Evaluation of the Loop-Mediated Isothermal Amplification Assay (LAMP) Eazyplex® Pneumocystis jirovecii
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Specimens
2.2. PCR
2.3. Evaluation of qPCR as Gold Standard
2.4. Statistical Analysis
3. Results
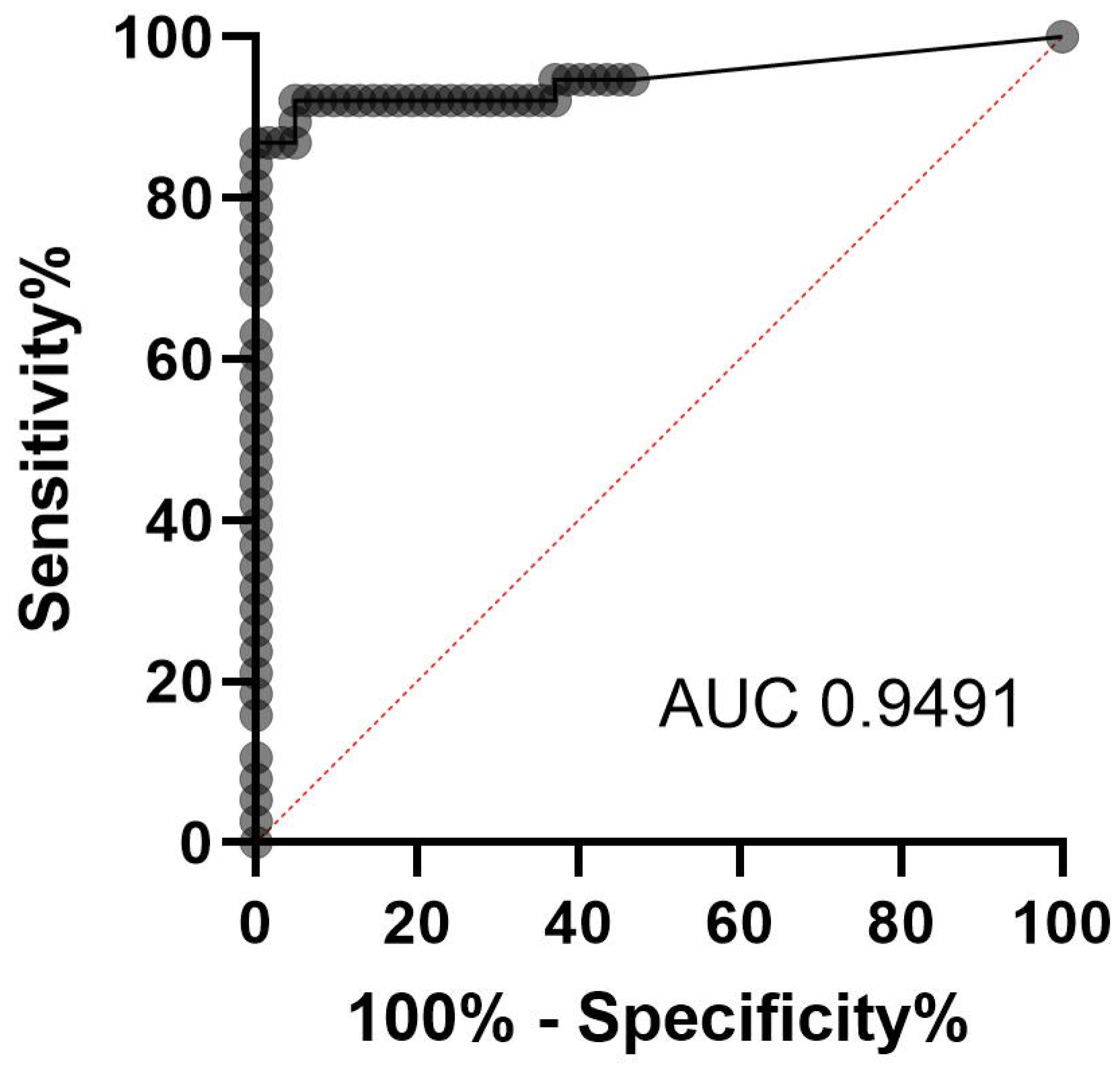
3.1. Evaluation of qPCR as Gold Standard
3.2. Characteristics of the Patients from Five Distinct Centers
3.3. Preanalytical Evaluation of the LAMP
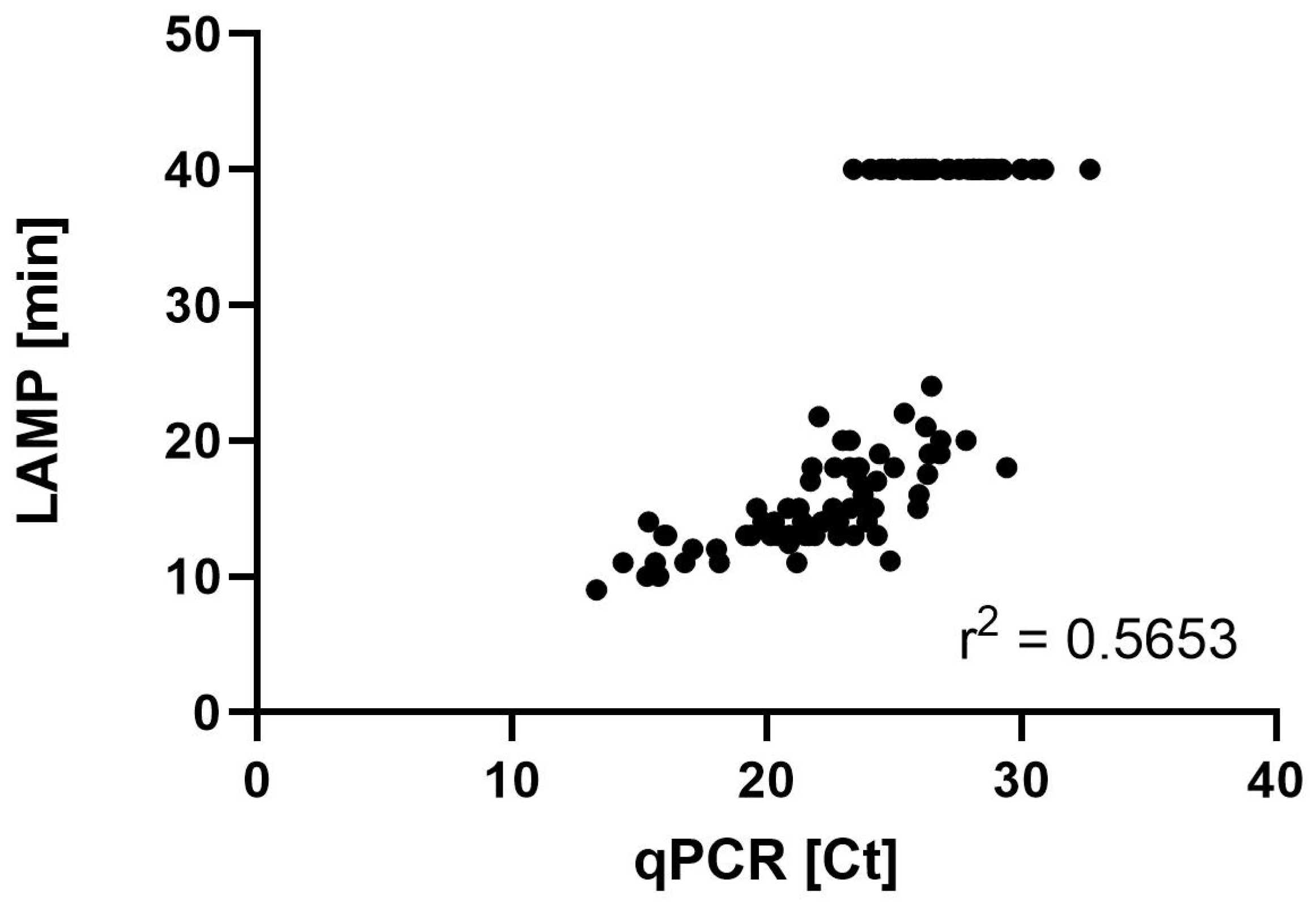
3.4. LAMP and qPCR Results
4. Discussion
Limitation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LAMP | loop-mediated isothermal amplification assay |
| P. jirovecii | Pneumocystis jirovecii |
| PcP | Pneumocystis pneumonia |
| NPV | negative predictive value |
| PPV | positive predictive value |
| r2 | squared Pearson correlation coefficient |
| IFA | immunofluorescence assays |
| BAL | bronchoalveolar lavage |
| qPCR | quantitative real-time PCR |
| cox2 | mitochondrial gene cytochrome c oxidase subunit 2 |
| mtLSU | multicopy mitochondrial large-subunit rRNA (mtLSU) gene |
| EORTC/MSGERC | European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium |
| BS | bronchial secretions |
| Ct | cycle threshold |
| TTP | time to positivity |
| min | minutes |
| ROC | receiver operating characteristic |
| LOD | limit of detection |
| F | Freiburg |
| M | Munster |
| N | Nuremberg |
| ER | Erlangen |
| E | Essen |
| SO | solid tumor |
| HIV | Human Immunodeficiency Virus |
| AD | autoimmune disease |
| ST | solid organ transplantation |
| HM | hematological malignancy |
| O | Others |
References
- Fishman, J.A. Pneumocystis jiroveci. Semin. Respir. Crit. Care Med. 2020, 41, 141–157. [Google Scholar] [CrossRef]
- Hitzenbichler, F.; Mohr, A.; Salzberger, B. Pneumocystis jirovecii pneumonia-an opportunistic infection undergoing change. Internist 2019, 60, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, J.P.; Chen, S.C.; Kauffman, C.A.; Steinbach, W.J.; Baddley, J.W.; Verweij, P.E.; Clancy, C.J.; Wingard, J.R.; Lockhart, S.R.; Groll, A.H.; et al. Revision and Update of the Consensus Definitions of Invasive Fungal Disease From the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin. Infect. Dis. 2020, 71, 1367–1376. [Google Scholar] [CrossRef]
