Genome-Wide Identification and Analysis of Chitinase GH18 Gene Family in Valsa mali
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Growth
2.2. Plant Growth
2.3. Fungal Infection
2.4. Plasmid Construction
2.5. Expression and Purification of VmGH18-4 and VmGH18-10
2.6. Genome-Wide Identification of GH18 Gene Family of V. mali
2.7. Phylogenetic Relationships, Gene Structure, and Conserved Motif Analysis
2.8. Chromosomal Location Analysis
2.9. RNA Isolation and Real-Time Quantitative PCR Analysis
3. Results
3.1. Genome-Wide Identification and Characterization of VmGH18 Genes in V. mali
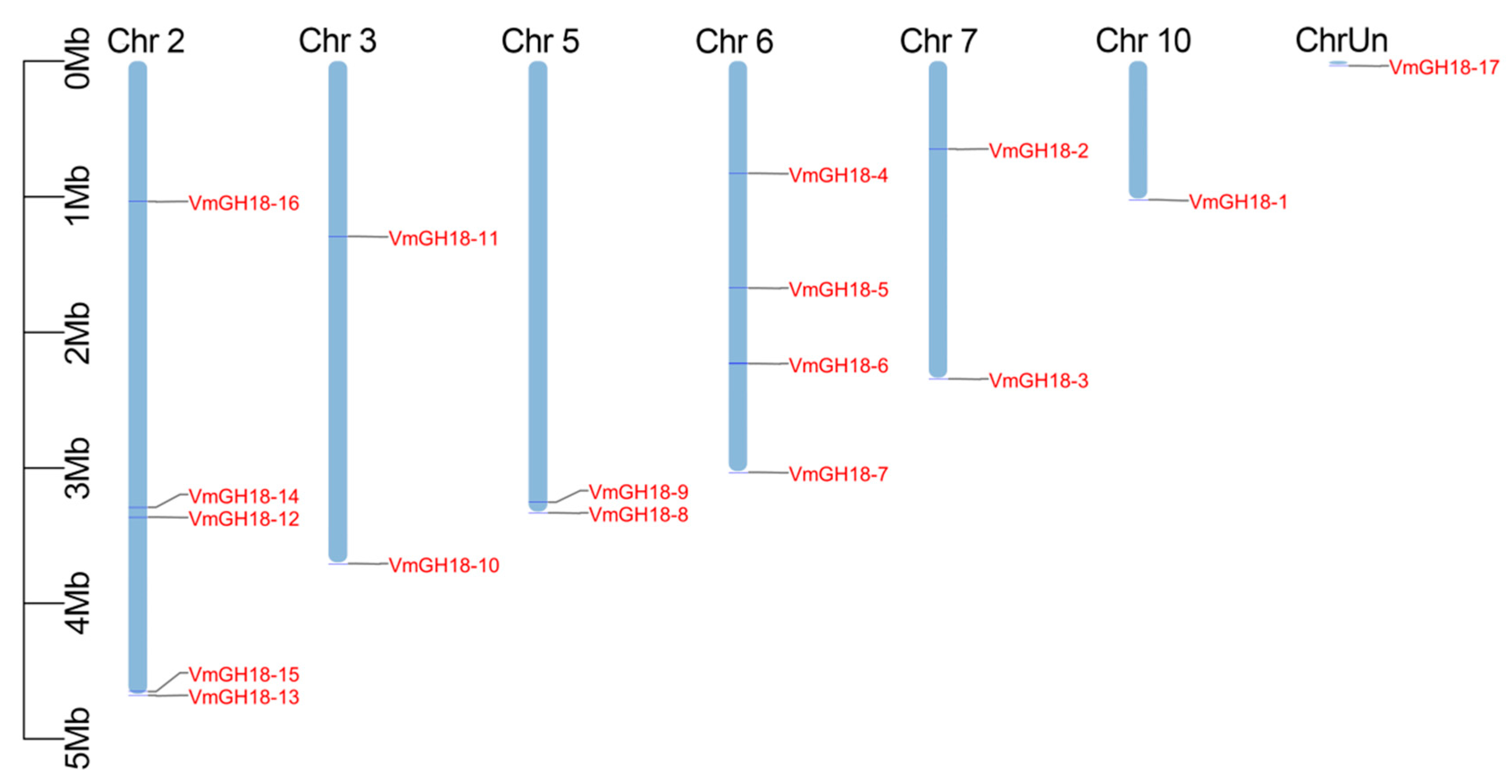
3.2. Chromosomal Location of VmGH18
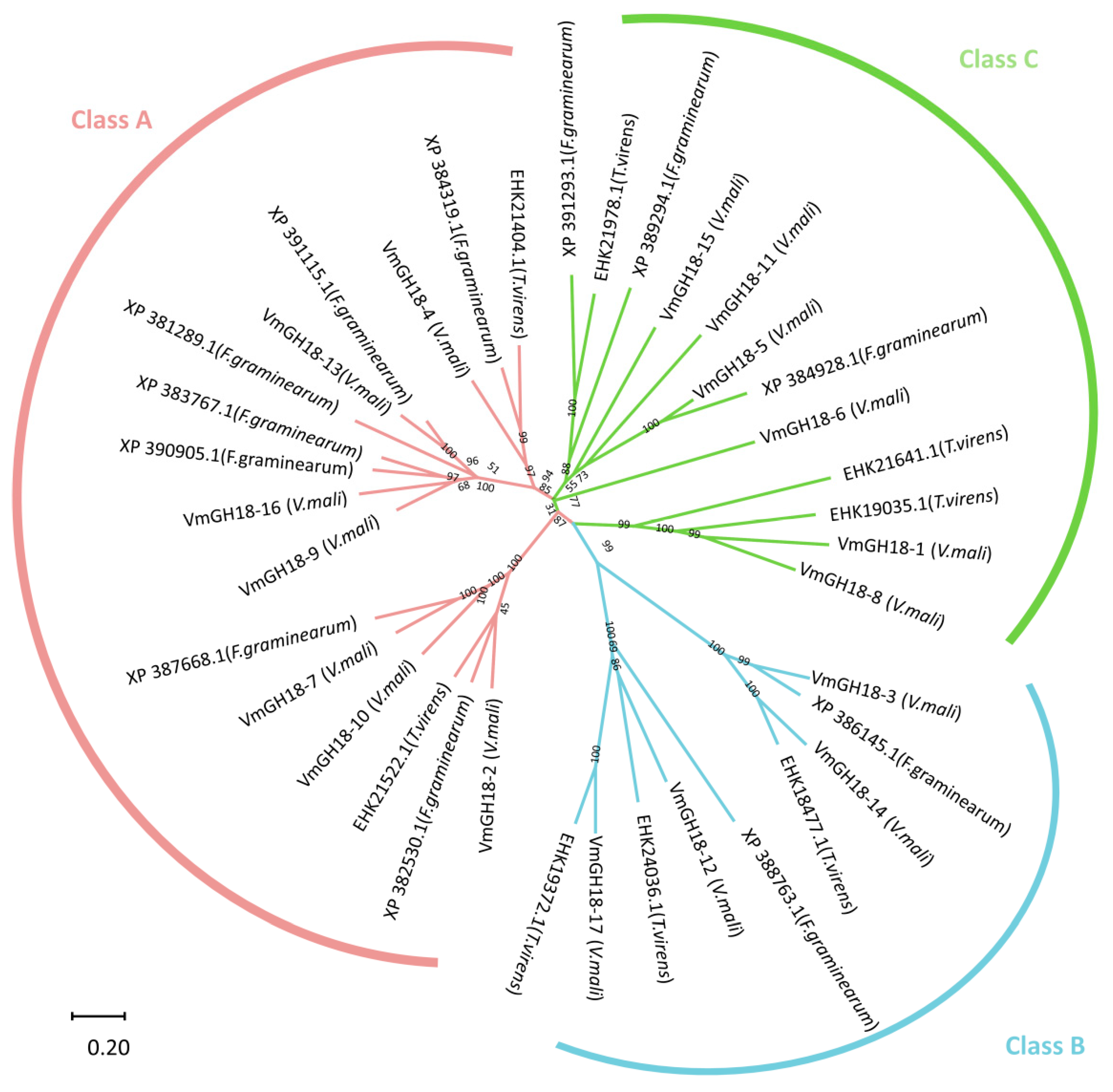
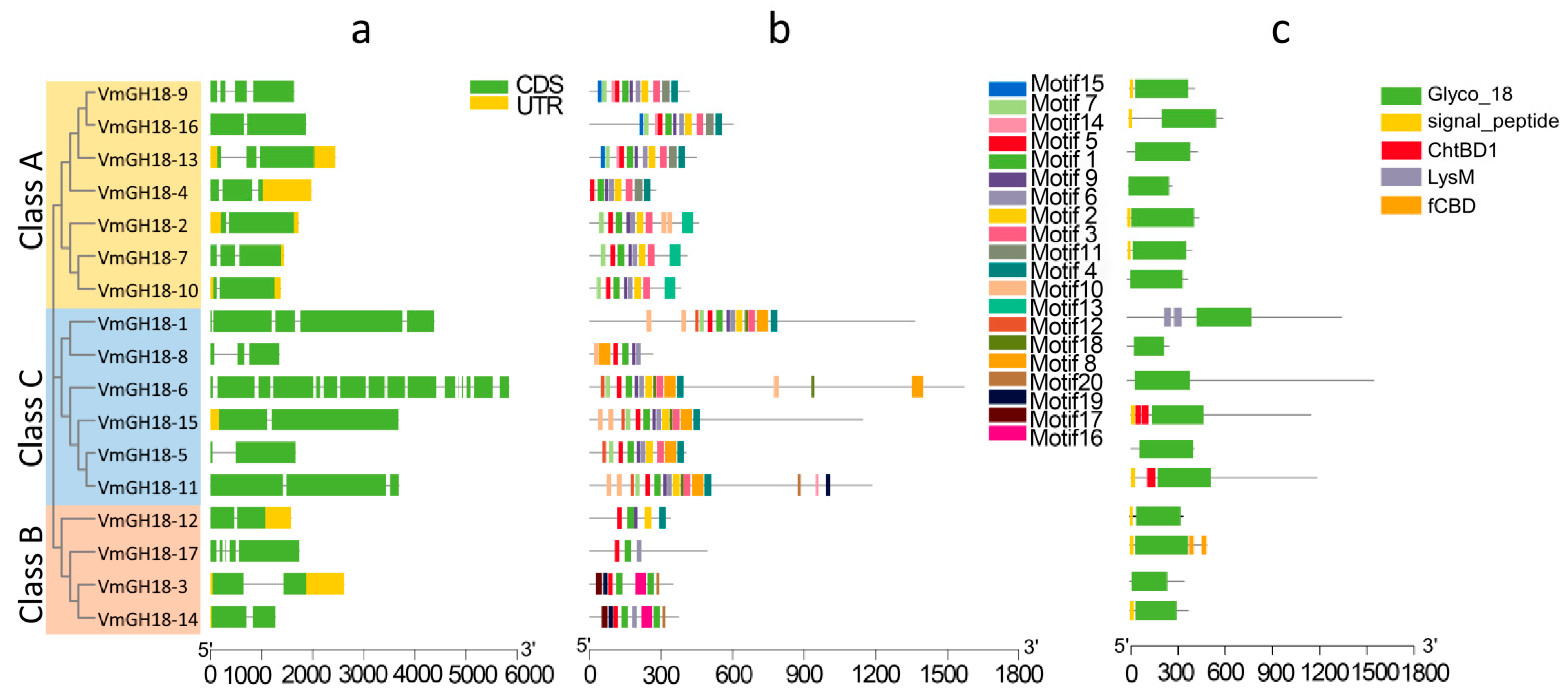
3.3. Gene Structure and Conserved Motif Analyses
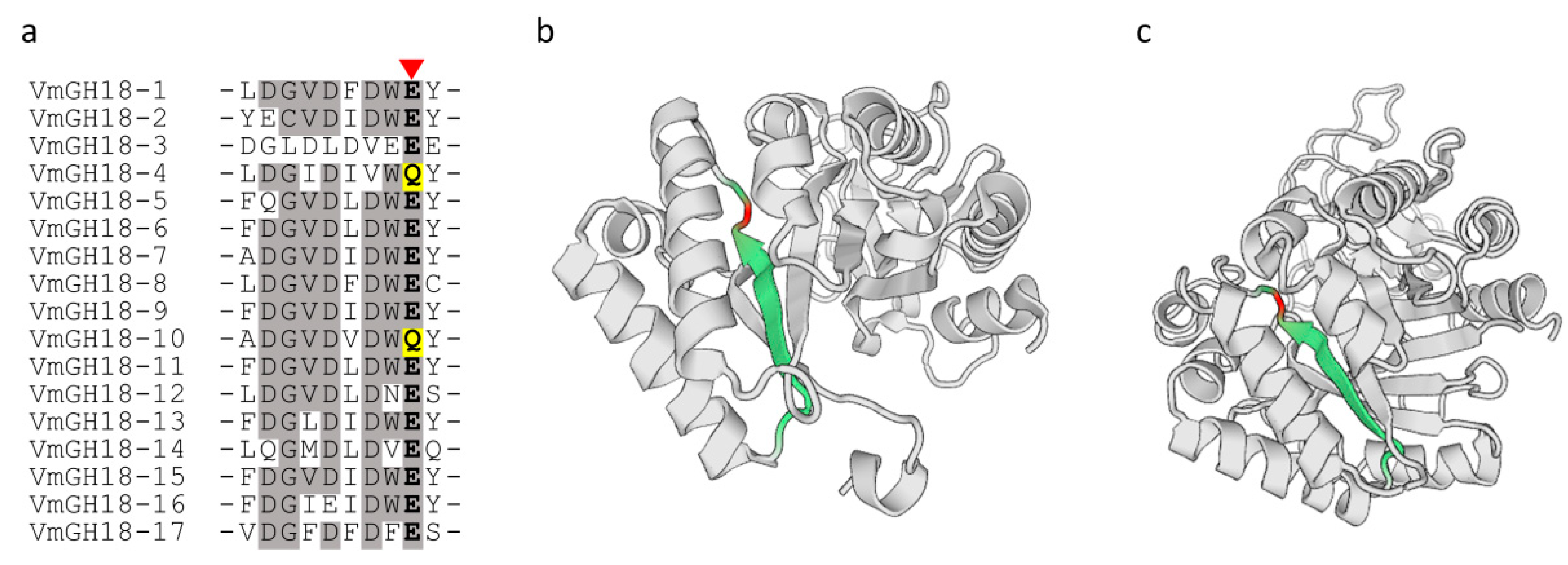
3.4. Conserved Domains and Active Site Analysis of VmGH18s
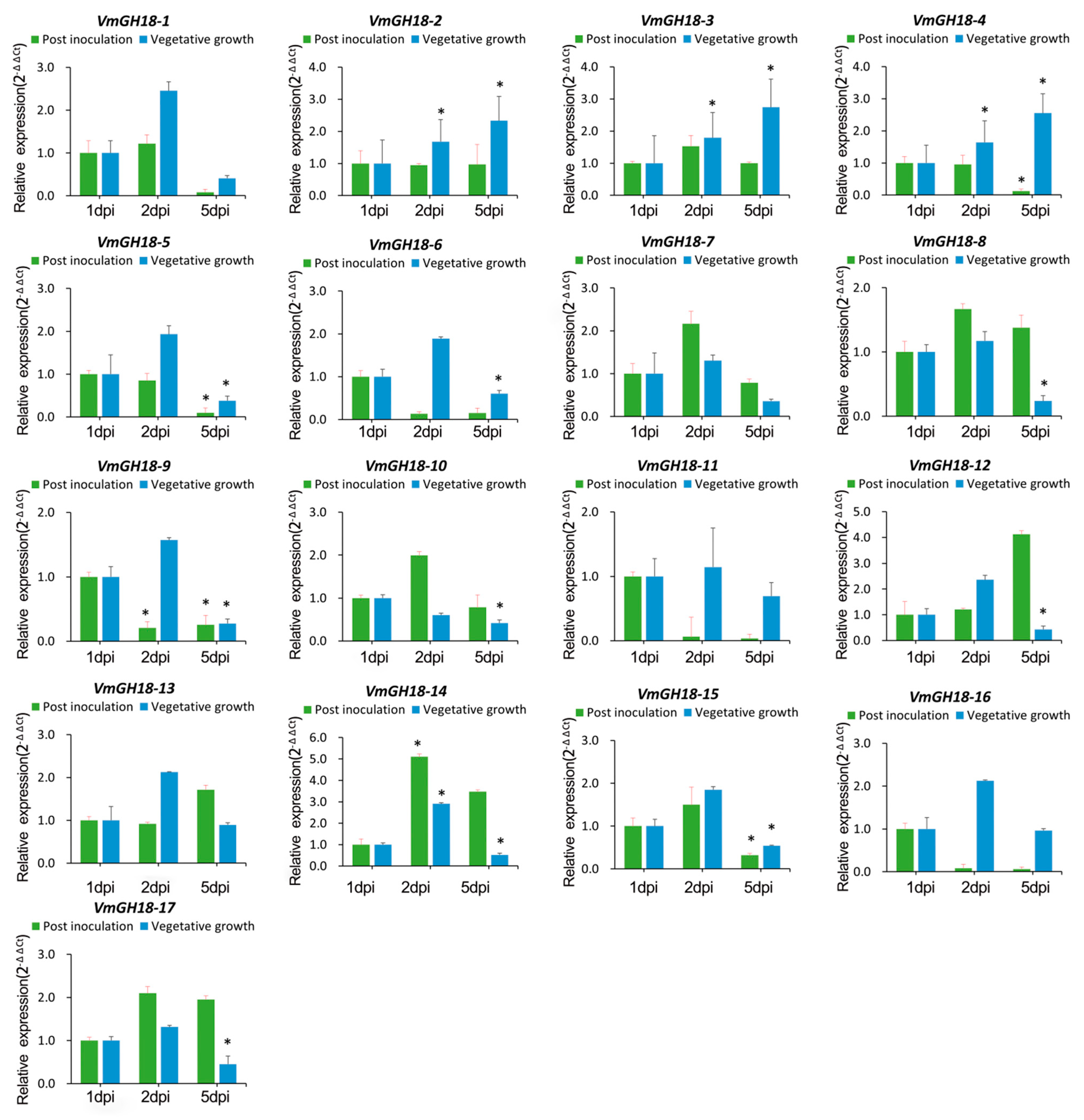
3.5. Gene Expression Profile of VmGH18
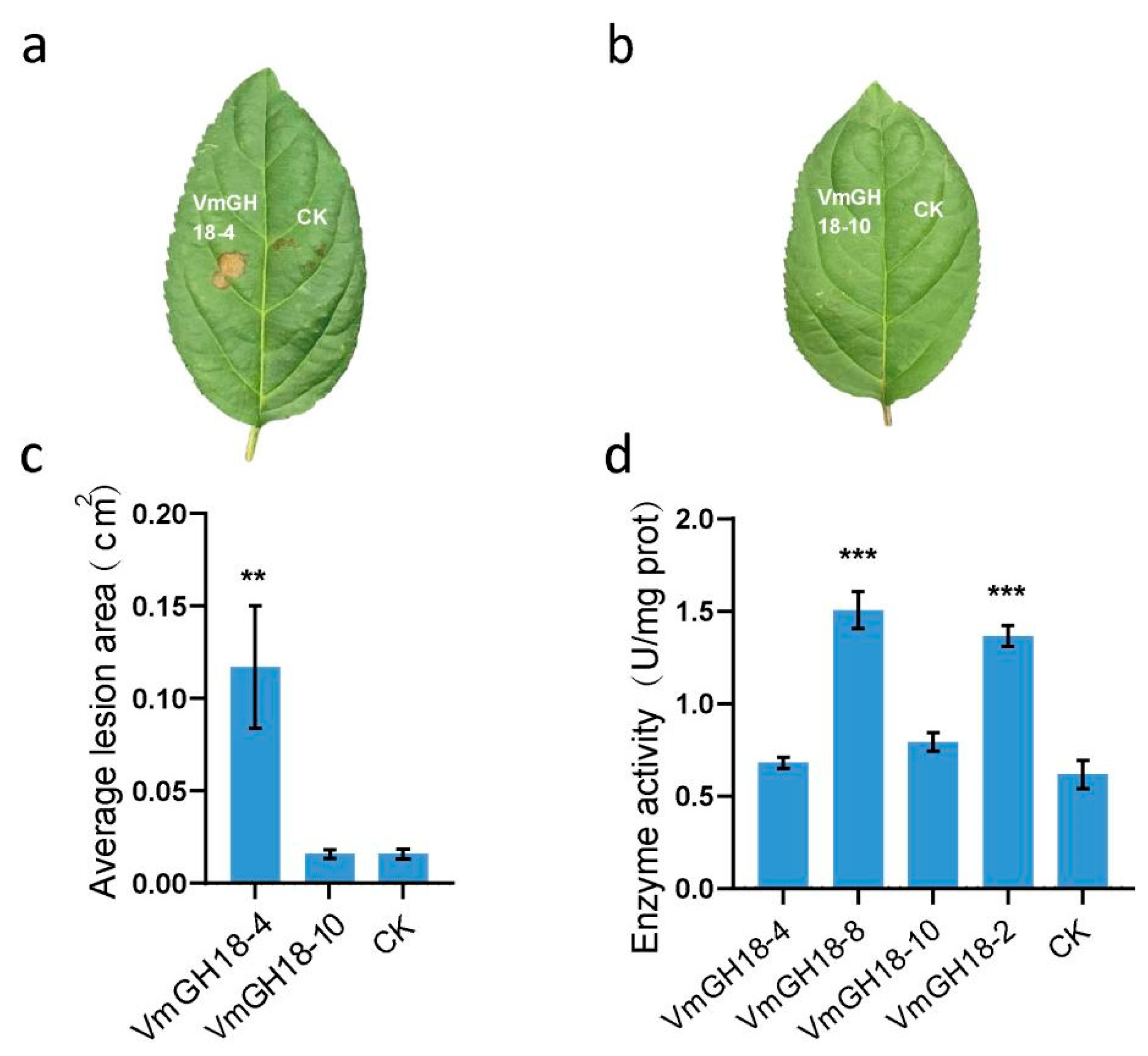
3.6. VmGH18-4 but Not VmGH18-10 Is an Elicitor of Plant Cell Death
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Synowiecki, J.; Al-Khateeb, N.A. Production, Properties, and Some New Applications of Chitin and Its Derivatives. Crit. Rev. Food Sci. Nutr. 2003, 43, 145–171. [Google Scholar] [CrossRef] [PubMed]
- Berini, F.; Katz, C.; Gruzdev, N.; Casartelli, M.; Tettamanti, G.; Marinelli, F. Microbial and viral chitinases: Attractive biopesticides for integrated pest management. Biotechnol. Adv. 2018, 36, 818–838. [Google Scholar] [PubMed]
- Merzendorfer, H.; Zimoch, L. Chitin metabolism in insects: Structure, function and regulation of chitin synthases and chitinases. J. Exp. Biol. 2003, 206, 4393–4412. [Google Scholar] [PubMed]
- Sánchez-Vallet, A.; Mesters, J.R.; Thomma, B.P.H.J. The battle for chitin recognition in plant-microbe interactions. FEMS Microbiol. Rev. 2015, 39, 171–183. [Google Scholar] [CrossRef]
- Horsch, M.; Mayer, C.; Sennhauser, U.; Rast, D.M. Beta-N-acetylhexosaminidase: A target for the design of antifungal agents. Pharmacol. Ther. 1997, 76, 187–218. [Google Scholar]
