Diversity of Alternaria Section Nimbya in Iran, with the Description of Eight New Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Isolates
2.2. Morphological Observations
2.3. DNA Extraction and PCR Amplification
2.4. Sequence Alignment and Phylogenetic Analyses
2.5. Genealogical Concordance Phylogenetic Species Recognition Analysis
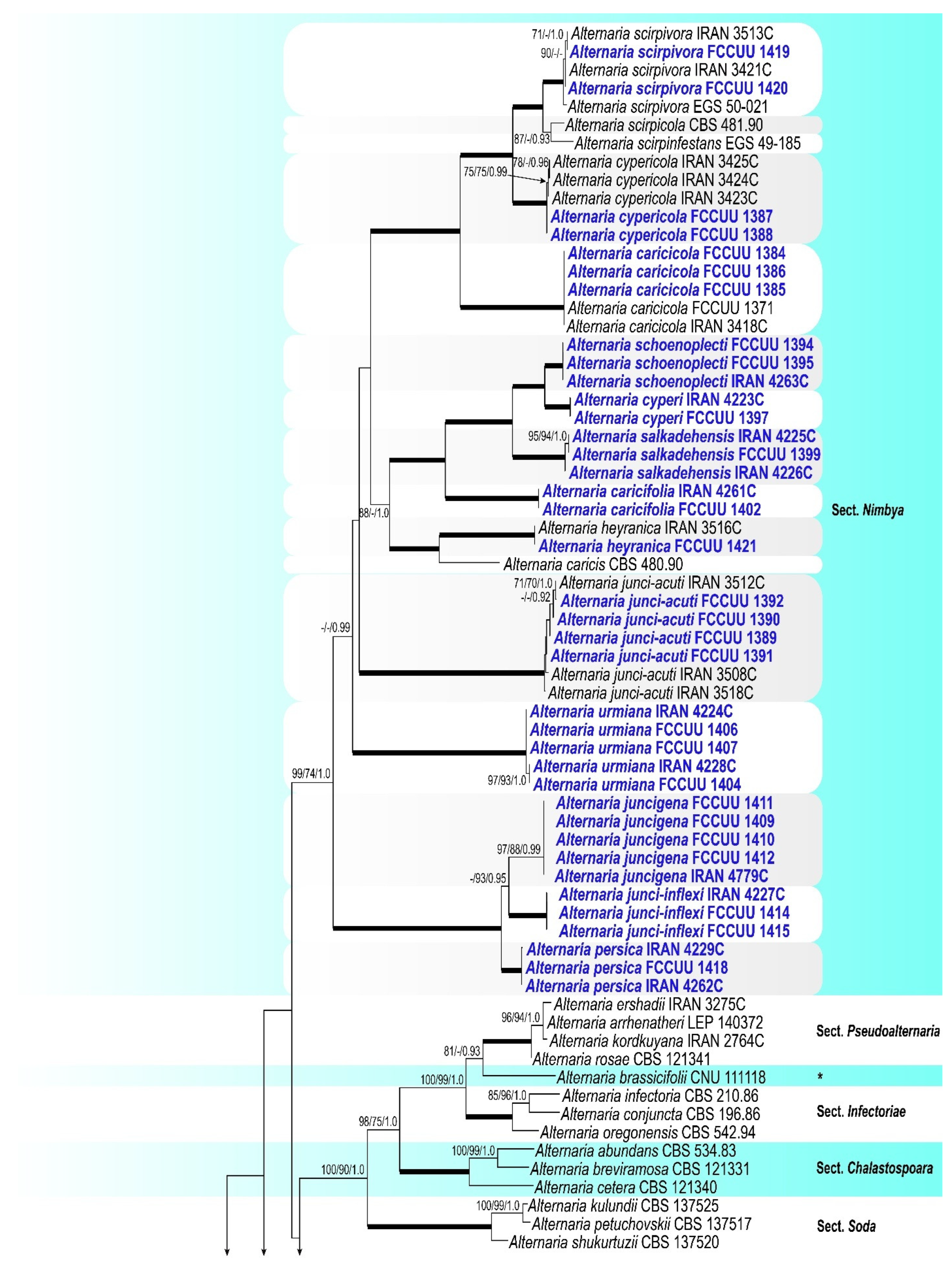
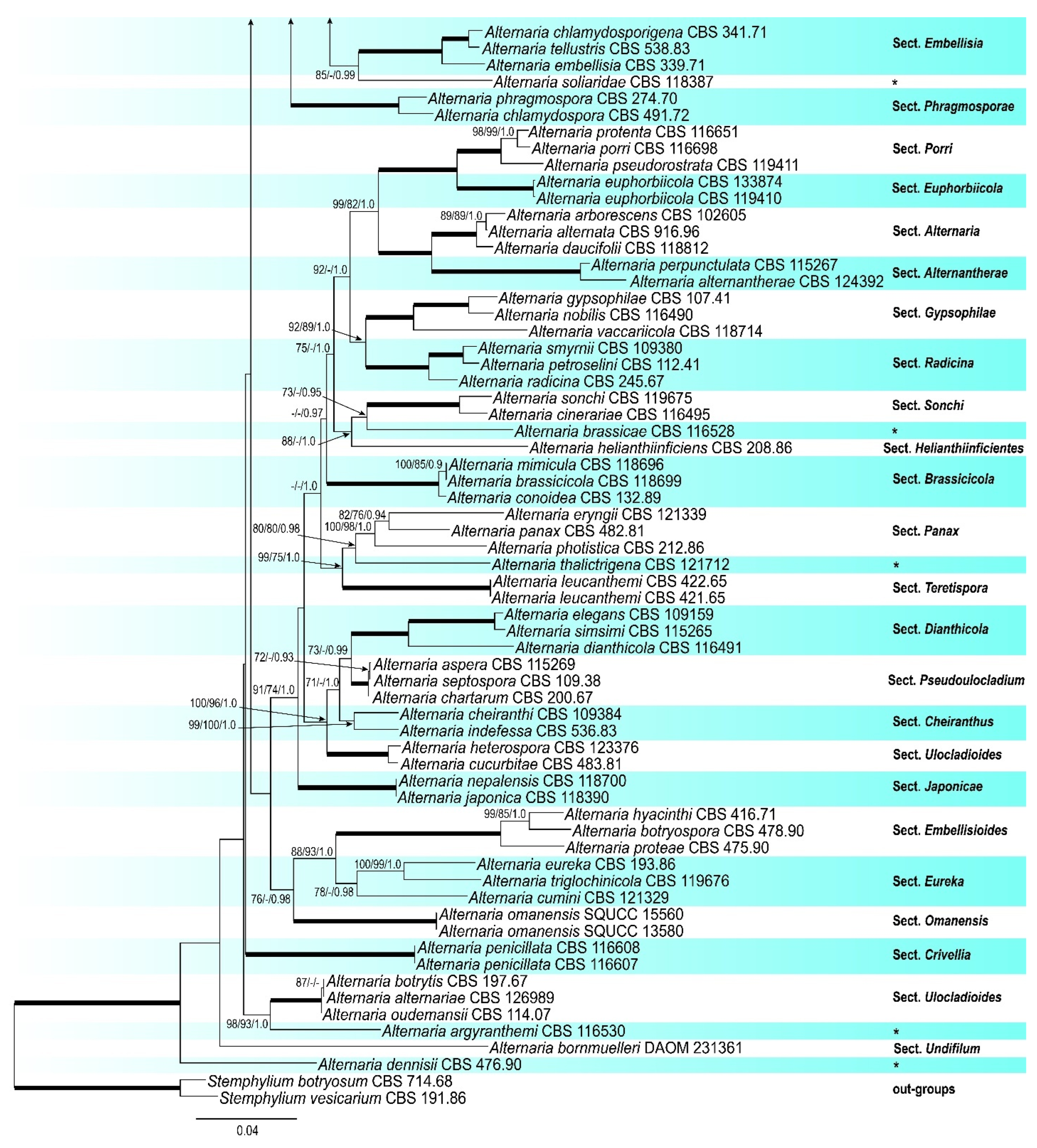
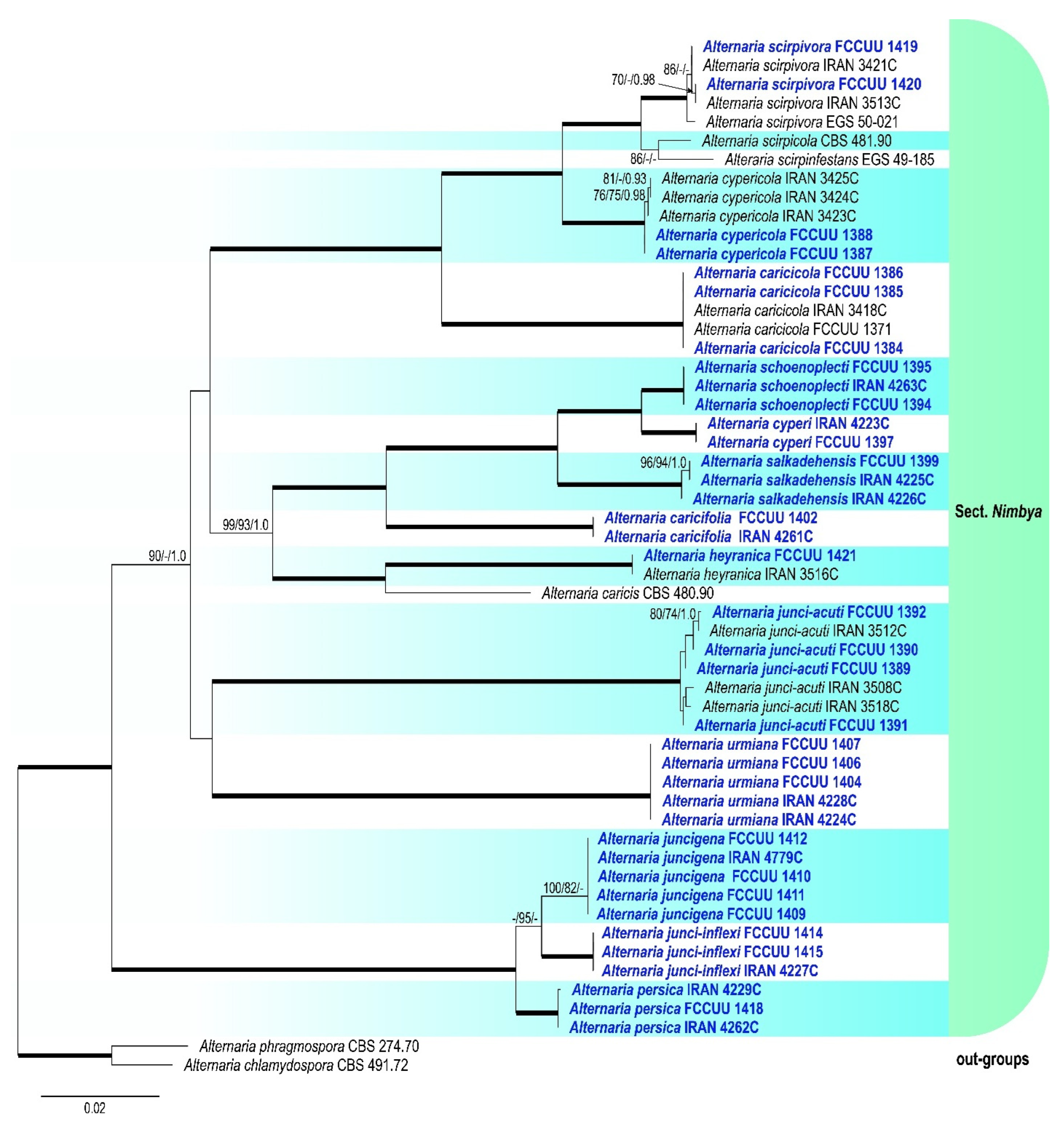
3. Results
3.1. Phylogenetic Analyses
3.2. Taxonomy
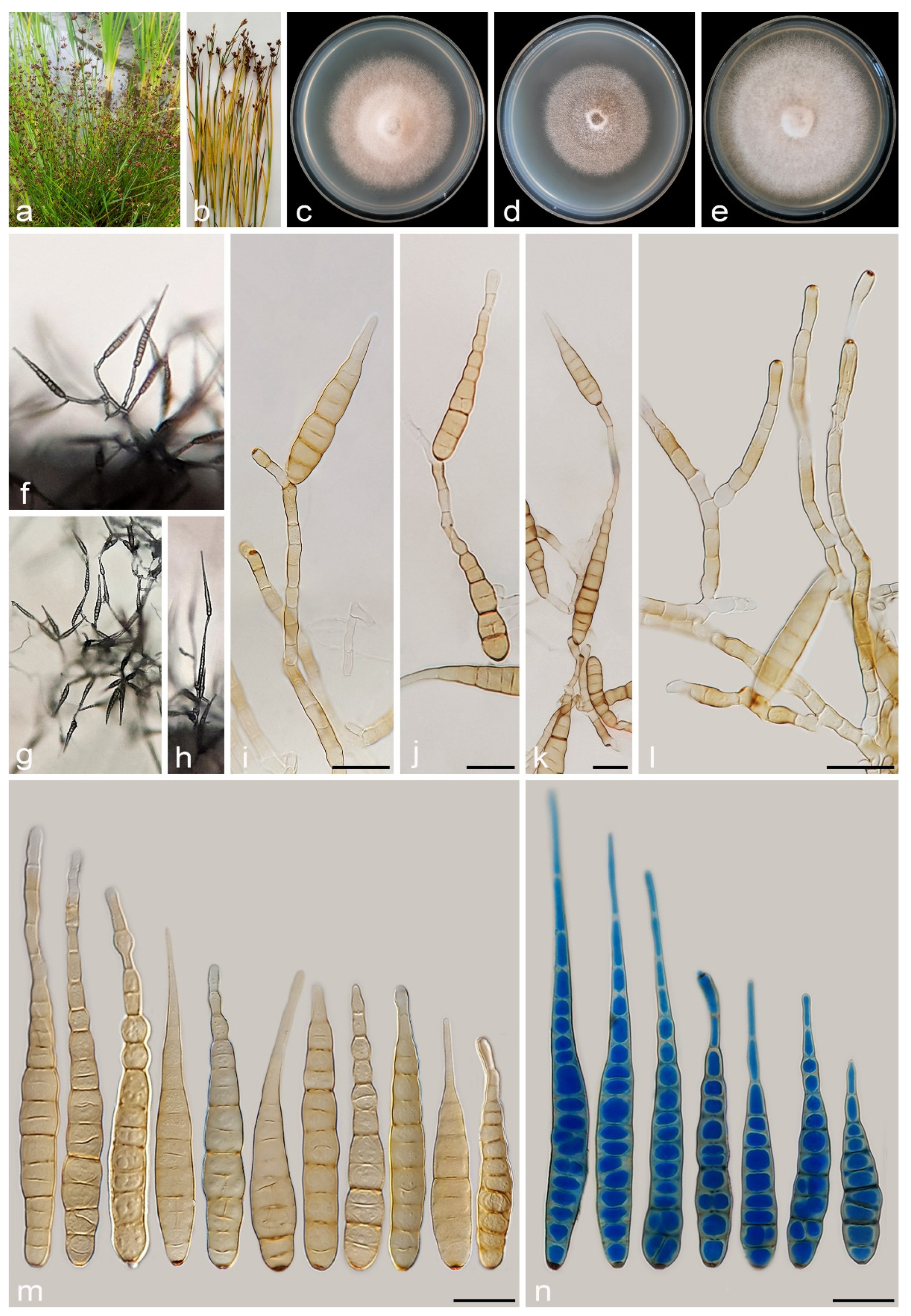
3.2.1. Alternaria caricifolia A. Ahmadpour, Y. Ghosta, Z. Alavi, F. Alavi and A. Poursafar, sp. nov. (Figure 4 and Figure 5)


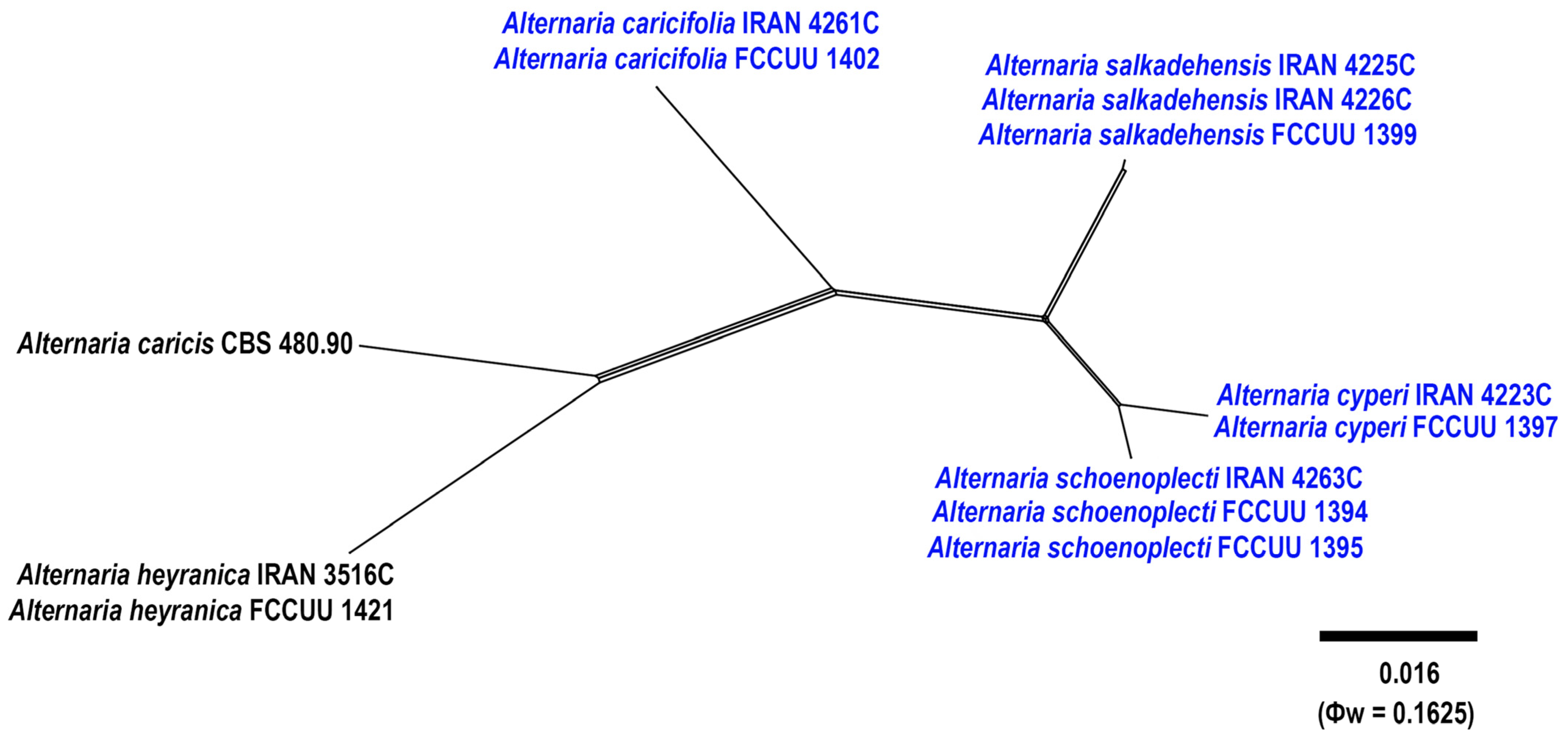
3.2.2. Alternaria cyperi A. Ahmadpour, Y. Ghosta, Z. Alavi, F. Alavi and A. Poursafar, sp. nov. (Figure 7)

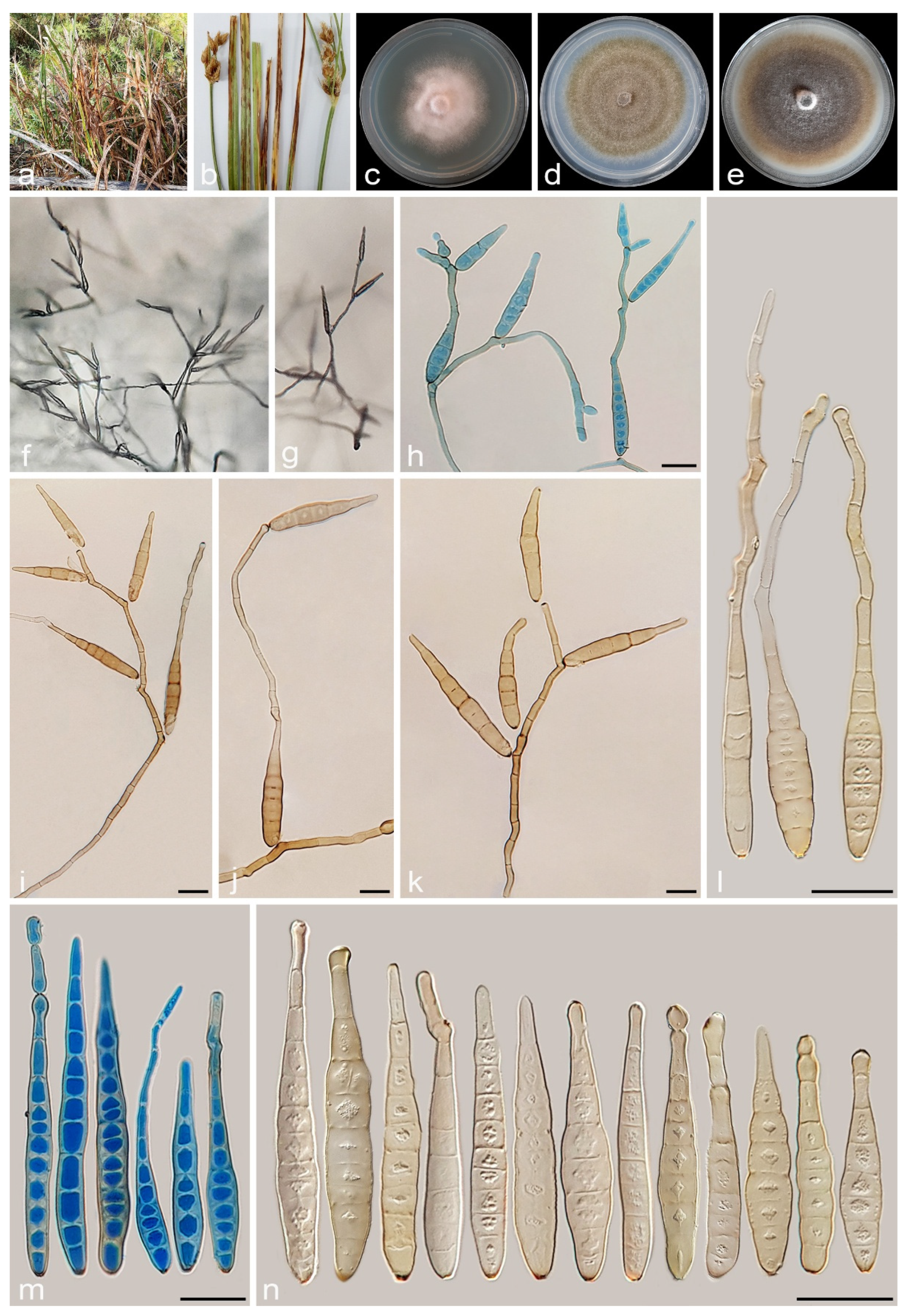
3.2.3. Alternaria juncigena A. Ahmadpour, Y. Ghosta, Z. Alavi, F. Alavi and A. Poursafar, sp. nov. (Figure 8)
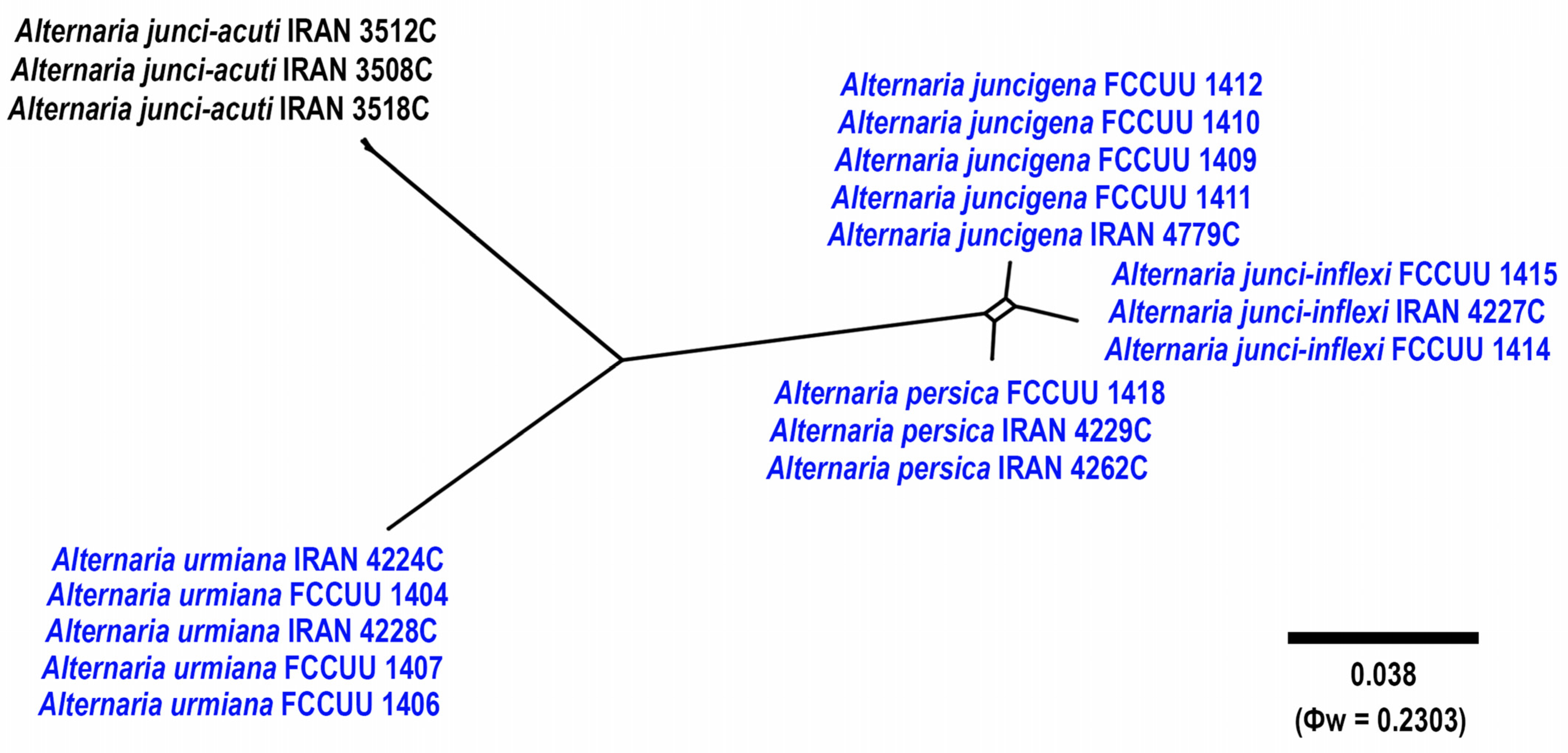
3.2.4. Alternaria junci-inflexi A. Ahmadpour, Y. Ghosta, Z. Alavi, F. Alavi and A. Poursafar, sp. nov. (Figure 10)

3.2.5. Alternaria persica A. Ahmadpour, Y. Ghosta, Z. Alavi, F. Alavi and A. Poursafar, sp. nov. (Figure 11)

3.2.6. Alternaria salkadehensis A. Ahmadpour, Y. Ghosta, Z. Alavi, F. Alavi and A. Poursafar, sp. nov. (Figure 12)
3.2.7. Alternaria schoenoplecti A. Ahmadpour, Y. Ghosta, Z. Alavi, F. Alavi and A. Poursafar, sp. nov. (Figure 13)

3.2.8. Alternaria urmiana A. Ahmadpour, Y. Ghosta, Z. Alavi, F. Alavi and A. Poursafar, sp. nov. (Figure 14)

3.2.9. Alternaria caricicola Ahmadpour, Phytotaxa 405(2): 69. 2019 [19]
3.2.10. Alternaria cypericola Ahmadpour, Poursafar and Ghosta, Mycologia 113: 1078. 2021 [20]
3.2.11. Alternaria heyranica Ahmadpour, Poursafar and Ghosta, Mycologia 113: 1078. 2021 [20]
3.2.12. Alternaria junci-acuti Ahmadpour, Poursafar and Ghosta, Mycologia 113: 1080. 2021 [20]
3.2.13. Alternaria scirpivora (E.G. Simmons and D.A. Johnson) Woudenb. and Crous, Stud. Mycol. 75: 198. 2013 [17]