- Maschmeyer, G.; Helweg-Larsen, J.; Pagano, L.; Robin, C.; Cordonnier, C.; Schellongowski, P. ECIL guidelines for treatment of Pneumocystis jirovecii pneumonia in non-HIV-infected haematology patients. J. Antimicrob. Chemother. 2016, 71, 2405–2413. [Google Scholar] [CrossRef] [PubMed]
- Alanio, A.; Hauser, P.M.; Lagrou, K.; Melchers, W.J.; Helweg-Larsen, J.; Matos, O.; Cesaro, S.; Maschmeyer, G.; Einsele, H.; Donnelly, J.P.; et al. ECIL guidelines for the diagnosis of Pneumocystis jirovecii pneumonia in patients with haematological malignancies and stem cell transplant recipients. J. Antimicrob. Chemother. 2016, 71, 2386–2396. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo Cartagena, A.; Asowata, O.E.; Ng, D.; Babady, N.E. An overview of the laboratory diagnosis of Pneumocystis jirovecii pneumonia. J. Clin. Microbiol. 2025, 63, e0036124. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Lagrou, K.; Chen, S.; Masur, H.; Viscoli, C.; Decker, C.F.; Pagano, L.; Groll, A.H. Pneumocystis jirovecii Disease: Basis for the Revised EORTC/MSGERC Invasive Fungal Disease Definitions in Individuals Without Human Immunodeficiency Virus. Clin. Infect. Dis. 2021, 72, S114–S120. [Google Scholar] [CrossRef]
- Scharmann, U.; Kirchhoff, L.; Schmidt, D.; Buer, J.; Steinmann, J.; Rath, P.M. Evaluation of a commercial Loop-mediated Isothermal Amplification (LAMP) assay for rapid detection of Pneumocystis jirovecii. Mycoses 2020, 63, 1107–1114. [Google Scholar] [CrossRef]
- Wakefield, A.E.; Pixley, F.J.; Banerji, S.; Sinclair, K.; Miller, R.F.; Moxon, E.R.; Hopkin, J.M. Detection of Pneumocystis carinii with DNA amplification. Lancet 1990, 336, 451–453. [Google Scholar] [CrossRef]
- Azoulay, E.; Bergeron, A.; Chevret, S.; Bele, N.; Schlemmer, B.; Menotti, J. Polymerase chain reaction for diagnosing pneumocystis pneumonia in non-HIV immunocompromised patients with pulmonary infiltrates. Chest 2009, 135, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Oren, I.; Hardak, E.; Finkelstein, R.; Yigla, M.; Sprecher, H. Polymerase chain reaction-based detection of Pneumocystis jirovecii in bronchoalveolar lavage fluid for the diagnosis of Pneumocystis pneumonia. Am. J. Med. Sci. 2011, 342, 182–185. [Google Scholar] [CrossRef]
- Thomas, C.F., Jr.; Limper, A.H. Pneumocystis pneumonia. N. Engl. J. Med. 2004, 350, 2487–2498. [Google Scholar] [CrossRef] [PubMed]
