Advancing Dermatomycosis Diagnosis: Evaluating a Microarray-Based Platform for Rapid and Accurate Fungal Detection—A Pilot Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Samples
2.2. Standard Mycological Assays
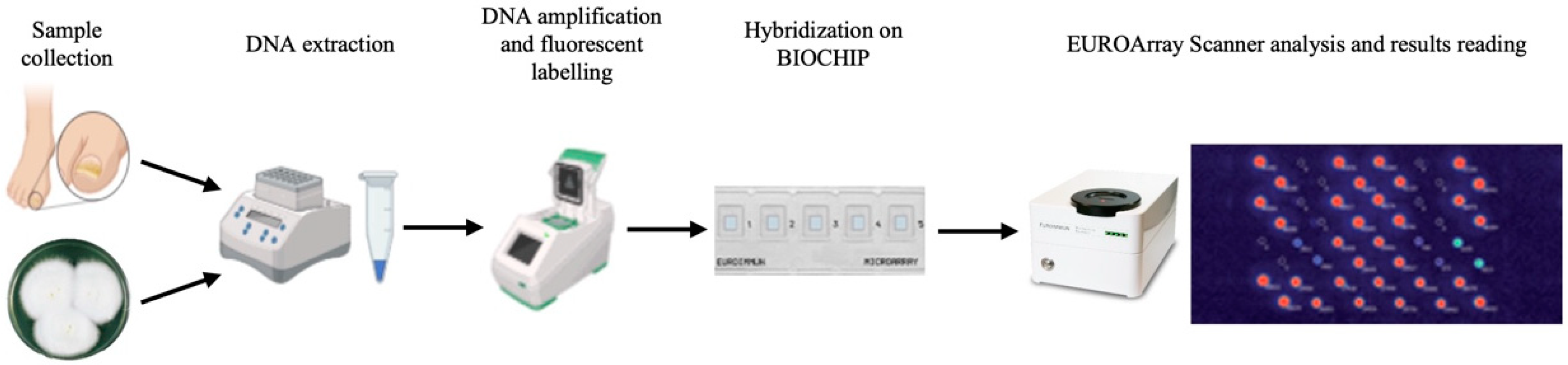
2.3. EUROArray Dermatomycosis Platform Assay
2.4. Analysis of Results
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Biswas, M.C.; Mukherjee, K.; Ghosh, S.; Roy-Chowdhury, M.; Acharya, K. Natural Products of Plant Origin: An Emerging Therapeutic For Dermatomycosis. Int. J. Dermatol. 2024, 63, 858–872. [Google Scholar] [CrossRef]
- Barac, A.; Stjepanovic, M.; Krajisnik, S.; Stevanovic, G.; Paglietti, B.; Milosevic, B. Dermatophytes: Update on clinical epidemiology and treatment. Mycopathologia 2024, 189, 101. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Wang, T.; Cooper, E.A.; Summerbell, R.C.; Piguet, V.; Tosti, A.; Piraccini, B.M. A Comprehensive Review of Nondermatophyte Mould Onychomycosis: Epidemiology, Diagnosis and Management. J. Eur. Acad. Dermatol. Venereol. 2024, 38, 480–495. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.A.; Mirza, R.; Sattar, A.; Khan, Y.; Khan, S.A. Unveiling Onychomycosis: Pathogenesis, Diagnosis, and Innovative Treatment Strategies. Microb. Pathog. 2025, 198, 107111. [Google Scholar] [CrossRef] [PubMed]
- Dubljanin, E.; Džamić, A.; Vujčić, I.; Grujičić, S.; Arsenijević, V.A.; Mitrović, S.; Čalovski, I.Č. Epidemiology of Onychomycosis in Serbia: A Laboratory-Based Survey and Risk Factor Identification. Mycoses 2017, 60, 25–32. [Google Scholar] [CrossRef]
- Gupta, A.K.; Gupta, G.; Jain, H.C.; Lynde, C.W.; Foley, K.A.; Daigle, D.; Cooper, E.A.; Summerbell, R.C. The Prevalence of Unsuspected Onychomycosis and Its Causative Organisms in a Multicentre Canadian Sample of 30 000 Patients Visiting Physicians’ Offices. J. Eur. Acad. Dermatol. Venereol. 2016, 30, 1567–1572. [Google Scholar] [CrossRef]
- Maraki, S.; Mavromanolaki, V.E. Epidemiology of Onychomycosis in Crete, Greece: A 12-Year Study. Mycoses 2016, 59, 798–802. [Google Scholar] [CrossRef]
- Gupta, C.; Jongman, M.; Das, S.; Snehaa, K.; Bhattacharya, S.N.; Seyedmousavi, S.; van Diepeningen, A.D. Genotyping and In Vitro Antifungal Susceptibility Testing of Fusarium Isolates from Onychomycosis in India. Mycopathologia 2016, 181, 497–504. [Google Scholar] [CrossRef]
- Totri, C.R.; Feldstein, S.; Admani, S.; Friedlander, S.F.; Eichenfield, L.F. Epidemiologic Analysis of Onychomycosis in the San Diego Pediatric Population. Pediatr. Dermatol. 2017, 34, 46–49. [Google Scholar] [CrossRef]
- Leung, A.K.C.; Lam, J.M.; Leong, K.F.; Hon, K.L.; Barankin, B.; Leung, A.A.M.; Wong, A.H.C. Onychomycosis: An Updated Review. Recent. Pat. Inflamm. Allergy Drug Discov. 2020, 14, 32–45. [Google Scholar] [CrossRef]
- Papini, M.; Piraccini, B.M.; Difonzo, E.; Brunoro, A. Epidemiology of Onychomycosis in Italy: Prevalence Data and Risk Factor Identification. Mycoses 2015, 58, 659–664. [Google Scholar] [CrossRef] [PubMed]
- Avner, S.; Nir, N.; Henri, T. Fifth Toenail Clinical Response to Systemic Antifungal Therapy Is Not a Marker of Successful Therapy for Other Toenails with Onychomycosis. J. Eur. Acad. Dermatol. Venereol. 2006, 20, 1194–1196. [Google Scholar] [CrossRef]
- Sigurgeirsson, B.; Steingrímsson, Ö. Risk Factors Associated with Onychomycosis. J. Eur. Acad. Dermatol. Venereol. 2004, 18, 48–51. [Google Scholar] [CrossRef]