- Prakash, N.A.U.; Jayanthi, M.; Sabarinathan, R.; Kangueane, P.; Mathew, L.; Sekar, K. Evolution, Homology Conservation, and Identification of Unique Sequence Signatures in GH19 Family Chitinases. J. Mol. Evol. 2010, 70, 466–478. [Google Scholar] [CrossRef]
- Fülöp, L.; Ponyi, T. Classification of glycosyl hydrolases based on structural homology. J. Univers. Sci. Online 2015, 2, 1–9. [Google Scholar]
- Gaber, Y.; Mekasha, S.; Vaaje-Kolstad, G.; Eijsink, V.G.H.; Fraaije, M.W. Characterization of a chitinase from the cellulolytic actinomycete Thermobifida fusca. Biochim. Biophys. Acta BBA Proteins Proteom. 2016, 1864, 1253–1259. [Google Scholar] [CrossRef]
- Tian, L.; Weixing, Z.; Jing, W.; Yong, Z.; Yanwei, D.; Mingbo, Q.; Qing, Y. The deduced role of a chitinase containing two nonsynergistic catalytic domains. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 30–40. [Google Scholar] [CrossRef]
- Suzuki, K.; Taiyoji, M.; Sugawara, N.; Nikaidou, N.; Henrissat, B.; Watanabe, T. The third chitinase gene (chiC) of Serratia marcescens 2170 and the relationship of its product to other bacterial chitinases. Biochem. J. 1999, 343 Pt 3, 587–596. [Google Scholar]
- Karlsson, M.; Stenlid, J. Comparative Evolutionary Histories of the Fungal Chitinase Gene Family Reveal Non-Random Size Expansions and Contractions due to Adaptive Natural Selection. Evol. Bioinform. Online 2008, 4, 47–60. [Google Scholar] [PubMed]
- Benítez, T.; Rincón, A.M.; Limón, M.C.; Codón, A.C. Biocontrol mechanisms of Trichoderma strains. Int. Microbiol. 2004, 7, 249–260. [Google Scholar] [PubMed]
- Gruber, S.; Vaaje-Kolstad, G.; Matarese, F.; López-Mondéjar, R.; Kubicek, C.P.; Seidl-Seiboth, V. Analysis of subgroup C of fungal chitinases containing chitin-binding and LysM modules in the mycoparasite Trichoderma atroviride. Glycobiology 2010, 21, 122–133. [Google Scholar]
- Yoshihito, K.; Toki, T.; Tomoyuki, N.; Takayuki, O.; Tamo, F. Structure, mechanism, and phylogeny of LysM-chitinase conjugates specifically found in fern plants. Plant Sci. 2022, 321, 111310. [Google Scholar]
