Resistance to Azoles in Candida parapsilosis Isolates from Spain Is Associated with an Impairment in Filamentation and Biofilm Formation
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Antifungal Susceptibility Testing in Microdilution Plates (EUCAST Protocol)
2.3. Growth Curves
2.4. Real Time Microscopy
2.5. Biofilm Formation and Measurement with XTT
2.6. Agar Invasiveness
2.7. Microfluidic Experiments
2.8. Susceptibility to Disinfectants
2.9. Statistical Analysis
3. Results
3.1. Antifungal Susceptibility of FNS C. parapsilosis Isolates
3.2. Characterization of Growth of Susceptible and FNS Strains
3.3. Pseudohyphae Formation
3.4. Biofilm Formation
3.5. Agar Invasiveness
3.6. Adhesion Under Dynamic Conditions
3.7. Susceptibility of C. parapsilosis to Clinical Disinfectants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Govrins, M.; Lass-Florl, C. Candida parapsilosis complex in the clinical setting. Nat. Rev. Microbiol. 2024, 22, 46–59. [Google Scholar] [CrossRef] [PubMed]
- Puig-Asensio, M.; Padilla, B.; Garnacho-Montero, J.; Zaragoza, O.; Aguado, J.M.; Zaragoza, R.; Montejo, M.; Munoz, P.; Ruiz-Camps, I.; Cuenca-Estrella, M.; et al. Epidemiology and predictive factors for early and late mortality in Candida bloodstream infections: A population-based surveillance in Spain. Clin. Microbiol. Infect. 2014, 20, O245–O254. [Google Scholar] [CrossRef] [PubMed]
- Larkin, E.L.; Dharmaiah, S.; Ghannoum, M.A. Biofilms and beyond: Expanding echinocandin utility. J. Antimicrob. Chemother. 2018, 73, i73–i81. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, D.M.; Mikherjee, P.K.; Clark, T.A.; Pujol, C.; Chandra, J.; Hajjeh, R.A.; Warnock, D.W.; Soil, D.R.; Ghannoum, M.A. Candida parapsilosis characterization in an outbreak setting. Emerg. Infect. Dis. 2004, 10, 1074–1081. [Google Scholar] [CrossRef]
- Toth, R.; Nosek, J.; Mora-Montes, H.M.; Gabaldon, T.; Bliss, J.M.; Nosanchuk, J.D.; Turner, S.A.; Butler, G.; Vagvolgyi, C.; Gacser, A. Candida parapsilosis: From Genes to the Bedside. Clin. Microbiol. Rev. 2019, 32, e00111-18. [Google Scholar] [CrossRef]
- Pfaller, M.A.; Diekema, D.J.; Turnidge, J.D.; Castanheira, M.; Jones, R.N. Twenty Years of the SENTRY Antifungal Surveillance Program: Results for Candida Species From 1997-2016. Open Forum Infect. Dis. 2019, 6, S79–S94. [Google Scholar] [CrossRef]
- Lamoth, F.; Lockhart, S.R.; Berkow, E.L.; Calandra, T. Changes in the epidemiological landscape of invasive candidiasis. J. Antimicrob. Chemother. 2018, 73, i4–i13. [Google Scholar] [CrossRef]