- 1 Conidia with true beaks ………………………………………………………………………………………………………… 2
- 1′ Conidia without true beaks, but with apical cell extension ………………………………………………………………… 5
- 2 Beak long, more than 100 µm, chlamydospores present ……………………….……….…………………………………… 3
- 2′ Beak short, less than 100 µm, chlamydospores absent …………….………………………………………………………… 4
- 3 Chlamydospores bulbous, hyaline to light brown …………………….….….………………………….………… A. heyranica
- 3′ Chlamydospores not bulbous, brown to dark brown ……………………….………………………………….. A. junci-acuti
- 4 Conidia 87.5–205 × 10–15 μm, 7–14 distosepta ……………………….……………………………………………. A. caricicola
- 4′ Conidia 87.5–155 × 10–15 μm, 6–12 distosepta ……………….………………………….……….………………. A. cypericola
- 5 Conidia strongly curved, C shaped, on Schoenoplectus ………….……….……………….…………………… A. schoenoplecti
- 5′ Conidia straight or slightly curved, not C shape………………………………….………………………………………….. 6
- 6 Secondary conidiophores long with 2–5 geniculations ………………………………….…………………………………… 7
- 6′ Secondary conidiophores short, without geniculations ………………….…………………………………………………. 8
- 7 Primary conidiophores long, up to 300 µm, longer apical cell extension (up to 45 μm) ……………………………………………………………………….………………………………………… A. salkadehensis
- 7′ Primary conidiophores short, up to 110 µm, shorter apical cell extension (up to 15 μm) ……………………………………………………………….………………………………………………………… A. cyperi
- 8 Conidia mostly solitary, rarely in chains of 2 conidia ………………………………………………………………………… 9
- 8′ Conidia solitary or mostly in chains of 2–4(–8) conidia ………………….…….…………………………………………… 10
- 9 Conidium body short and wide (35–55 × 12–15 µm), conidial surface smooth or verrucose, without ascomata ………………………………………………………………………………………….……………………… A. urmiana
- 9′ Conidium body long and narrow (35–100 × 5–9 µm), conidial surface smooth, without longisepta and with ascomata ……………………………………………………………………………………….…………….………… A. caricifolia
- 9″ Conidium body moderate size (21–80 × 8–12 µm), conidial surface smooth or verrucose, with 1–3 longisepta, without ascomata ……………………………………………….…….……………………………………………………… A. junci-inflexi