- Eijsink, V.; Vaaje-Kolstad, G.; V?Rum, K.M.; Horn, S.J. Towards new enzymes for biofuels: Lessons from chitinase research. Trends Biotechnol. 2008, 26, 228–235. [Google Scholar]
- Yamazaki, H.; Yamazaki, D.; Takaya, N.; Takagi, M.; Ohta, A.; Horiuchi, H. A chitinase gene, chiB, involved in the autolytic process of Aspergillus nidulans. Curr. Genet. 2007, 51, 89–98. [Google Scholar]
- Shin, K.-S.; Kwon, N.-J.; Kim, Y.H.; Park, H.-S.; Kwon, G.-S.; Yu, J.-H. Differential Roles of the ChiB Chitinase in Autolysis and Cell Death of Aspergillus nidulans. Eukaryot. Cell 2009, 8, 738–746. [Google Scholar] [CrossRef]
- Dünkler, A.; Walther, A.; Specht, C.A.; Wendland, J. Candida albicans CHT3 encodes the functional homolog of the Cts1 chitinase of Saccharomyces cerevisiae. Fungal Genet. Biol. 2005, 42, 935–947. [Google Scholar]
- Kuranda, M.J.; Robbins, P.W. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J. Biol. Chem. 1991, 266, 19758–19767. [Google Scholar]
- Adams, D.J. Fungal cell wall chitinases and glucanases. Microbiology 2004, 150, 2029–2035. [Google Scholar]
- Seidl, V. Chitinases of filamentous fungi: A large group of diverse proteins with multiple physiological functions. Fungal Biol. Rev. 2008, 22, 36–42. [Google Scholar]
- Li, D.C. Review of Fungal Chitinases. Mycopathologia 2006, 161, 345. [Google Scholar]
- Gan, Z.; Yang, J.; Tao, N.; Liang, L.; Mi, Q.; Li, J.; Zhang, K.Q. Cloning of the gene Lecanicillium psalliotae chitinase Lpchi1 and identification of its potential role in the biocontrol of root-knot nematode Meloidogyne incognita. Appl. Microbiol. Biotechnol. 2007, 76, 1309–1317. [Google Scholar] [PubMed]
- Alcazar-Fuoli, L.; Clavaud, C.; Lamarre, C.; Aimanianda, V.; Seidl-Seiboth, V.; Mellado, E.; Latgé, J.P. Functional analysis of the fungal/plant class chitinase family in Aspergillus fumigatus. Fungal Genet. Biol. 2011, 48, 418–429. [Google Scholar] [CrossRef]
- Emri, T.; Molnár, Z.; Szilágyi, M.; Pócsi, I. Regulation of autolysis in aspergillus nidulans. Appl. Biochem. Biotechnol. 2008, 151, 211–220. [Google Scholar] [CrossRef]