- Mamali, V.; Siopi, M.; Charpantidis, S.; Samonis, G.; Tsakris, A.; Vrioni, G.; On Behalf Of The Candi-Candi, N. Increasing Incidence and Shifting Epidemiology of Candidemia in Greece: Results from the First Nationwide 10-Year Survey. J. Fungi 2022, 8, 116. [Google Scholar] [CrossRef]
- Anuradha, S.; Samaddar, A.; Maurya, A.; Hada, V.; Narula, H.; Shrimali, T.; Gupta, N.; Kumar, P.; Singh, K.; Nag, V.L. Analysis of Blood Culture Data Influences Future Epidemiology of Bloodstream Infections: A 5-year Retrospective Study at a Tertiary Care Hospital in India. Indian J. Crit. Care Med. 2021, 25, 1258–1262. [Google Scholar] [CrossRef]
- Pammi, M.; Holland, L.; Butler, G.; Gacser, A.; Bliss, J.M. Candida parapsilosis is a significant neonatal pathogen: A systematic review and meta-analysis. Pediatr. Infect. Dis. J. 2013, 32, e206–e216. [Google Scholar] [CrossRef]
- Branco, J.; Miranda, I.M.; Rodrigues, A.G. Candida parapsilosis Virulence and Antifungal Resistance Mechanisms: A Comprehensive Review of Key Determinants. J. Fungi 2023, 9, 80. [Google Scholar] [CrossRef] [PubMed]
- Arevalo-Jaimes, B.V.; Admella, J.; Blanco-Cabra, N.; Torrents, E. Culture media influences Candida parapsilosis growth, susceptibility, and virulence. Front. Cell Infect. Microbiol. 2023, 13, 1323619. [Google Scholar] [CrossRef] [PubMed]
- Daneshnia, F.; de Almeida Junior, J.N.; Ilkit, M.; Lombardi, L.; Perry, A.M.; Gao, M.; Nobile, C.J.; Egger, M.; Perlin, D.S.; Zhai, B.; et al. Worldwide emergence of fluconazole-resistant Candida parapsilosis: Current framework and future research roadmap. Lancet Microbe 2023, 4, e470–e480. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Kim, Y.J.; Yong, D.; Byun, J.H.; Kim, T.S.; Chang, Y.S.; Choi, M.J.; Byeon, S.A.; Won, E.J.; Kim, S.H.; et al. Fluconazole-Resistant Candida parapsilosis Bloodstream Isolates with Y132F Mutation in ERG11 Gene, South Korea. Emerg. Infect. Dis. 2018, 24, 1768–1770. [Google Scholar] [CrossRef]
- Rodrigues, D.K.B.; Bonfietti, L.X.; Garcia, R.A.; Araujo, M.R.; Rodrigues, J.S.; Gimenes, V.M.F.; Melhem, M.S.C. Antifungal susceptibility profile of Candida clinical isolates from 22 hospitals of Sao Paulo State, Brazil. Braz. J. Med. Biol. Res. 2021, 54, e10928. [Google Scholar] [CrossRef]
- Castanheira, M.; Deshpande, L.M.; Messer, S.A.; Rhomberg, P.R.; Pfaller, M.A. Analysis of global antifungal surveillance results reveals predominance of Erg11 Y132F alteration among azole-resistant Candida parapsilosis and Candida tropicalis and country-specific isolate dissemination. Int. J. Antimicrob. Agents 2020, 55, 105799. [Google Scholar] [CrossRef]