- 10 Conidia mostly in chains of 2–8, conidium body small (20–60 × 5–8 µm) ……………………………………. A. scirpivora
- 10′ Conidia mostly in chains of 2–3, conidium body medium (55–85 × 8–11 µm), conidial surface smooth or verrucose ………………………………………………………………….……………….……….…………….……… A. persica
- 10″ Conidia mostly in chains of 2–4, conidium body large (40–110 × 10–16 µm), conidial surface smooth ……………………………………………………………………….………………………………………… A. junicigena
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Simmons, E.G. Macrospora Fuckel (Pleosporales) and related anamorphs. Sydowia 1989, 41, 314–329. [Google Scholar]
- Simmons, E.G. Alternaria an Identification Manual; CBS Fungal Biodiversity Centre: Utrecht, The Netherlands, 2007. [Google Scholar]
- Simmons, E.G. Alternaria themes and variations (112–144). Mycotaxon 1995, 55, 55–163. [Google Scholar]
- Simmons, E.G. Alternaria themes and variations (151–223). Mycotaxon 1997, 65, 1–91. [Google Scholar]
- Simmons, E.G. Alternaria themes and variations (244–286) species on Solanaceae. Mycotaxon 2000, 75, 1–115. [Google Scholar]
- Simmons, E.G. Novel dematiaceous hyphomycetes. Stud. Mycol. 2004, 50, 109–118. [Google Scholar]
- Chen, W.Q.; Lin, X.F.; Zhang, T.Y. A new species of Nimbya. Mycosystema 1997, 16, 106–108. [Google Scholar]
- Johnson, D.A.; Simmons, E.G.; Miller, J.S.; Stewart, E.L. Taxonomy and pathology of Macrospora/Nimbya on some North American bulrushes (Scirpus spp.). Mycotaxon 2002, 84, 413–428. [Google Scholar]
- Zhao, G.Z.; Zhang, T.Y. Notes on dictyosporous hyphomycetes from China VII. The genus Nimbya. Fungal Divers. 2005, 19, 201–215. [Google Scholar]
- Pryor, B.M.; Bigelow, D.M. Molecular characterization of Embellisia and Nimbya species and their relationship to Alternaria, Ulocladium and Stemphylium. Mycologia 2003, 95, 1141–1154. [Google Scholar] [CrossRef]
- Wang, Y.; Geng, Y.; Ma, J.; Wang, Q.; Zhang, X.G. Sinomyces: A new genus of anamorphic Pleosporaceae. Fungal Biol. 2011, 115, 188–195. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Park, M.S.; Pryor, B.M. Nimbya and Embellisia revisited, with nov. comb For Alternaria celosiae and A. perpunctulata. Mycol. Prog. 2012, 11, 799–815. [Google Scholar] [CrossRef]