- Gupta, A.K.; Taborda, P.; Taborda, V.; Gilmour, J.; Rachlis, A.; Salit, I.; Gupta, M.A.; MacDonald, P.; Cooper, E.A.; Summerbell, R.C. Epidemiology and Prevalence of Onychomycosis in HIV-Positive Individuals. Int. J. Dermatol. 2000, 39, 746–753. [Google Scholar] [CrossRef]
- García-Romero, M.T.; Lopez-Aguilar, E.; Arenas, R. Onychomycosis in Immunosuppressed Children Receiving Chemotherapy. Pediatr. Dermatol. 2014, 31, 618–620. [Google Scholar] [CrossRef]
- Gupta, A.K.; Daigle, D.; Foley, K.A. The Prevalence of Culture-Confirmed Toenail Onychomycosis in at-Risk Patient Populations. J. Eur. Acad. Dermatol. Venereol. 2015, 29, 1039–1044. [Google Scholar] [CrossRef] [PubMed]
- Klaassen, K.M.G.; Dulak, M.G.; Van De Kerkhof, P.C.M.; Pasch, M.C. The Prevalence of Onychomycosis in Psoriatic Patients: A Systematic Review. J. Eur. Acad. Dermatol. Venereol. 2014, 28, 533–541. [Google Scholar] [CrossRef]
- Svejgaard, E.L.; Nilsson, J. Onychomycosis in Denmark: Prevalence of Fungal Nail Infection in General Practice. Mycoses 2004, 47, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Versteeg, S.G.; Shear, N.H. Onychomycosis in the 21st Century: An Update on Diagnosis, Epidemiology, and Treatment. J. Cutan. Med. Surg. 2017, 21, 525–539. [Google Scholar] [CrossRef]
- Bombace, F.; Iovene, M.R.; Galdiero, M.; Martora, F.; Nicoletti, G.F.; D’Andrea, M.; Della Pepa, M.E.; Vitiello, M. Non-Dermatophytic Onychomycosis Diagnostic Criteria: An Unresolved Question. Mycoses 2016, 59, 558–565. [Google Scholar] [CrossRef]
- Wilsmann-Theis, D.; Sareika, F.; Bieber, T.; Schmid-Wendtner, M.H.; Wenzel, J. New Reasons for Histopathological Nail-Clipping Examination in the Diagnosis of Onychomycosis. J. Eur. Acad. Dermatol. Venereol. 2011, 25, 235–237. [Google Scholar] [CrossRef] [PubMed]
- Ghannoum, M.; Mukherjee, P.; Isham, N.; Markinson, B.; Del Rosso, J.; Leal, L. Examining the Importance of Laboratory and Diagnostic Testing When Treating and Diagnosing Onychomycosis. Int. J. Dermatol. 2018, 57, 131–138. [Google Scholar] [CrossRef] [PubMed]
- D’Hue, Z.; Perkins, S.M.; Billings, S.D. GMS Is Superior to PAS for Diagnosis of Onychomycosis. J. Cutan. Pathol. 2008, 35, 745–747. [Google Scholar] [CrossRef]
- Barak, O.; Asarch, A.; Horn, T. PAS Is Optimal for Diagnosing Onychomycosis. J. Cutan. Pathol. 2010, 37, 1038–1040. [Google Scholar] [CrossRef]
- Wickes, B.L.; Wiederhold, N.P. Molecular Diagnostics in Medical Mycology. Nat. Commun. 2018, 9, 5135. [Google Scholar] [CrossRef]
- Wickes, B.L.; Romanelli, A.M. Diagnostic Mycology: Xtreme Challenges. J. Clin. Microbiol. 2020, 58, e01345-19. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistlerr, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple Evolutionary Origins of the Fungus Causing Panama Disease of Banana: Concordant Evidence from Nuclear and Mitochondrial Gene Genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef] [PubMed]
- Zarrin, M.; Rashidnia, Z.; Faramarzi, S.; Harooni, L. Rapid Identification of Aspergillus fumigatus Using Βeta-Tubulin and RodletA Genes. Open Access Maced. J. Med. Sci. 2017, 5, 848–851. [Google Scholar] [CrossRef]
- Knoll, M.A.; Steixner, S.; Lass-Flörl, C. How to Use Direct Microscopy for Diagnosing Fungal Infections. Clin. Microbiol. Infect. 2023, 29, 1031–1038. [Google Scholar] [CrossRef]