- Hubler, C.M.; Carvalhaes, C.G.; Castanheira, M. Recent increase in fluconazole-nonsusceptible Candida parapsilosis in a global surveillance with the expansion of the Erg11 Y132F genotype and a rapid detection method for this alteration. Diagn. Microbiol. Infect. Dis. 2023, 106, 115957. [Google Scholar] [CrossRef]
- Ceballos-Garzon, A.; Penuela, A.; Valderrama-Beltran, S.; Vargas-Casanova, Y.; Ariza, B.; Parra-Giraldo, C.M. Emergence and circulation of azole-resistant C. albicans, C. auris and C. parapsilosis bloodstream isolates carrying Y132F, K143R or T220L Erg11p substitutions in Colombia. Front. Cell Infect. Microbiol. 2023, 13, 1136217. [Google Scholar] [CrossRef]
- Singh, A.; Singh, P.K.; de Groot, T.; Kumar, A.; Mathur, P.; Tarai, B.; Sachdeva, N.; Upadhyaya, G.; Sarma, S.; Meis, J.F.; et al. Emergence of clonal fluconazole-resistant Candida parapsilosis clinical isolates in a multicentre laboratory-based surveillance study in India. J. Antimicrob. Chemother. 2019, 74, 1260–1268. [Google Scholar] [CrossRef]
- McTaggart, L.R.; Eshaghi, A.; Hota, S.; Poutanen, S.M.; Johnstone, J.; De Luca, D.G.; Bharat, A.; Patel, S.N.; Kus, J.V. First Canadian report of transmission of fluconazole-resistant Candida parapsilosis within two hospital networks confirmed by genomic analysis. J. Clin. Microbiol. 2024, 62, e0116123. [Google Scholar] [CrossRef]
- Alcoceba, E.; Gomez, A.; Lara-Esbri, P.; Oliver, A.; Beltran, A.F.; Ayestaran, I.; Munoz, P.; Escribano, P.; Guinea, J. Fluconazole-resistant Candida parapsilosis clonally related genotypes: First report proving the presence of endemic isolates harbouring the Y132F ERG11 gene substitution in Spain. Clin. Microbiol. Infect. 2022, 28, 1113–1119. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Daneshnia, F.; Hilmioglu-Polat, S.; Fang, W.; Yasar, M.; Polat, F.; Metin, D.Y.; Rigole, P.; Coenye, T.; Ilkit, M.; et al. First Report of Candidemia Clonal Outbreak Caused by Emerging Fluconazole-Resistant Candida parapsilosis Isolates Harboring Y132F and/or Y132F+K143R in Turkey. Antimicrob. Agents Chemother. 2020, 64, e01020. [Google Scholar] [CrossRef] [PubMed]
- Arastehfar, A.; Hilmioglu-Polat, S.; Daneshnia, F.; Pan, W.; Hafez, A.; Fang, W.; Liao, W.; Sahbudak-Bal, Z.; Metin, D.Y.; Junior, J.N.A.; et al. Clonal Candidemia Outbreak by Candida parapsilosis Carrying Y132F in Turkey: Evolution of a Persisting Challenge. Front. Cell Infect. Microbiol. 2021, 11, 676177. [Google Scholar] [CrossRef] [PubMed]