- Goterris, L.; Mancebo Fernandez, M.A.; Aguilar-Company, J.; Falco, V.; Ruiz-Camps, I.; Martin-Gomez, M.T. Molecular Diagnosis of Pneumocystis jirovecii Pneumonia by Use of Oral Wash Samples in Immunocompromised Patients: Usefulness and Importance of the DNA Target. J. Clin. Microbiol. 2019, 57, e01287-19. [Google Scholar] [CrossRef] [PubMed]
- Veintimilla, C.; Alvarez-Uria, A.; Martin-Rabadan, P.; Valerio, M.; Machado, M.; Padilla, B.; Alonso, R.; Diez, C.; Munoz, P.; Marin, M. Pneumocystis jirovecii Pneumonia Diagnostic Approach: Real-Life Experience in a Tertiary Centre. J. Fungi 2023, 9, 414. [Google Scholar] [CrossRef]
- Uemura, N.; Makimura, K.; Onozaki, M.; Otsuka, Y.; Shibuya, Y.; Yazaki, H.; Kikuchi, Y.; Abe, S.; Kudoh, S. Development of a loop-mediated isothermal amplification method for diagnosing Pneumocystis pneumonia. J. Med. Microbiol. 2008, 57, 50–57. [Google Scholar] [CrossRef]
- Singh, P.; Singh, S.; Mirdha, B.R.; Guleria, R.; Agarwal, S.K.; Mohan, A. Evaluation of Loop-Mediated Isothermal Amplification Assay for the Detection of Pneumocystis jirovecii in Immunocompromised Patients. Mol. Biol. Int. 2015, 2015, 819091. [Google Scholar] [CrossRef]
- Xu, L.; Kong, J. A multiplexed nucleic acid microsystem for point-of-care detection of HIV co-infection with MTB and PCP. Talanta 2013, 117, 532–535. [Google Scholar] [CrossRef]
- Nakashima, K.; Aoshima, M.; Ohkuni, Y.; Hoshino, E.; Hashimoto, K.; Otsuka, Y. Loop-mediated isothermal amplification method for diagnosing Pneumocystis pneumonia in HIV-uninfected immunocompromised patients with pulmonary infiltrates. J. Infect. Chemother. 2014, 20, 757–761. [Google Scholar] [CrossRef]
- Kawano, S.; Maeda, T.; Suzuki, T.; Abe, T.; Mikita, K.; Hamakawa, Y.; Ono, T.; Sonehara, W.; Miyahira, Y.; Kawana, A. Loop-mediated isothermal amplification with the Procedure for Ultra Rapid Extraction kit for the diagnosis of pneumocystis pneumonia. J. Infect. Chemother. 2015, 21, 224–226. [Google Scholar] [CrossRef]
- Wang, D.D.; Zheng, M.Q.; Zhang, N.; An, C.L. Investigation of Pneumocystis jirovecii colonization in patients with chronic pulmonary diseases in the People’s Republic of China. Int. J. Chronic Obstr. Pulm. Dis. 2015, 10, 2079–2085. [Google Scholar] [CrossRef]