- Uhrlaß, S.; Verma, S.B.; Gräser, Y.; Rezaei-Matehkolaei, A.; Hatami, M.; Schaller, M.; Nenoff, P. Trichophyton indotineae—An Emerging Pathogen Causing Recalcitrant Dermatophytoses in India and Worldwide—A Multidimensional Perspective. J. Fungi 2022, 8, 757. [Google Scholar] [CrossRef]
| Strains from Indicated Species (No. of Tested) | Results by Strains, Expressed as |
|---|---|
| No. of Identified | |
| On-panel species | |
| Candida albicans (2) | 2 |
| Candida parapsilosis (3) | 3 |
| Fusarium oxysporum (2) | 2 |
| Fusarium solani (3) | 2 a |
| Microsporum audouinii (1) | 1 |
| Microsporum canis (2) | 2 |
| Microsporum gypseum (2) | 2 |
| Nannizia fulva (1) | 1 |
| Trichophyton erinacei (2) | 2 |
| Trichophyton interdigitale (6) | 6 |
| Trichophyton mentagrophytes (5) | 5 |
| Trichophyton rubrum (3) | 3 |
| Trichophyton tonsurans (1) | 1 |
| Trichophyton violaceum (1) | 1 |
| Total species (34) | 33 |
| Off-panel species | |
| Aspergillus fumigatus (1) | 0 |
| Fusarium delphinoides (1) | 0 |
| Penicillium chrysogenum (2) | 0 |
| Trichophyton indotineae (2) | Both identifed as T. mentagrophytes |
| Total species (6) | 0 |
| Fungal Species | Identified by the EUROArray Platform (No. of Positive Samples) | Identified by Culture (No. of Positive Samples) |
|---|---|---|
| Dermatophyte (universal) | 1 | Not applicable |
| Alternaria alternata | 0 a | 4 |
| Aspergillus niger | 0 a | 1 |
| Candida albicans | 1 | 2 |
| Candida parapsilosis | 11 | 5 |
| Candida guilliermondii | 1 | 1 |
| Fusarium solani | 2 | 3 |
| Nannizia gypsea | 2 | 0 |
| Rhizopus arrhizus | 0 a | 1 |
| Rhodotorula mucilaginosa | 0 a | 1 |
| Trichophyton interdigitale | 2 | 1 |
| Trichophyton indotinae | 1 b | 1 |
| Trichosporon mucoides | 0 a | 1 |
| Trichophyton rubrum | 9 | 8 |
| EUROArray Results Evaluated According to Those from the Comparator (Microscopy and/or Culture) a | |
|---|---|
| Metric | Value |
| True positives (n) | 22 |
| False negatives (n) | 6 |
| False positives (n) | 7 |
| True negatives (n) | 77 |
| Results evaluated in total (n) | 112 |
| Sensitivity (%) | 78.6 |
| Specificity (%) | 91.7 |
| Positive predictive value (%) | 75.9 |
| Negative predictive value (%) | 92.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ivagnes, V.; De Carolis, E.; Magrì, C.; Torelli, R.; Posteraro, B.; Sanguinetti, M. Advancing Dermatomycosis Diagnosis: Evaluating a Microarray-Based Platform for Rapid and Accurate Fungal Detection—A Pilot Study. J. Fungi 2025, 11, 234. https://doi.org/10.3390/jof11030234
Ivagnes V, De Carolis E, Magrì C, Torelli R, Posteraro B, Sanguinetti M. Advancing Dermatomycosis Diagnosis: Evaluating a Microarray-Based Platform for Rapid and Accurate Fungal Detection—A Pilot Study. Journal of Fungi. 2025; 11(3):234. https://doi.org/10.3390/jof11030234
Chicago/Turabian StyleIvagnes, Vittorio, Elena De Carolis, Carlotta Magrì, Riccardo Torelli, Brunella Posteraro, and Maurizio Sanguinetti. 2025. "Advancing Dermatomycosis Diagnosis: Evaluating a Microarray-Based Platform for Rapid and Accurate Fungal Detection—A Pilot Study" Journal of Fungi 11, no. 3: 234. https://doi.org/10.3390/jof11030234
APA StyleIvagnes, V., De Carolis, E., Magrì, C., Torelli, R., Posteraro, B., & Sanguinetti, M. (2025). Advancing Dermatomycosis Diagnosis: Evaluating a Microarray-Based Platform for Rapid and Accurate Fungal Detection—A Pilot Study. Journal of Fungi, 11(3), 234. https://doi.org/10.3390/jof11030234










