CrossRef]
- Riccioni, C.; Belfiori, B.; Rubini, A.; Passeri, V.; Arcioni, S.; Paolocci, F. Tuber melanosporum Outcrosses: Analysis of the Genetic Diversity within and among Its Natural Populations under This New Scenario. New Phytol. 2008, 180, 466–478. [Google Scholar] [CrossRef]
- Milenković, M.; Grebenc, T.; Marković, M.; Ivančević, B. Tuber petrophilum, a New Truffle Species from Serbia. Mycotaxon 2016, 130, 1141–1152. [Google Scholar] [CrossRef]
- Slavova, M.; Leonardi, M.; Iotti, M.; Paz-Conde, A.; Assyov, B.; Pacioni, G. Phylogenetic and morphological evidence supports the recently described Tuber thracicum. Plant Biosyst. 2025, 159, 1–9. [Google Scholar] [CrossRef]
- Boutahir, S.; Iotti, M.; Piattoni, F.; Zambonelli, A. Morphological and Molecular Characterization of Tuber oligospermum Mycorrhizas. Afr. J. Agric. Res. 2013, 8, 4081–4087. [Google Scholar] [CrossRef]
- Bonuso, E.; Zambonelli, A.; Bergemann, S.E.; Iotti, M.; Garbelotto, M. Multilocus Phylogenetic and Coalescent Analyses Identify Two Cryptic Species in the Italian Bianchetto Truffle, Tuber borchii Vittad. Conserv. Genet. 2010, 11, 1453–1466. [Google Scholar] [CrossRef]
- Tedersoo, L.; Hansen, K.; Perry, B.A.; Kjøller, R. Molecular and Morphological Diversity of Pezizalean Ectomycorrhiza. New Phytol. 2006, 170, 581–596. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Hardy, G.S.J.; Barber, P.A.; Alvarado, P.; Barnes, C.W.; Buchanan, P.K.; Heykoop, M.; Moreno, G.; et al. Fungal Planet Description Sheets: 558–624. Persoonia 2017, 38, 380–381. [Google Scholar] [CrossRef]
- Alvarado, P.; Moreno, G.; Manjón, J.L. A New Tuber without Spore Ornamentation. Tuber melosporum comb. nov. Bol. Soc. Micol. Madr. 2012, 36, 194. [Google Scholar]
- Crous, P.W.; Wingfield, M.J.; Chooi, Y.H.; Gilchrist, C.L.; Lacey, E.; Pitt, J.I.; Roets, F.; Swart, W.J.; Cano-Lira, J.F.; Valenzuela-Lopez, N.; et al. Fungal Planet Description Sheets: 1042–1111. Persoonia 2020, 44, 301–459. [Google Scholar] [CrossRef] [PubMed]
- Iotti, M.; Amicucci, A.; Bonito, G.; Bonuso, E.; Stocchi, V.; Zambonelli, A. Selection of a Set of Specific Primers for the Identification of Tuber rufum: A Truffle Species with High Genetic Variability. FEMS Microbiol. Lett. 2007, 277, 223–231. [Google Scholar] [CrossRef]
- Osmundson, T.W.; Robert, V.A.; Schoch, C.L.; Baker, L.J.; Smith, A.; Robich, G.; Garbelotto, M.M. Filling Gaps in Biodiversity Knowledge for Macrofungi: Contributions and Assessment of an Herbarium Collection DNA Barcode Sequencing Project. PLoS ONE 2013, 8, e62419. [Google Scholar] [CrossRef]
- Leonardi, M.; Paz-Conde, A.; Guevara, G.; Salvi, D.; Pacioni, G. Two New Species of Tuber Previously Reported as Tuber malacodermum. Mycologia 2019, 111, 676–689. [Google Scholar] [CrossRef]
- Bonito, G.; Trappe, J.M.; Rawlinson, P.; Vilgalys, R. Improved Resolution of Major Clades within Tuber and Taxonomy of Species within the Tuber gibbosum Complex. Mycologia 2010, 102, 1042–1057. [Google Scholar] [CrossRef]
- Crous, P.W.; Cowan, D.A.; Maggs-Kölling, G.; Yilmaz, N.; Thangavel, R.; Wingfield, M.J.; Groenewald, J.Z. Fungal Planet Description Sheets: 1182–1283. Persoonia 2021, 46, 313. [Google Scholar] [CrossRef]