- Corzo-Leon, D.E.; Peacock, M.; Rodriguez-Zulueta, P.; Salazar-Tamayo, G.J.; MacCallum, D.M. General hospital outbreak of invasive candidiasis due to azole-resistant Candida parapsilosis associated with an Erg11 Y132F mutation. Med. Mycol. 2021, 59, 664–671. [Google Scholar] [CrossRef]
- Diaz-Garcia, J.; Gomez, A.; Alcala, L.; Reigadas, E.; Sanchez-Carrillo, C.; Perez-Ayala, A.; Gomez-Garcia de la Pedrosa, E.; Gonzalez-Romo, F.; Merino-Amador, P.; Cuetara, M.S.; et al. Evidence of Fluconazole-Resistant Candida parapsilosis Genotypes Spreading across Hospitals Located in Madrid, Spain and Harboring the Y132F ERG11p Substitution. Antimicrob. Agents Chemother. 2022, 66, e0071022. [Google Scholar] [CrossRef]
- Thomaz, D.Y.; de Almeida, J.N., Jr.; Sejas, O.N.E.; Del Negro, G.M.B.; Carvalho, G.; Gimenes, V.M.F.; de Souza, M.E.B.; Arastehfar, A.; Camargo, C.H.; Motta, A.L.; et al. Environmental Clonal Spread of Azole-Resistant Candida parapsilosis with Erg11-Y132F Mutation Causing a Large Candidemia Outbreak in a Brazilian Cancer Referral Center. J. Fungi 2021, 7, 259. [Google Scholar] [CrossRef]
- Fekkar, A.; Blaize, M.; Bougle, A.; Normand, A.C.; Raoelina, A.; Kornblum, D.; Kamus, L.; Piarroux, R.; Imbert, S. Hospital outbreak of fluconazole-resistant Candida parapsilosis: Arguments for clonal transmission and long-term persistence. Antimicrob. Agents Chemother. 2021, 65, e02036-20. [Google Scholar] [CrossRef]
- Kim, T.Y.; Huh, H.J.; Lee, G.Y.; Choi, M.J.; Yu, H.J.; Cho, S.Y.; Chang, Y.S.; Kim, Y.J.; Shin, J.H.; Lee, N.Y. Evolution of Fluconazole Resistance Mechanisms and Clonal Types of Candida parapsilosis Isolates from a Tertiary Care Hospital in South Korea. Antimicrob. Agents Chemother. 2022, 66, e0088922. [Google Scholar] [CrossRef]
- Trevijano-Contador, N.; Torres-Cano, A.; Carballo-Gonzalez, C.; Puig-Asensio, M.; Martin-Gomez, M.T.; Jimenez-Martinez, E.; Romero, D.; Nuvials, F.X.; Olmos-Arenas, R.; Moreto-Castellsague, M.C.; et al. Global Emergence of Resistance to Fluconazole and Voriconazole in Candida parapsilosis in Tertiary Hospitals in Spain During the COVID-19 Pandemic. Open Forum Infect. Dis. 2022, 9, ofac605. [Google Scholar] [CrossRef]