- Simmons, E.G. Alternaria taxonomy: Current status, viewpoint, challenge. In Alternaria Biology, Plant Diseases and Metabolites; Chelkowski, J., Visconti, A., Eds.; Elsevier Science Publishers: Amsterdam, The Netherlands, 1992; pp. 1–35. [Google Scholar]
- Lawrence, D.P.; Gannibal, P.B.; Peever, T.L.; Pryor, B.M. The sections of Alternaria: Formalizing species-groups concepts. Mycologia 2013, 105, 530–546. [Google Scholar] [CrossRef] [PubMed]
- Neergaard, P. Danish Species of Alternaria and Stemphylium; Oxford University Press: London, UK, 1945. [Google Scholar]
- Joly, P. Le Genre Alternaria. Encyclopédie Mycologique XXXIII; P. Lechevalier: Paris, France, 1964. [Google Scholar]
- Woudenberg, J.H.C.; Groenewald, J.Z.; Binder, M.; Crous, P.W. Alternaria redefined. Stud. Mycol. 2013, 75, 171–212. [Google Scholar] [CrossRef] [PubMed]
- Gannibal, P.B. Distribution of Alternaria species among sections. 4. Species formerly assigned to genus Nimbya. Mycotaxon 2018, 133, 37–43. [Google Scholar] [CrossRef]
- Ahmadpour, A. Alternaria caricicola, a new species of Alternaria in the section Nimbya from Iran. Phytotaxa 2019, 405, 65–73. [Google Scholar] [CrossRef]
- Ahmadpour, A.; Ghosta, Y.; Poursafar, A. Novel species of Alternaria section Nimbya from Iran as revealed by morphological and molecular data. Mycologia 2021, 113, 1073–1088. [Google Scholar] [CrossRef]
- Bouchenak-Khelladi, Y.; Muaysa, A.M.; Linder, H.P. A revised evolutionary history of Poales: Origins and diversification. Bot. J. Linn. Soc. 2014, 175, 4–16. [Google Scholar] [CrossRef]
- Elliott, T.L.; Spalink, D.; Larridon, I.; Zuntini, A.R.; Escudero, M.; Hackel, J.; Barrett, R.L.; Martín-Bravo, S.; Márquez-Corro, J.I.; Mendoza, C.G.; et al. Global analysis of Poales diversification—Parallel evolution in space and time into open and closed habitats. New Phytol. 2023, 242, 727–743. [Google Scholar] [CrossRef]