- Xue, T.; Ma, Z.; Liu, F.; Du, W.; He, L.; Wang, J.; An, C. Pneumocystis jirovecii colonization and its association with pulmonary diseases: A multicenter study based on a modified loop-mediated isothermal amplification assay. BMC Pulm. Med. 2020, 20, 70. [Google Scholar] [CrossRef] [PubMed]
- Huber, T.; Serr, A.; Geissdorfer, W.; Hess, C.; Lynker-Assmus, C.; von Loewenich, F.D.; Bogdan, C.; Held, J. Evaluation of the Amplex eazyplex Loop-Mediated Isothermal Amplification Assay for Rapid Diagnosis of Pneumocystis jirovecii Pneumonia. J. Clin. Microbiol. 2020, 58, e01739-20. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.W.Y.; Ho, Y.I.I.; Wong, A.H.; Leung, E.C.M.; Lee, A.L.H.; Chow, V.C.Y. Comparison of PneumID real-time PCR assay with Amplex eazyplex LAMP assay for laboratory diagnosis of Pneumocystis jirovecii Pneumonia. Med. Mycol. 2022, 60, myac043. [Google Scholar] [CrossRef]
- Perret, T.; Kritikos, A.; Hauser, P.M.; Guiver, M.; Coste, A.T.; Jaton, K.; Lamoth, F. Ability of quantitative PCR to discriminate Pneumocystis jirovecii pneumonia from colonization. J. Med. Microbiol. 2020, 69, 705–711. [Google Scholar] [CrossRef]
| Parameter | Results |
|---|---|
| No. of patients | 95 |
| Age (mean ± SD) (year) | 59 ± 16.5 |
| Sex (%) | |
| Male | 62 (65.3) |
| Female | 33 (34.7) |
| Underlying disease (no. of patients (%)) | |
| HIV/AIDS | 25 (26.3) |
| Solid tumor | 20 (21.1) |
| Hematologic malignancy | 20 (21.1) |
| Organ transplant | 16 (16.8) |
| Kidney | 11 (11.6) |
| Liver | 3 (3.2) |
| Heart | 1 (1.1) |
| Lung | 1 (1.1) |
| Immunological disorder | 7 (7.4) |
| Other | 7 (7.4) |
| Patient ID | Diagnosis | Categorization EORTC | qPCR (Ct) | Fungal Load (Copies/mL) | LAMP (TTP in min) |
|---|---|---|---|---|---|
| F-1 | SO | probable | 16.81 | 2.60 × 107 | 11.87 |
| F-2 | SO | proven | 18.04 | 1.00 × 107 | 12.37 |
| F-3 | HIV | possible | 20.89 | 1.10 × 106 | 13.30 |
| F-4 | SO | probable | 19.62 | 3.00 × 106 | 15.32 |
| F-5 | HIV | proven | 21.73 | 5.90 × 105 | 17.68 |
| F-6 | AD | proven | 22.23 | 4.00 × 105 | 14.15 |
| F-7 | AD | probable | 23.27 | 1.80 × 105 | 20.75 |
| F-8 | ST | probable | 22.98 | 2.20 × 105 | 20.48 |
| F-9 | HM | probable | 28.16 | 4.20 × 103 | negative |
| F-10 | HIV | possible | 27.08 | 9.50 × 103 | negative |
| F-11 | SO | proven | 26.39 | 1.60 × 104 | 19.98 |
| F-12 | AD | possible | 32.70 | 1.70 × 102 | negative |