- Diaz-Garcia, J.; Machado, M.; Alcala, L.; Reigadas, E.; Perez-Ayala, A.; Gomez-Garcia de la Pedrosa, E.; Gonzalez-Romo, F.; Cuetara, M.S.; Garcia-Esteban, C.; Quiles-Melero, I.; et al. Trends in antifungal resistance in Candida from a multicenter study conducted in Madrid (CANDIMAD study): Fluconazole-resistant C. parapsilosis spreading has gained traction in 2022. Antimicrob. Agents Chemother. 2023, 67, e0098623. [Google Scholar] [CrossRef]
- Souza, A.C.; Fuchs, B.B.; Pinhati, H.M.; Siqueira, R.A.; Hagen, F.; Meis, J.F.; Mylonakis, E.; Colombo, A.L. Candida parapsilosis Resistance to Fluconazole: Molecular Mechanisms and In Vivo Impact in Infected Galleria mellonella Larvae. Antimicrob. Agents Chemother. 2015, 59, 6581–6587. [Google Scholar] [CrossRef] [PubMed]
- Demirci-Duarte, S.; Arikan-Akdagli, S.; Gulmez, D. Species distribution, azole resistance and related molecular mechanisms in invasive Candida parapsilosis complex isolates: Increase in fluconazole resistance in 21 years. Mycoses 2021, 64, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Asadzadeh, M.; Ahmad, S.; Al-Sweih, N.; Khan, Z. Epidemiology and Molecular Basis of Resistance to Fluconazole Among Clinical Candida parapsilosis Isolates in Kuwait. Microb. Drug Resist. 2017, 23, 966–972. [Google Scholar] [CrossRef] [PubMed]
- Magobo, R.E.; Lockhart, S.R.; Govender, N.P. Fluconazole-resistant Candida parapsilosis strains with a Y132F substitution in the ERG11 gene causing invasive infections in a neonatal unit, South Africa. Mycoses 2020, 63, 471–477. [Google Scholar] [CrossRef]
- Silva, A.P.; Miranda, I.M.; Guida, A.; Synnott, J.; Rocha, R.; Silva, R.; Amorim, A.; Pina-Vaz, C.; Butler, G.; Rodrigues, A.G. Transcriptional profiling of azole-resistant Candida parapsilosis strains. Antimicrob. Agents Chemother. 2011, 55, 3546–3556. [Google Scholar] [CrossRef]
- Doorley, L.A.; Rybak, J.M.; Berkow, E.L.; Zhang, Q.; Morschhauser, J.; Rogers, P.D. Candida parapsilosis Mdr1B and Cdr1B Are Drivers of Mrr1-Mediated Clinical Fluconazole Resistance. Antimicrob. Agents Chemother. 2022, 66, e0028922. [Google Scholar] [CrossRef]
- Berkow, E.L.; Manigaba, K.; Parker, J.E.; Barker, K.S.; Kelly, S.L.; Rogers, P.D. Multidrug Transporters and Alterations in Sterol Biosynthesis Contribute to Azole Antifungal Resistance in Candida parapsilosis. Antimicrob. Agents Chemother. 2015, 59, 5942–5950. [Google Scholar] [CrossRef]
- Doorley, L.A.; Barker, K.S.; Zhang, Q.; Rybak, J.M.; Rogers, P.D. Mutations in TAC1 and ERG11 are major drivers of triazole antifungal resistance in clinical isolates of Candida parapsilosis. Clin. Microbiol. Infect. 2023, 29, 1602.e1-1602.e7. [Google Scholar] [CrossRef]