- Larridon, I. A linear classification of Cyperaceae. Kew Bull. 2022, 77, 309–315. [Google Scholar] [CrossRef]
- Mishra, S.; Tripathi, A.; Tripathi, D.K.; Chauhan, D.K. Role of sedges (Cyperaceae) in wetlands, environmental cleaning and as food material: Possibilities and future perspectives. In Plant-Environment Interaction: Responses and Approaches to Mitigate Stress; Azooz, M.M., Ahmad, P., Eds.; John Wiley & Sons, Ltd.: Bognor Regis, UK, 2016; pp. 327–338. [Google Scholar]
- Rameshkumar, K.B.; Sruthy, B.; Viji, A.R.; Dhruvan, T. Diversity of Cyperaceae Plants in South India: Phytochemical Perspective; KSCSTE-Jawaharlal Nehru Tropical Botanic Garden and Research Institute: Palode, India, 2022. [Google Scholar]
- Bryson, C.T.; Carter, R. The significance of Cyperaceae as weeds. In Sedges: Uses, Diversity, and Systematics of the Cyperaceae; Naczi, R.C.F., Ford, B.A., Eds.; Missouri Botanical Garden Press: St. Louis, MO, USA, 2008; pp. 15–101. [Google Scholar]
- Rodrigues, M.J.; Gangadhar, K.N.; Zengin, G.; Mollica, A.; Varela, J.; Barreira, L.; Custódio, L. Juncaceae species as sources of innovative bioactive compounds for the food industry: In vitro antioxidant activity, neuroprotective properties and in silico studies. Food Chem. Toxicol. 2017, 107, 590–596. [Google Scholar] [CrossRef]
- Park, S.H.; Kang, J.W.; Park, G.H.; Jo, S.J.; Min, K.W.; Lee, K.S.; Park, H.B. Isolation and identification of antifungal metabolites from Juncus torreyi. Nat. Prod. Sci. 2024, 30, 161–166. [Google Scholar] [CrossRef]
- Nirenberg, H.I. Untersuchungen über die morphologische und biologische Differenzierung in der Fusarium-Section Liseola. Mitt. Biol. Bundesanst. Land-Und Forstwirtsch. Berl.-Dahl. 1976, 169, 1–117. [Google Scholar]
- Rayner, R.W. A Mycological Colour Chart; Commonwealth Mycological Institute: Kew, UK, 1970. [Google Scholar]
- Crous, P.W.; Gams, W.; Stalpers, J.A.; Robert, V.; Stegehuis, G. MycoBank: An online initiative to launch mycology into the 21st century. Stud. Mycol. 2004, 50, 19–22. [Google Scholar]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Shinsky, J.J., White, T.J., Eds.; Elsevier: Berlin/Heidelberg, Germany, 1990; pp. 315–322. [Google Scholar]