- Pcdcsi, I.; Leiter, c.; Kwon, N.-J.; Shin, K.-S.; Kwon, G.-S.; Pusztahelyi, T.; Emri, T.; Abuknesha, R.A.; Price, R.G.; Yu, J.-H. Asexual sporulation signalling regulates autolysis of Aspergillus nidulans via modulating the chitinase ChiB production. J. Appl. Microbiol. 2009, 107, 514–523. [Google Scholar] [CrossRef]
- Ihrmark, K.; Asmail, N.; Ubhayasekera, W.; Melin, P.; Stenlid, J.; Karlsson, M. Comparative Molecular Evolution of Trichoderma Chitinases in Response to Mycoparasitic Interactions. Evol. Bioinform. 2010, 6, 1–26. [Google Scholar]
- Lorencini, F.G.; Andrea, S.V.; Toledo, T.; Vital, D.; Costa, D.; Oliveira, F.; Thomma, B.; Guimar?Es, P.; Lima, T. Suppression of Plant Immunity by Fungal Chitinase-like Effectors. Curr. Biol. 2018, 28, 3023–3030. [Google Scholar]
- Alhoraibi, H.; Bigeard, J.; Rayapuram, N.; Colcombet, J.; Hirt, H. Plant Immunity: The MTI-ETI Model and Beyond. Curr. Issues Mol. Biol. 2019, 30, 39–58. [Google Scholar] [CrossRef]
- Jaswal, R.; Kiran, K.; Rajarammohan, S.; Dubey, H.; Singh, P.K.; Sharma, Y.; Deshmukh, R.; Sonah, H.; Gupta, N.; Sharma, T.R. Effector Biology of Biotrophic Plant Fungal Pathogens: Current Advances and Future Prospects. Microbiol. Res. 2020, 241, 126567. [Google Scholar] [CrossRef]
- Bruno Pok Man, N.; Pingtao, D.; Jonathan, D.G.J. Channeling plant immunity. Cell 2021, 184, 3358–3360. [Google Scholar] [CrossRef]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal Effectors and Plant Susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [PubMed]
- Pusztahelyi, T. Chitin and chitin-related compounds in plant-fungal interactions. Mycology 2018, 9, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Jesús, M.; Diego, R.; Jesús, H.; Michael, T.; Antonio, D.V.; Alejandro, P.G. Effectors With Chitinase Activity (EWCAs), a family of conserved, secreted fungal chitinases that suppress chitin-triggered immunity. Plant Cell 2021, 33, 1319–1340. [Google Scholar]
- Abe, K.; Kotoda, N.; Kato, H.; Soejima, J.I. Genetic studies on resistance to Valsa canker in apple: Genetic variance and breeding values estimated from intra- and inter-specific hybrid progeny populations. Tree Genet. Genomes 2011, 7, 363–372. [Google Scholar]