- Grossman, N.T.; Pham, C.D.; Cleveland, A.A.; Lockhart, S.R. Molecular mechanisms of fluconazole resistance in Candida parapsilosis isolates from a U.S. surveillance system. Antimicrob. Agents Chemother. 2015, 59, 1030–1037. [Google Scholar] [CrossRef]
- Thomaz, D.Y.; de Almeida, J.N., Jr.; Lima, G.M.E.; Nunes, M.O.; Camargo, C.H.; Grenfell, R.C.; Benard, G.; Del Negro, G.M.B. An Azole-Resistant Candida parapsilosis Outbreak: Clonal Persistence in the Intensive Care Unit of a Brazilian Teaching Hospital. Front. Microbiol. 2018, 9, 2997. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gefland, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990. [Google Scholar]
- Arendrup, M.C.; Meletiadis, J.; Mouton, J.W.; Guinea, J.; Cuenca-Estrella, M.; Lagrou, K.; Howard, S.J.; Mouton, J.; Arikan-Akdagli, S.; Barchiesi, F.; et al. EUCAST technical note on isavuconazole breakpoints for Aspergillus, itraconazole breakpoints for Candida and updates for the antifungal susceptibility testing method documents. Clin. Microbiol. Infect. 2016, 22, 571.e1–571.e4. [Google Scholar] [CrossRef] [PubMed]
- Ramage, G.; Vandewalle, K.; Wickes, B.L.; Lopez-Ribot, J.L. Characteristics of biofilm formation by Candida albicans. Rev. Iberoam. Micol. 2001, 18, 163–170. [Google Scholar] [PubMed]
- Homann, O.R.; Dea, J.; Noble, S.M.; Johnson, A.D. A phenotypic profile of the Candida albicans regulatory network. PLoS Genet. 2009, 5, e1000783. [Google Scholar] [CrossRef]
- Naseem, S.; Douglas, L.M.; Konopka, J.B. Candida albicans Agar Invasion Assays. Bio Protoc. 2020, 10, e3730. [Google Scholar] [CrossRef]
- Fu, L.; Le, T.; Liu, Z.; Wang, L.; Guo, H.; Yang, J.; Chen, Q.; Hu, J. Different efficacies of common disinfection methods against Candida auris and other Candida species. J. Infect. Public. Health 2020, 13, 730–736. [Google Scholar] [CrossRef]
- Snelders, E.; Camps, S.M.; Karawajczyk, A.; Schaftenaar, G.; Kema, G.H.; van der Lee, H.A.; Klaassen, C.H.; Melchers, W.J.; Verweij, P.E. Triazole fungicides can induce cross-resistance to medical triazoles in Aspergillus fumigatus. PLoS ONE 2012, 7, e31801. [Google Scholar] [CrossRef]
- Verweij, P.E.; Lucas, J.A.; Arendrup, M.C.; Bowyer, P.; Brinkmann, A.J.F.; Denning, D.; Dyer, P.; Fisher, M.C.; Geenen, P.L.; Gisi, U.; et al. The one health problem of azole resistance in Aspergillus fumigatus: Current insights and future research agenda. Fungal Biol. Rev. 2020, 34, 202–214. [Google Scholar] [CrossRef]