| M-13 | SO | probable | 19.85 | 2.50 × 106 | 14.85 |
| M-14 | ST | probable | 21.90 | 5.20 × 105 | 13.93 |
| M-15 | HIV | possible | 19.18 | 4.20 × 106 | 13.78 |
| M-16 | ST | probable | 27.92 | 5.20 × 103 | negative |
| M-17 | SO | probable | 24.78 | 7.40 × 104 | negative |
| N-18 | O | possible | 23.31 | 2.00 × 105 | 15.33 |
| N-19 | O | possible | 21.79 | 6.30 × 105 | 18.17 |
| N-20 | HIV | possible | 19.39 | 3.80 × 106 | 13.65 |
| N-21 | ST | probable | 24.93 | 5.90 × 104 | negative |
| N-22 | SO | probable | 22.16 | 4.70 × 105 | 14.05 |
| N-23 | HIV | possible | 15.77 | 5.80 × 107 | 10.47 |
| F-24 | HIV | proven | 15.31 | 8.20 × 107 | 10.32 |
| F-25 | HIV | possible | 13.33 | 3.80 × 108 | 9.90 |
| ER-26 | ST | probable | 22.61 | 2.70 × 105 | 15.75 |
| ER-27 | ST | probable | 25.86 | 2.30 × 104 | negative |
| ER-27a | 25.87 | 2.30 × 104 | negative | ||
| ER-28 | HM | possible | 24.51 | 6.40 × 104 | negative |
| ER-29 | ST | probable | 28.12 | 4.20 × 103 | negative |
| ER-30 | ST | probable | 25.57 | 2.90 × 104 | negative |
| ER-31 | ST | probable | 26.81 | 1.10 × 104 | 19.75 |
| ER-32 | ST | probable | 23.45 | 1.40 × 105 | 13.75 |
| ER-32a | 23.95 | 9.80 × 104 | 14.00 | ||
| ER-32b | 28.09 | 4.30 × 103 | negative | ||
| ER-32c | 22.80 | 2.30 × 105 | 13.75 | ||
| ER-33 | SO | probable | 21.52 | 6.10 × 105 | 13.00 |
| ER-34 | ST | not classifiable | 30.88 | 5.00 × 102 | negative |
| ER-35 | ST | not classifiable | 22.68 | 2.60 × 105 | 18.00 |
| ER-36 | HM | not classifiable | 24.07 | 9.00 × 104 | negative |
| ER-37 | HM | probable | 23.78 | 1.10 × 105 | 16.25 |
| ER-38 | O | probable | 21.66 | 5.50 × 105 | 13.78 |
| ER-38a | 27.56 | 6.50 × 103 | negative | ||
| ER-38b | 24.92 | 4.70 × 104 | negative | ||
| ER-39 | SO | not classifiable | 29.22 | 1.80 × 103 | negative |
| ER-40 | HM | possible | 26.48 | 1.50 × 104 | 24.75 |
| ER-41 | AD | not classifiable | 28.85 | 2.50 × 103 | negative |
| ER-42 | HM | possible | 17.09 | 1.70 × 107 | 12.67 |
| ER-42a | 23.26 | 1.90 × 105 | 18.73 | ||
| ER-42b | 26.55 | 1.50 × 104 | negative | ||
| ER-43 | AD | not classifiable | 26.38 | 1.70 × 104 | negative |
| ER-44 | O | probable | 15.36 | 8.70 × 107 | 14.52 |
| ER-45 | ST | not classifiable | 23.63 | 1.40 × 105 | 18.12 |
| ER-46 | HM | not classifiable | 26.25 | 1.90 × 104 | 26.00 |
| ER-47 | O | probable | 30.00 | 1.00 × 103 | negative |