- Ke, X.; Yin, Z.; Song, N.; Dai, Q.; Voegele, R.T.; Liu, Y.; Wang, H.; Gao, X.; Kang, Z.; Huang, L. Transcriptome profiling to identify genes involved in pathogenicity of Valsa mali on apple tree. Fungal Genet. Biol. 2014, 68, 31–38. [Google Scholar]
- Feng, H.; Wang, C.; He, Y.; Tang, L.; Han, P.; Liang, J.; Huang, L. Apple Valsa canker: Insights into pathogenesis and disease control. Phytopathol. Res. 2023, 5, 45. [Google Scholar] [CrossRef]
- Meng, X.-l.; Qi, X.-h.; Han, Z.-y.; Guo, Y.-b.; Wang, Y.-n.; Hu, T.-l.; Wang, L.-m.; Cao, K.-q.; Wang, S.-t. Latent Infection of Valsa mali in the Seeds, Seedlings and Twigs of Crabapple and Apple Trees is a Potential Inoculum Source of Valsa Canker. Sci. Rep. 2019, 9, 7738. [Google Scholar] [CrossRef]
- Liu, X.; Li, X.; Bozorov, T.A.; Ma, R.; Ma, J.; Zhang, Y.; Yang, H.; Li, L.; Zhang, D. Characterization and pathogenicity of six Cytospora strains causing stem canker of wild apple in the Tianshan Forest, China. For. Pathol. 2020, 50, e12587. [Google Scholar]
- Liu, B.; Peng, L.X. Studies on Tissue Culture System of Malus sieversii. J. Tianjin Agric. Univ. 2011, 18, 3. [Google Scholar] [CrossRef]
- Jaina, M.; Sara, C.; Lowri, W.; Matloob, Q.; Gustavoa, S.; Sonnhammer, E.; Tosatto, S.; Lisanna, P.; Shriya, R.; Richardson, L.J. Pfam: The protein families database in 2021. Nucleic Acids Res. 2020, 49, D412–D419. [Google Scholar]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [PubMed]
- Ivica, L.; Supriya, K.; Peer, B. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2020, 49, D458–D460. [Google Scholar]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [PubMed]
- Bateman, A.; Martin, M.-J.; Orchard, S.; Magrane, M.; Agivetova, R.; Ahmad, S.; Alpi, E.; Bowler-Barnett, E.H.; Britto, R.; Bursteinas, B.; et al. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar]
- Yin, Z.; Ke, X.; Huang, D.; Gao, X.; Voegele, R.T.; Kang, Z.; Huang, L. Validation of reference genes for gene expression analysis in Valsa mali var. mali using real-time quantitative PCR. World J. Microbiol. Biotechnol. 2013, 29, 1563–1571. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR. Methods 2002, 25, 402–408. [Google Scholar]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2012, 40, D290–D301. [Google Scholar]