- Berbee, M.L.; Pirseyedi, M.; Hubbard, S. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 1999, 91, 964–977. [Google Scholar] [CrossRef]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- Sung, G.H.; Sung, J.M.; Hywel-Jones, N.L.; Spatafora, J.W. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 2007, 44, 1204–1223. [Google Scholar] [CrossRef]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef]
- Hong, S.G.; Cramer, R.A.; Lawrence, C.B.; Pryor, B.M. Alt a 1 allergen homologs from Alternaria and related taxa: Analysis of phylogenetic content and secondary structure. Fungal Genet. Biol. 2005, 42, 119–129. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Woudenberg, J.H.C.; Truter, M.; Groenewald, J.Z.; Crous, P.W. Large–spored Alternaria pathogens in section Porri disentangled. Stud. Mycol. 2014, 79, 1–47. [Google Scholar] [CrossRef]
- Woudenberg, J.H.C.; Seidl, M.F.; Groenewald, J.Z.; de Vries, M.; Stielow, J.B.; Thomma, B.P.H.J.; Crous, P.W. Alternaria section Alternaria: Species, formae speciales or pathotypes? Stud. Mycol. 2015, 82, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Al Ghafri, A.A.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Al-Saady, N.A.; Al-Sadi, A.M. A new section and a new species of Alternaria from Oman. Phytotaxa 2019, 405, 279–289. [Google Scholar] [CrossRef]
- Nishikawa, J.; Nakashima, C. Japanese species of Alternaria and their species boundaries based on host range. Fungal Syst. Evol. 2020, 5, 197–281. [Google Scholar] [CrossRef] [PubMed]
- Gannibal, P.B.; Orina, A.S.; Gasich, E.L. A new section for Alternaria helianthiinficiens found on sunflower and new asteraceous hosts in Russia. Mycol. Prog. 2022, 21, 34. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 108, 1160–1166. [Google Scholar] [CrossRef]
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.61. 2019. Available online: http://www.mesquiteproject.org (accessed on 20 February 2025).