- Sharma, C.; Kadosh, D. Perspective on the origin, resistance, and spread of the emerging human fungal pathogen Candida auris. PLoS Pathog. 2023, 19, e1011190. [Google Scholar] [CrossRef]
- Glushakova, A.; Kachalkin, A. Wild and partially synanthropic bird yeast diversity, in vitro virulence, and antifungal susceptibility of Candida parapsilosis and Candida tropicalis strains isolated from feces. Int. Microbiol. 2024, 27, 883–897. [Google Scholar] [CrossRef]
- Glushakova, A.; Kachalkin, A.; Rodionova, E. The role of fruits as reservoirs for resistant and virulent strains of opportunistic yeasts. World J. Microbiol. Biotechnol. 2023, 39, 313. [Google Scholar] [CrossRef]
- Caggiano, G.; Fioriti, S.; Morroni, G.; Apollonio, F.; Triggiano, F.; D’Achille, G.; Stefanizzi, P.; Dalfino, L.; Ronga, L.; Mosca, A.; et al. Genotypic and phenotypic characteristics of Candida parapsilosis bloodstream isolates: Health Care Associated Infections in a teaching Hospital in Italy. J. Infect. Public. Health 2024, 17, 967–974. [Google Scholar] [CrossRef] [PubMed]
- Byun, J.H.; Won, E.J.; Cho, H.W.; Kim, D.; Lee, H.; Kim, S.H.; Choi, M.J.; Byun, S.A.; Lee, G.Y.; Kee, S.J.; et al. Detection and Characterization of Two Phenotypes of Candida parapsilosis in South Korea: Clinical Features and Microbiological Findings. Microbiol. Spectr. 2023, 11, e0006623. [Google Scholar] [CrossRef] [PubMed]
- Stefanek, M.; Garaiova, M.; Valcek, A.; Jordao, L.; Bujdakova, H. Comparative Analysis of Two Candida parapsilosis Isolates Originating from the Same Patient Harbouring the Y132F and R398I Mutations in the ERG11 Gene. Cells 2023, 12, 1579. [Google Scholar] [CrossRef]
- Daneshnia, F.; Floyd, D.J.; Ryan, A.P.; Ghahfarokhy, P.M.; Ebadati, A.; Jusuf, S.; Munoz, J.; Jeffries, N.E.; Elizabeth Yvanovich, E.; Apostolopoulou, A.; et al. Evaluation of outbreak persistence caused by multidrug-resistant and echinocandin-resistant Candida parapsilosis using multidimensional experimental and epidemiological approaches. Emerg. Microbes Infect. 2024, 13, 2322655. [Google Scholar] [CrossRef]
- Jiang, C.; Dong, D.; Yu, B.; Cai, G.; Wang, X.; Ji, Y.; Peng, Y. Mechanisms of azole resistance in 52 clinical isolates of Candida tropicalis in China. J. Antimicrob. Chemother. 2013, 68, 778–785. [Google Scholar] [CrossRef]
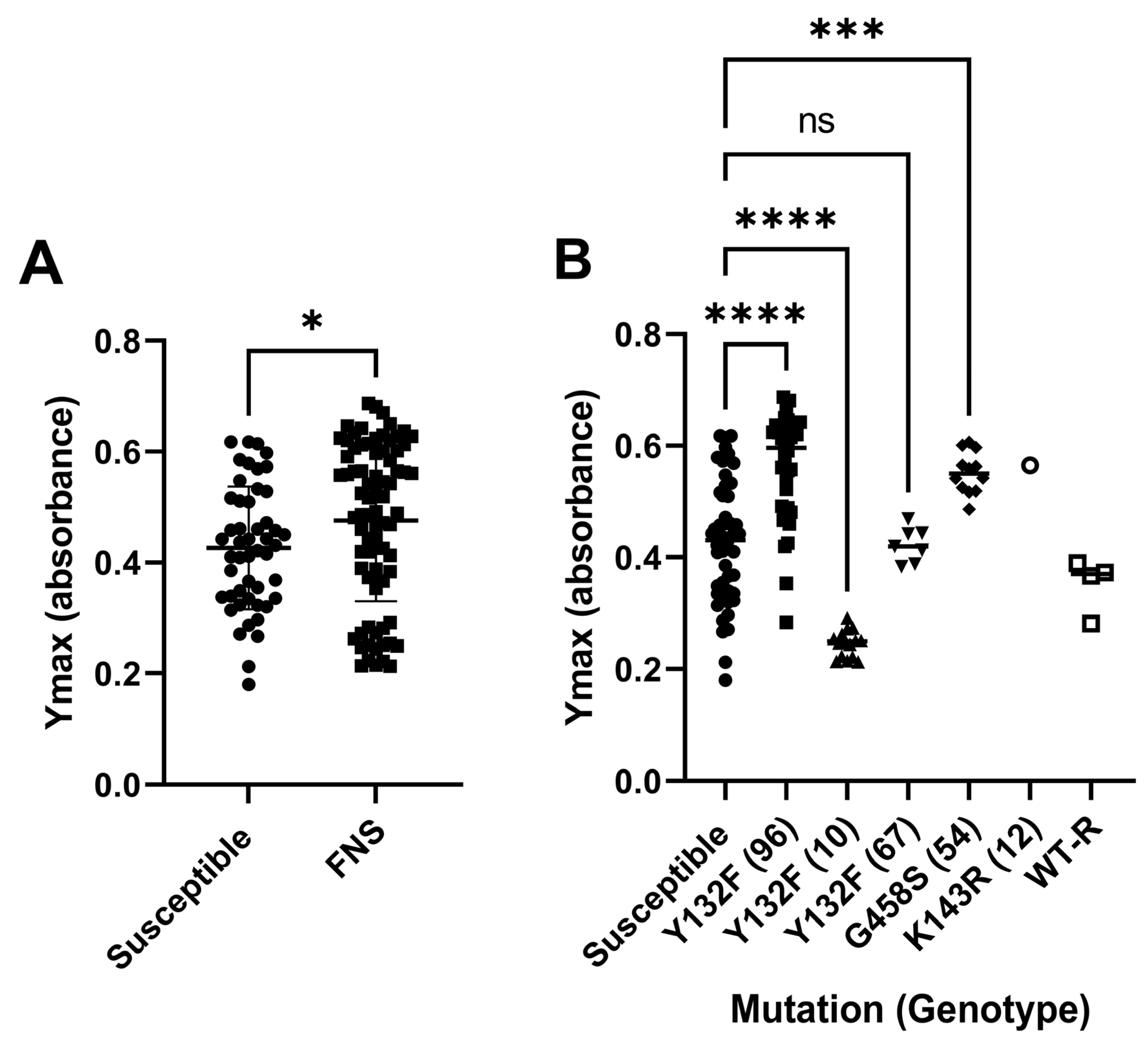
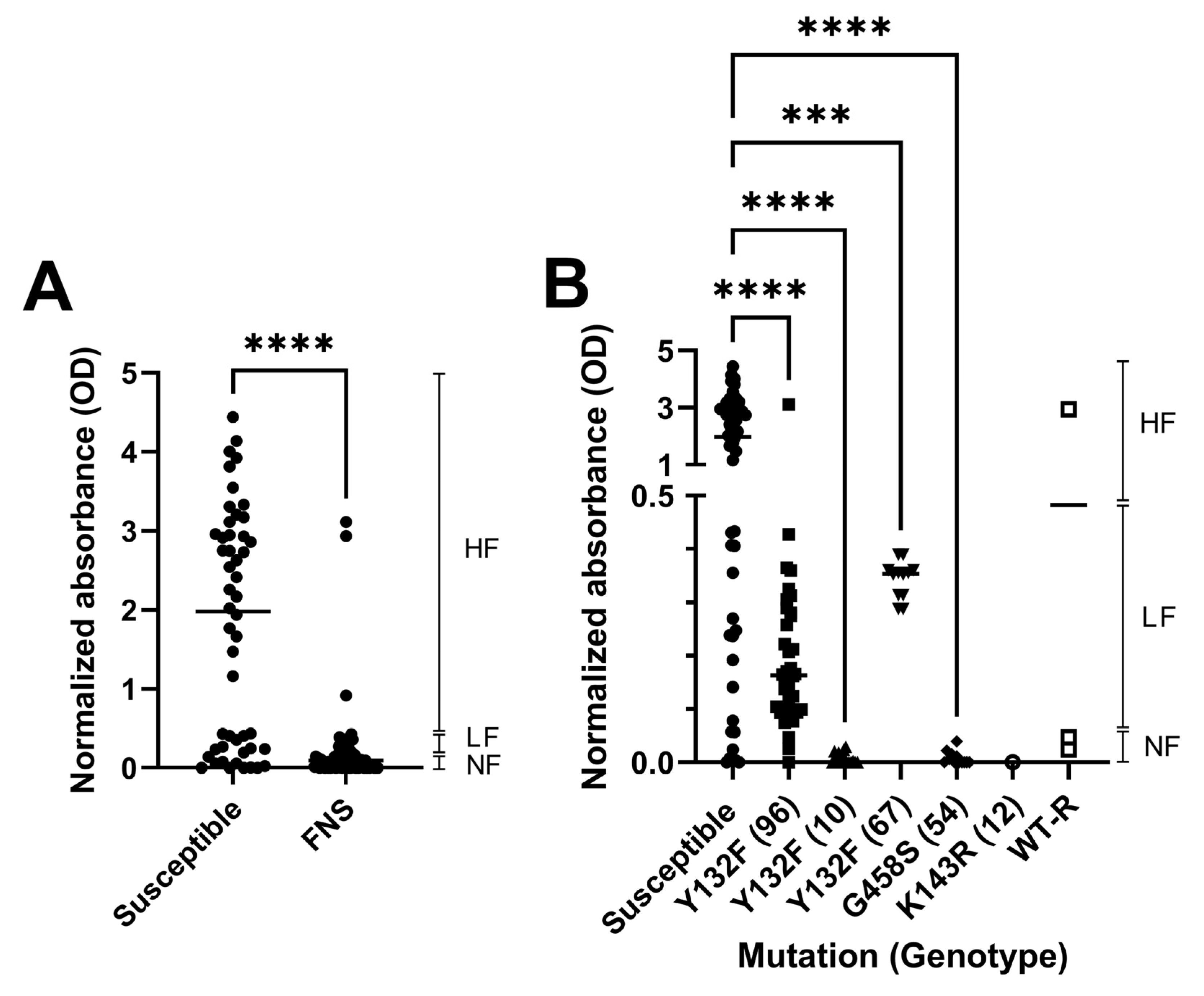

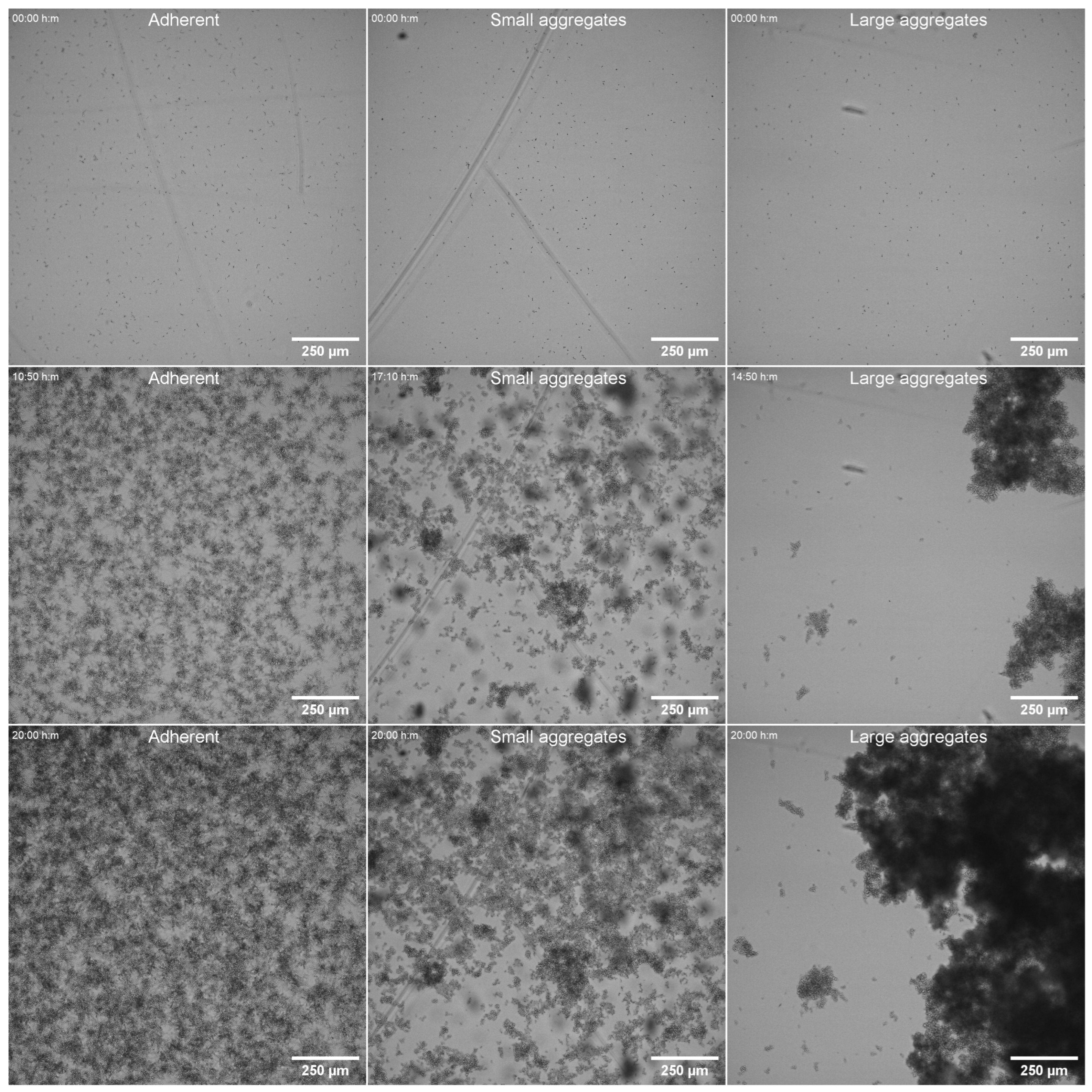

| Susceptible (n) | FNS Non Susceptible (n) | |||||
|---|---|---|---|---|---|---|