| ER-48 | ST | probable | 25.38 | 3.70 × 104 | negative |
| ER-48a | 28.33 | 3.70 × 103 | negative | ||
| ER-49 | HM | not classifiable | 28.67 | 2.80 × 103 | negative |
| ER-50 | AD | possible | 15.63 | 7.10 × 107 | 11.18 |
| ER-50a | 16.08 | 5.00 × 107 | 13.20 | ||
| ER-51 | O | no PCP expected | 24.35 | 8.10 × 104 | 13.37 |
| ER-52 | HIV | not classifiable | 14.36 | 1.90 × 108 | 11.22 |
| ER-53 | ST | probable | 26.25 | 1.90 × 104 | 21.58 |
| E-54 | HIV | possible | 28.14 | 3.10 × 103 | negative |
| E-55 | SO | probable | 23.57 | 1.10 × 105 | 17.00 |
| E-56 | HIV | possible | 27.14 | 6.90 × 103 | negative |
| E-57 | HIV | possible | 20.61 | 1.20 × 106 | 13.00 |
| E-57a | 22.06 | 3.80 × 105 | 522.00 | ||
| E-58 | HIV | possible | 21.28 | 6.70 × 105 | 15.00 |
| E-59 | SO | probable | 24.86 | 4.40 × 104 | 11.25 |
| E-60 | SO | probable | 20.87 | 9.70 × 105 | 12.75 |
| E-61 | SO | probable | 21.41 | 6.20 × 105 | 14.00 |
| E-62 | SO | probable | 24.20 | 7.10 × 104 | 15.00 |
| E-63 | O | possible | 26.33 | 1.60 × 104 | 17.00 |
| E-64 | HM | probable | 28.63 | 2.40 × 103 | negative |
| E-65 | SO | probable | 24.44 | 6.60 × 104 | 19.00 |
| E-66 | HM | probable | 21.71 | 5.30 × 105 | 13.00 |
| E-67 | HIV | probable | 25.42 | 3.10 × 104 | 22.25 |
| E-68 | HIV | probable | 23.43 | 1.50 × 105 | 13.25 |
| E-69 | ST | probable | 25.98 | 2.00 × 104 | 16.50 |
| E-70 | HIV | possible | 15.97 | 1.17 × 108 | 13.25 |
| E-70a | 23.41 | 1.40 × 105 | negative | ||
| E-71 | HM | possible | 28.39 | 6.50 × 103 | negative |
| E-72 | HM | probable | 27.16 | 1.70 × 104 | negative |
| E-73 | HIV | possible | 29.26 | 3.30 × 103 | negative |
| E-74 | HIV | possible | 20.29 | 3.90 × 106 | 14.75 |
| E-75 | HIV | possible | 23.82 | 3.40 × 105 | 15.25 |
| E-76 | HIV | possible | 25.95 | 6.30 × 104 | 15.38 |
| E-77 | ST | probable | 29.44 | 4.10 × 103 | 18.03 |
| E-77a | 23.30 | 5.00 × 105 | 15.70 | ||
| E-78 | HM | probable | 26.06 | 6.60 × 104 | negative |
| E-79 | HM | probable | 21.20 | 7.50 × 105 | 11.57 |
| E-80 | HM | no classifiable | 27.84 | 4.80 × 103 | 20.10 |
| E-81 | HM | probable | 30.02 | 2.60 × 103 | negative |
| E-82 | ST | possible | 30.52 | 1.80 × 103 | negative |
| E-83 | HIV | possible | 20.16 | 3.90 × 106 | 13.02 |
| E-84 | HIV | probable | 25.00 | 8.80 × 104 | 18.02 |