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nylander, J.A.A. MrModeltest v2.0. Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. The CIPRES Science Gateway: Enabling High-Impact Science for Phylogenetics Researchers with Limited Resources. In Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the Extreme to the Campus and Beyond (ACM), Chicago, IL, USA, 16–20 July 2012. [Google Scholar]
- Swofford, D.L. Paup: Phylogenetic Analysis Using Parsimony (and Other Methods) 4.0. B5. Version 4.0b10; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]
- Rambaut, A. FigTree, a Graphical Viewer of Phylogenetic Trees. 2019. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 20 February 2025).
- Quaedvlieg, W.; Binder, M.; Groenewald, J.Z.; Summerell, B.A.; Carnegie, A.J.; Burgess, T.I.; Crous, P.W. Introducing the consolidated species concept to resolve species in the Teratosphaeriaceae. Persoonia 2014, 33, 1–40. [Google Scholar] [CrossRef]
- Bruen, T.C.; Philippe, H.; Bryant, D. A simple and robust statistical test for detecting the presence of recombination. Genetics 2006, 172, 2665–2681. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Alavi, Z.; Ahmadpour, A.; Ghosta, Y. Sexual morph of Alternaria scirpivora (Alternaria section Nimbya) from Iran. Mycol. Iran. 2024, 11, 73–81. [Google Scholar]
- Farr, D.F.; Rossman, A.Y.; Castlebury, L.A. United States National Fungus Collections Fungus-Host Dataset. Available online: https://fungi.ars.usda.gov/ (accessed on 20 February 2025).
- Hashemlou, E.; Ghosta, Y.; Poursafar, A.; Azizi, R. Morphological and molecular identification of Alternaria hedjaroudei sp. nov., a new species in section Panax from Iran. Phytotaxa 2020, 438, 130–140. [Google Scholar] [CrossRef]
- Li, J.; Jiang, H.; Jeewon, R.; Hongsanan, S.; Bhat, D.J.; Tang, S.M.; Lumyong, S.; Mortimer, P.E.; Xu, J.C.; Camporesi, E.; et al. Alternaria: Update on species limits, evolution, multi-locus phylogeny, and classification. Stud. Fungi 2023, 7, 23. [Google Scholar] [CrossRef]
- Lucas, M.T.; Webster, J. Conidia of Pleospora scirpicola and P. valesiaca. Trans. Br. Mycol. Soc. 1964, 47, 247–256. [Google Scholar] [CrossRef]
- Arie, T.; Christiansen, S.K.; Yoder, O.C.; Turgeon, B.G. Efficient cloning of Ascomycete mating type genes by PCR amplification of the conserved MAT HMG Box. Fungal Genet. Biol. 1997, 21, 118–130. [Google Scholar] [CrossRef]
- Arie, T.; Kaneko, I.; Yoshida, T.; Noguchi, M.; Nomura, Y.; Yamaguchi, I. Mating-type genes from asexual phytopathogenic ascomycetes Fusarium oxysporum and Alternaria alternata. Mol. Plant Microb. Interact. 2000, 13, 1330–1339. [Google Scholar] [CrossRef]
- Berbee, M.L.; Payne, B.P.; Zhang, G.; Roberts, R.G.; Turgeon, B.G. Shared ITS DNA substitutions in isolates of opposite mating type reveal a recombining history for three presumed asexual species in the filamentous ascomycete genus Alternaria. Mycol. Res. 2003, 107, 169–182. [Google Scholar] [CrossRef]
- Meng, J.W.; Zhu, W.; He, M.H.; Wu, E.J.; Duan, G.H.; Xie, Y.K.; Jin, Y.J.; Yang, L.N.; Shang, L.P.; Zhan, J. Population genetic analysis reveals cryptic sex in the phytopathogenic fungus Alternaria alternata. Sci. Rep. 2015, 5, 18250. [Google Scholar] [CrossRef]
- Stewart, J.E.; Kawabe, M.; Abdo, Z.; Arie, T.; Peever, T.L. Contrasting codon usage patterns and purifying selection at the mating locus in putatively asexual Alternaria fungal species. PLoS ONE 2011, 6, e20083. [Google Scholar] [CrossRef]
- Gannibal, P.B.; Gomzhina, M.M. Revision of Alternaria sections Pseudoulocladium and Ulocladioides: Assessment of species boundaries, determination of mating-type loci, and identification of Russian strains. Mycologia 2024, 116, 744–763. [Google Scholar] [CrossRef]
- Sun, S.; Heitman, J. From two to one: Unipolar sexual reproduction. Fungal Biol. Rev. 2015, 29, 118–125. [Google Scholar] [CrossRef]
- Aung, S.L.L.; Liu, F.Y.; Gou, Y.N.; New, Z.M.; Yu, Z.H.; Deng, J.X. Morphological and phylogenetic analyses reveal two new Alternaria species (Pleosporales, Pleosporaceae) in Alternaria section from Cucurbitaceae plants in China. MycoKeys 2024, 107, 125–139. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Li, D.W.; Cui, W.L.; Huang, L. Seven new species of Alternaria (Pleosporales, Pleosporaceae) associated with Chinese fir, based on morphological and molecular evidence. MycoKeys 2024, 101, 1–44. [Google Scholar] [CrossRef] [PubMed]
- Simpson, D.A.; Yesson, C.; Culham, A.; Couch, C.A.; Musaya, A.M. Climate change and Cyperaceae. In Climate Change, Ecology and Systematics; Hodkinson, T., Jones, M., Waldren, S., Parnell, J., Eds.; Cambridge University Press: Cambridge, UK, 2011; pp. 439–456. [Google Scholar]
- Léveillé-Bourret, É.; Eggertson, Q.; Hambleton, S.; Starr, J.R. Cryptic diversity and significant cophylogenetic signal detected by DNA barcoding the rust fungi (Pucciniaceae) of Cyperaceae-Juncaceae. J. Syst. Evol. 2021, 59, 833–851. [Google Scholar] [CrossRef]