- Langner, T.; Goehre, V. Fungal chitinases: Function, regulation, and potential roles in plant/pathogen interactions. Curr. Genet. 2016, 62, 243–254. [Google Scholar] [CrossRef]
- Cecilia Gortari, M.; Alberto Hours, R. Fungal chitinases and their biological role in the antagonism onto nematode eggs. A review. Mycol. Prog. 2008, 7, 221–238. [Google Scholar] [CrossRef]
- Yin, Z.Y.; Ke, X.W.; Kang, Z.S.; Huang, L.L. Apple resistance responses against Valsa mali revealed by transcriptomics analyses. Physiol. Mol. Plant Pathol. 2016, 93, 85–92. [Google Scholar] [CrossRef]
- Yang, Y.; Sossah, F.L.; Li, Z.; Hyde, K.D.; Li, D.; Xiao, S.; Fu, Y.; Yuan, X.; Li, Y. Genome-Wide Identification and Analysis of Chitinase GH18 Gene Family in Mycogone perniciosa. Front. Microbiol. 2021, 11, 596719. [Google Scholar] [CrossRef] [PubMed]
- Gruber, S.; Kubicek, C.P.; Seidl-Seiboth, V. Differential Regulation of Orthologous Chitinase Genes in Mycoparasitic Trichoderma Species. Appl. Environ. Microbiol. 2011, 77, 7217–7226. [Google Scholar] [CrossRef]
- Pham, T.A.T.; Schwerdt, J.G.; Shirley, N.J.; Xing, X.; Hsieh, Y.S.Y.; Srivastava, V.; Bulone, V.; Little, A. Analysis of cell wall synthesis and metabolism during early germination of Blumeria graminis f. sp. hordei conidial cells induced in vitro. Cell Surf. 2019, 5, 100030. [Google Scholar] [CrossRef]
- Brunner, K.; Peterbauer, C.K.; Mach, R.L.; Lorito, M.; Zeilinger, S.; Kubicek, C.P. The Nag1 N-acetylglucosaminidase of Trichoderma atroviride is essential for chitinase induction by chitin and of major relevance to biocontrol. Curr. Genet. 2003, 43, 289–295. [Google Scholar] [CrossRef]
- Duenkler, A.; Jorde, S.; Wendland, J. An Ashbya gossypii cts2 mutant deficient in a sporulation-specific chitinase can be complemented by Candida albicans CHT4. Microbiol. Res. 2008, 163, 701–710. [Google Scholar] [CrossRef]
- Yin, Z.; Liu, H.; Li, Z.; Ke, X.; Dou, D. Genome sequence of Valsa canker pathogens uncovers a potential adaptation of colonization of woody bark. New Phytol. 2015, 208, 1202–1216. [Google Scholar] [CrossRef]