| AmB MIC | WT | Y132F (Heterozygous) | Y132F | G458S | K143R | WT |
| 0.06 | 2 | |||||
| 0.125 | 8 | 2 | 1 | |||
| 0.25 | 30 | 2 | 18 | 6 | 2 | |
| 0.5 | 14 | 10 | 23 | 5 | 1 | |
| 1 | 5 | 6 | 1 | 2 | ||
| Total | 59 | 12 | 49 | 12 | 1 | 5 |
| Min-Max | 0.06–1 | 0.25–0.5 | 0.125–1 | 0.125–0.5 | 1–1 | 0.25–1 |
| Mode | 0.25 | 0.5 | 0.5 | 0.25 | nd | nd |
| % of Strains | |||
|---|---|---|---|
| Susceptible (n = 59) | FNS (n = 79) | ||
| Morphology | Pseudohyphae | 73 | 3 |
| Yeast | 27 | 97 | |
| Biofilm | High-forming | 63 | 2 |
| Low-forming | 21 | 31 | |
| Non-forming | 16 | 67 | |
| Agar invasiveness | Invasion | 97 | 31 |
| No invasion | 3 | 69 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres-Cano, A.; de Armentia, C.; Roldán, A.; López-Peralta, E.; Manosalva, J.; Merino-Amador, P.; González-Romo, F.; Puig-Asensio, M.; Ardanuy, C.; Martín-Gómez, M.T.; et al. Resistance to Azoles in Candida parapsilosis Isolates from Spain Is Associated with an Impairment in Filamentation and Biofilm Formation. J. Fungi 2025, 11, 299. https://doi.org/10.3390/jof11040299
Torres-Cano A, de Armentia C, Roldán A, López-Peralta E, Manosalva J, Merino-Amador P, González-Romo F, Puig-Asensio M, Ardanuy C, Martín-Gómez MT, et al. Resistance to Azoles in Candida parapsilosis Isolates from Spain Is Associated with an Impairment in Filamentation and Biofilm Formation. Journal of Fungi. 2025; 11(4):299. https://doi.org/10.3390/jof11040299
Chicago/Turabian StyleTorres-Cano, Alba, Cristina de Armentia, Alejandra Roldán, Elena López-Peralta, Juliana Manosalva, Paloma Merino-Amador, Fernando González-Romo, Mireia Puig-Asensio, Carmen Ardanuy, María Teresa Martín-Gómez, and et al. 2025. "Resistance to Azoles in Candida parapsilosis Isolates from Spain Is Associated with an Impairment in Filamentation and Biofilm Formation" Journal of Fungi 11, no. 4: 299. https://doi.org/10.3390/jof11040299
APA StyleTorres-Cano, A., de Armentia, C., Roldán, A., López-Peralta, E., Manosalva, J., Merino-Amador, P., González-Romo, F., Puig-Asensio, M., Ardanuy, C., Martín-Gómez, M. T., Romero-Herrero, D., Pérez-Ayala, A., López-Lomba, M., Durán-Valle, M. T., Sánchez-Romero, I., Muñoz-Algarra, M., Roiz-Mesones, M. P., Lara-Plaza, I., Ruíz Pérez de Pipaón, M., ... Zaragoza, O. (2025). Resistance to Azoles in Candida parapsilosis Isolates from Spain Is Associated with an Impairment in Filamentation and Biofilm Formation. Journal of Fungi, 11(4), 299. https://doi.org/10.3390/jof11040299