- Chen, J.; Sun, L.H.; Su, Y.; Zhao, W.Q.; Liu, P.G. Choiromyces helanshanensis sp. nov., a New Species of Subterranean Truffle from China. Mycoscience 2016, 57, 279–286. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.-T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A Fast Online Phylogenetic Tool for Maximum Likelihood Analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian Evolutionary Analysis by Sampling Trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Mello, A.; Vizzini, A.; Longato, S.; Rollo, F.; Bonfante, P.; Trappe, J.M. Tuber borchii versus Tuber maculatum: Neotype Studies and DNA Analyses. Mycologia 2000, 92, 326–331. [Google Scholar] [CrossRef]
- Wedén, C.; Danell, E.; Tibell, L. Species Recognition in the Truffle Genus Tuber—The Synonyms Tuber aestivum and Tuber uncinatum. Environ. Microbiol. 2005, 7, 1535–1546. [Google Scholar] [CrossRef] [PubMed]
- Vizzini, A. Novitates. Miscellanea. Riv. Micol. 2008, 51, 63–66. [Google Scholar]
- Lancellotti, E.; Iotti, M.; Zambonelli, A.; Franceschini, A. The Puberulum Group sensu lato (Whitish Truffles). In True Truffle (Tuber spp.) in the World: Soil Ecology, Systematics and Biochemistry; Zambonelli, A., Bonito, G., Eds.; Springer: Cham, Switzerland, 2016; pp. 105–124. [Google Scholar] [CrossRef]
- Colgan, W., III; Trappe, J.M. NATS Truffle and Truffle-Like Fungi. 7. Tuber anniae sp. nov. (Ascomycota). Mycotaxon 1997, 64, 437–442. [Google Scholar]
- Wang, X.H.; Benucci, G.M.N.; Xie, X.D.; Bonito, G.; Leisola, M.; Liu, P.G.; Shamekh, S. Morphological, Mycorrhizal, and Molecular Characterization of Finnish Truffles Belonging to the Tuber anniae Species-Complex. Fungal Ecol. 2013, 6, 269–280. [Google Scholar] [CrossRef]
- Piña Páez, C.; Bonito, G.M.; Guevara-Guerrero, G.; Castellano, M.A.; Garibay-Orijel, R.; Trappe, J.M.; Rámirez, R.P. Description and Distribution of Tuber incognitum sp. nov. and Tuber anniae in the Transmexican Volcanic Belt. MycoKeys 2018, 41, 17–27. [Google Scholar] [CrossRef]
- Tan, Y.P.; Bishop-Hurley, S.L.; Shivas, R.G.; Cowan, D.A.; Maggs-Kölling, G.; Maharachchikumbura, S.S.; Marqués-Gálvez, J.E. Fungal Planet Description Sheets: 1436–1477. Persoonia 2022, 49, 261–350. [Google Scholar] [CrossRef]
- Tsampiras, L. Diversity of Macromycetes on Parnitha Mountain. Master’s Thesis, Department of Biology, University of Athens, Athens, Greece, 2011. (In Greek). [Google Scholar]
- Dimou, D.M.; Zervakis, G.I.; Polemis, E. Mycodiversity Studies in Selected Ecosystems of Greece: 4. Macrofungi from Abies cephalonica Forests and Other Intermixed Tree Species (Oxya Mountain, Central Greece). Mycotaxon 2008, 104, 39–42. [Google Scholar]
- Athanasiou, Z. Mushrooms–Identification Guide for 642 Species; Psychalou: Athens, Greece, 2010; 430p. (In Greek) [Google Scholar]
- Diamandis, S.; Perlerou, C.; Christopoulos, V. Hypogeous Fungi and Perspectives for Truffle Cultivation in Greece. In Proceedings of the XVI Congress of European Mycologists, Chalkidiki, Greece, 19–23 September 2011; pp. 160–163, (Abstract). [Google Scholar]
- Papadimitriou, A. Fungi of the Rhodopi Mountains National Park; Rodopi Mountain-Range National Park: Παρανέστι, Greece, 2015; 402p. (In Greek) [Google Scholar]
- Diamandis, S. Recent Records of Hypogeous Fungi in Greece. In Proceedings of the XV Congress of European Mycologists, Saint Petersburg, Russia, 16–21 September 2007; p. 118, (Abstract). [Google Scholar]
- Konstantinidis, G. Mushrooms (Mushroom-Collector’s Photographic Guide), 2nd ed.; G. Konstantinidis: Grevena, Greece, 2014; 559p. (In Greek) [Google Scholar]
- 100 + 1 Mushrooms, The Research in Lesvos; Mycological Environmental Association “Mushroom Society of Lesvos”, Mythos Publications: Mytilene, Greece, 2013. (In Greek)
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).