- Jan, D.; Van de Peer, Y.; Vanessa, V. Nearby transposable elements impact plant stress gene regulatory networks: A meta-analysis in A. thaliana and S. lycopersicum. BMC Genom. 2022, 23, 18. [Google Scholar] [CrossRef]
- Min, Z.; Lei, L.; Xubo, L.; Yang, W.; Ying, L.; Qing, G.; Shulin, L.; Yuxin, S.; Xiaoma, T.; Di, Z.; et al. A Translocation Pathway for Vesicle-Mediated Unconventional Protein Secretion. Cell 2020, 181, 637–652. [Google Scholar] [CrossRef]
- Yang, C.; Yu, Y.Q.; Huang, J.K.; Meng, F.W.; Pang, J.H.; Zhao, Q.Q.; Islam, M.A.; Xu, N.; Tian, Y.; Liu, J. Binding of the Chitinase MoChia1 by a Rice Tetratricopeptide Repeat Protein Allows Free Chitin to Trigger Immune Responses. Plant Cell 2019, 31, 172–188. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Fan, D.; Li, Y.; Chen, Y.; Huang, L.; Yan, X. Transcriptome analysis of Valsa mali reveals its response mechanism to the biocontrol actinomycete Saccharothrix yanglingensis Hhs.015. BMC Microbiol. 2018, 18, 90. [Google Scholar] [CrossRef]
- Geum-Jae, J.; Fazlurrahman, K.; Nazia, T.; Young-Mog, K. Chitinases as key virulence factors in microbial pathogens: Understanding their role and potential as therapeutic targets. Int. J. Biol. Macromol. 2023, 249, 126021. [Google Scholar] [CrossRef]
- Zheng, J.; Xu, Y. A Review: Development of Plant Protection Methods and Advances in Pesticide Application Technology in Agro-Forestry Production. Agriculture 2023, 13, 2165. [Google Scholar] [CrossRef]
| Sequence | Protein Product | Protein Length | pI | MW (kDa) | Signal Peptide | Genomic Position | Gene Length |
|---|---|---|---|---|---|---|---|
| VmGH18-1 | KUI73695.1 | 1362 | 3.88 | 146.38 | / | Chr11:10206981025079 (−) | 4089 |
| VmGH18-2 | KUI71748.1 | 456 | 4.23 | 48.58 | SP (Sec/SPI) | Chr7:646626648056 (−) | 1371 |
| VmGH18-3 | KUI71641.1 | 349 | 4.52 | 38.3 | / | Chr7:23421462343974 (−) | 1050 |
| VmGH18-4 | KUI70699.1 | 278 | 8 | 29.73 | / | Chr6:827947828966 (−) | 837 |
| VmGH18-5 | KUI70476.1 | 404 | 4.17 | 44.39 | / | Chr6:16707121672374 (−) | 1215 |