| E-84a | HIV | 18.14 | 1.90 × 107 | 11.70 | |
| E-85 | HIV | probable | 20.84 | 2.30 × 106 | 15.43 |
| E-86 | HIV | probable | 20.46 | 4.70 × 106 | 13.70 |
| E-87 | AD | probable | 26.84 | 3.50 × 104 | 20.97 |
| E-88 | ST | possible | 28.76 | 1.70 × 103 | negative |
| E-89 | SO | probable | 25.24 | 2.70 × 104 | invalid |
| E-90 | HM | probable | 26.14 | 1.70 × 104 | negative |
| E-91 | HM | probable | 26.50 | 1.60 × 104 | negative |
| E-92 | ST | probable | 22.84 | 6.90 × 105 | 14.78 |
| E-93 | HM | probable | 28.94 | 2.30 × 103 | negative |
| E-94 | HM | probable | 24.32 | 8.30 × 104 | 17.42 |
| E-95 | HIV | probable | 16.78 | 2.60 × 107 | 11.02 |
| Storage Conditions | Sample 1 TTP [min] | Sample 2 TTP [min] | Sample 3 TTP [min] |
|---|---|---|---|
| Initial | 11:34 | 17:25 | 13:19 |
| after two years of storage in −20 °C | 14:04 | 16:37 | 15:29 |
| 24 h storage in 21 °C | 14:53 | 20:06 | 13:39 |
| 48 h storage in 21 °C | 14:54 | 20:16 | 12:19 |
| 72 h storage in 21 °C | 14:04 | 18:19 | 12:56 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scharmann, U.; Kirchhoff, L.; Buer, J.; Schuler, F.; Serr, A.; Rößler, S.; Held, J.; Szumlanski, T.; Steinmann, J.; Rath, P.-M. Evaluation of the Loop-Mediated Isothermal Amplification Assay (LAMP) Eazyplex® Pneumocystis jirovecii. J. Fungi 2025, 11, 300. https://doi.org/10.3390/jof11040300
Scharmann U, Kirchhoff L, Buer J, Schuler F, Serr A, Rößler S, Held J, Szumlanski T, Steinmann J, Rath P-M. Evaluation of the Loop-Mediated Isothermal Amplification Assay (LAMP) Eazyplex® Pneumocystis jirovecii. Journal of Fungi. 2025; 11(4):300. https://doi.org/10.3390/jof11040300
Chicago/Turabian StyleScharmann, Ulrike, Lisa Kirchhoff, Jan Buer, Franziska Schuler, Annerose Serr, Susann Rößler, Jürgen Held, Tobias Szumlanski, Joerg Steinmann, and Peter-Michael Rath. 2025. "Evaluation of the Loop-Mediated Isothermal Amplification Assay (LAMP) Eazyplex® Pneumocystis jirovecii" Journal of Fungi 11, no. 4: 300. https://doi.org/10.3390/jof11040300
APA StyleScharmann, U., Kirchhoff, L., Buer, J., Schuler, F., Serr, A., Rößler, S., Held, J., Szumlanski, T., Steinmann, J., & Rath, P.-M. (2025). Evaluation of the Loop-Mediated Isothermal Amplification Assay (LAMP) Eazyplex® Pneumocystis jirovecii. Journal of Fungi, 11(4), 300. https://doi.org/10.3390/jof11040300