| VmGH18-6 | KUI70319.1 | 1569 | 3.95 | 170.67 | / | Chr6:22273132233150 (−) | 4710 |
| VmGH18-7 | KUI70276.1 | 409 | 4.56 | 45.39 | SP (Sec/SPI) | Chr6:30330583034434(+) | 1230 |
| VmGH18-8 | KUI69933.1 | 265 | 4.53 | 28.94 | / | Chr5:33309903332333 (−) | 798 |
| VmGH18-9 | KUI69708.1 | 418 | 4 | 45.06 | SP (Sec/SPI) | Chr5:32510833252716 (−) | 1257 |
| VmGH18-10 | KUI67677.1 | 382 | 8.39 | 42.63 | / | Chr3:37058953707093 (−) | 1149 |
| VmGH18-11 | KUI67400.1 | 1183 | 4.68 | 127.29 | SP (Sec/SPI) | Chr3:12911601294848 (−) | 3552 |
| VmGH18-12 | KUI66967.1 | 338 | 4.32 | 35.75 | SP (Sec/SPI) | Chr2:33626333363702(+) | 1017 |
| VmGH18-13 | KUI66706.1 | 447 | 7.47 | 50.47 | / | Chr2:46765024678403 (−) | 1344 |
| VmGH18-14 | KUI66287.1 | 373 | 3.85 | 40.99 | SP (Sec/SPI) | Chr2:32909873292234 (−) | 1122 |
| VmGH18-15 | KUI66075.1 | 1145 | 4.85 | 123.78 | SP (Sec/SPI) | Chr2:46476994651222 (−) | 3438 |
| VmGH18-16 | KUI65769.1 | 602 | 4.08 | 64.63 | SP (Sec/SPI) | Chr2:10324881034351(+) | 1809 |
| VmGH18-17 | KUI64200.1 | 491 | 3.63 | 51.53 | SP (Sec/SPI) | ChrUn:3131033037 (−) | 1476 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kahar, G.; Haxim, Y.; Zhang, X.; Liu, X.; Liu, H.; Wen, X.; Li, X.; Zhang, D. Genome-Wide Identification and Analysis of Chitinase GH18 Gene Family in Valsa mali. J. Fungi 2025, 11, 290. https://doi.org/10.3390/jof11040290
Kahar G, Haxim Y, Zhang X, Liu X, Liu H, Wen X, Li X, Zhang D. Genome-Wide Identification and Analysis of Chitinase GH18 Gene Family in Valsa mali. Journal of Fungi. 2025; 11(4):290. https://doi.org/10.3390/jof11040290
Chicago/Turabian StyleKahar, Gulnaz, Yakupjan Haxim, Xuechun Zhang, Xiaojie Liu, Huawei Liu, Xuejing Wen, Xiaoshuang Li, and Daoyuan Zhang. 2025. "Genome-Wide Identification and Analysis of Chitinase GH18 Gene Family in Valsa mali" Journal of Fungi 11, no. 4: 290. https://doi.org/10.3390/jof11040290
APA StyleKahar, G., Haxim, Y., Zhang, X., Liu, X., Liu, H., Wen, X., Li, X., & Zhang, D. (2025). Genome-Wide Identification and Analysis of Chitinase GH18 Gene Family in Valsa mali. Journal of Fungi, 11(4), 290. https://doi.org/10.3390/jof11040290