- Mannaf, M.N.H.A.; Juraimi, A.S.; Motmainna, M.; Ya, N.N.C.; Su, A.S.M.; Roslim, M.H.M.; Ahmad, A.; Noor, N.M. Detection of sedge weeds infestation in wetland rice cultivation using hyperspectral images and artificial intelligence: A review. Pertanika J. Sci. Technol. 2024, 32, 1317–1334. [Google Scholar] [CrossRef]
- Shi, B.; Osunkoya, O.O.; Chadha, A.; Florentine, S.K.; Dhileepan, K. Biology, ecology and management of the invasive navua sedge (Cyperus aromaticus)—A global review. Plants 2021, 10, 1851. [Google Scholar] [CrossRef]
- Cai, X.; Gu, M. Bio-herbicides in organic horticulture. Horticulturae 2016, 2, 3. [Google Scholar] [CrossRef]
- Feng, L.; Hu, L.; Bo, J.; Ji, M.; Ze, S.; Ding, Y.; Yang, B.; Zhao, N. Identification and biological characteristics of Alternaria gossypina as a promising biocontrol agent for the control of Mikania micrantha. J. Fungi 2024, 10, 691. [Google Scholar] [CrossRef]
- Hoagland, R.E.; Boyette, C.D. Effects of the fungal bioherbicide, Alternaria cassia on peroxidase, pectinolytic and proteolytic activities in sicklepod seedlings. J. Fungi 2021, 7, 1032. [Google Scholar] [CrossRef]
- Pantović, J.G.; Sečanski, M.; Gordanić, S.; Todosijević, Š.L. Weed biological control with fungi-based bioherbicides. Acta Agric. Serb. 2023, 28, 23–37. [Google Scholar] [CrossRef]

| Loci | Primer Name | Primer Sequence (5′–3′) | Direction | Reference |
|---|---|---|---|---|
| ITS | ITS1 | TCCGTAGGTGAACCTGCGG | Forward | [32] |
| ITS4 | TCCTCCGCTTATTGATATGC | Reverse | ||
| GAPDH | gpd1 | CAACGGCTTCGGTCGCATTG | Forward | [33] |
| gpd2 | GCCAAGCAGTTGGTTGTG | Reverse | ||
| TEF1 | EF1-728F | CATCGAGAAGTTCGAGAAGG | Forward | [34] |
| EF1-986R | TACTTGAAGGAACCCTTACC | Reverse | ||
| RPB2 | RPB2-5F2 | GGGGWGAYCAGAAGAAGGC | Forward | [35] |
| RPB2-7cR | CCCATRGCTTGTYYRCCCAT | Reverse | [36] | |
| Alt a 1 | Alt-for | ATGCAGTTCAC CACCATCGC | Forward | [37] |
| Alt-rev | ACGAGGGTGAY GTAGGCGTC | Reverse | ||
| ISSR | ISSR5 | (GA)5YC | - | - |
| Species Name | Section | Collection No. | Country | Host/Substrate | GenBank Accession Numbers | ||||
|---|---|---|---|---|---|---|---|---|---|
| ITS | GAPDH | TEF1 | RPB2 | Alt a 1 | |||||
| Alternaria abundans | Chalastospora | CBS 534.83 | New Zealand | Fragaria sp. | JN383485 | KC584154 | KC584707 | KC584448 | JN383503 |
| A. alternantherae | Althernantherae | CBS 124392 | China | Solanum melongena | KC584179 | KC584096 | KC584633 | KC584374 | KP123846 |
| A. alternariae | Ulocladium | CBS 126989 | USA | Daucus carota | AF229485 | AY278815 | KC584730 | KC584470 | AY563316 |
| A. alternata | Alternaria | CBS 916.96 | India | Arachis hypogaea | AF347031 | AY278808 | KC584634 | KC584375 | AY563301 |
| A. arborescens | Alternaria | CBS 102605 | USA | Solanum lycopersicum | AF347033 | AY278810 | KC584636 | KC584377 | AY563303 |
| A. argyranthemi | - | CBS 116530 | New Zealand | Argyranthemum sp. | KC584181 | KC584098 | KC584637 | KC584378 | - |
| A. arrhenatheri | Pseudoalternaria | LEP 140372 | USA | Arrhenatherum elatius | JQ693677 | JQ693635 | - | - | - |
| A. aspera | Pseudoulocladium | CBS 115269 | Japan | Pistacia vera | KC584242 | KC584166 | KC584734 | KC584474 | KF533899 |
| A. bornmuelleri | Undifilum | DAOM 231361 | Austria | Securigera varia | FJ357317 | FJ357305 | KC584751 | KC584491 | JN383516 |
| A. botryospora | Embellisioides | CBS_478.90 | New Zealand | Leptinella dioica | AY278844 | AY278831 | KC584720 | KC584461 | AY563324 |
| A. botrytis | Ulocladium | CBS 197.67 | USA | Contaminant | KC584243 | KC584168 | KC584736 | KC584476 | - |
| A. brassicae | - | CBS 116528 | USA | Brassica oleracea | KC584185 | KC584102 | KC584641 | KC584382 | - |
| A. brassicicola | Brassicicola | CBS 118699 | USA | Brassica oleracea | JX499031 | KC584103 | KC584642 | KC584383 | - |
| A. brassicifolii | - | CNU 111118 | Korea | Brassica rapa L. subsp. pekinensis | JQ317188 | KM821537 | - | - | - |
| A. breviramosa | Chalastospora | CBS 121331 | Australia | Triticum sp. | FJ839608 | KC584148 | KC584700 | KC584442 | - |
| A. caricicola | Nimbya | IRAN 3418C | Iran | Carex sp. | MK508871 | MK505392 | MT187265 | MT187279 | MT187233 |
| A. caricicola | Nimbya | FCCUU 1371 | Iran | Carex sp. | MK508872 | MK505393 | MT187266 | MT187280 | MT187234 |
| A. caricicola | Nimbya | FCCUU 1384 | Iran | Cyperus sp. | OM349100 | OM640668 | OM640735 | OM640719 | OM616893 |
| A. caricicola | Nimbya | FCCUU 1385 | Iran | Cyperus sp. | OM349115 | OM640669 | OM640734 | OM640718 | OM616894 |
| A. caricicola | Nimbya | FCCUU 1386 | Iran | Cyperus sp. | OM349116 | OM640670 | OM640736 | OM640720 | OM616895 |
| A. caricifolia | Nimbya | IRAN 4261C | Iran | Cyperus sp. | OM349113 | OM640666 | OM640748 | OM640714 | OM616920 |
| A. caricifolia | Nimbya | FCCUU 1402 | Iran | Cyperus sp. | OM349114 | OM640667 | OM640749 | OM640715 | OM616921 |
| A. caricis | Nimbya | CBS 480.90 | USA | Carex hoodii | AY278839 | AY278826 | KC584726 | KC584467 | AY563321 |
| A. cetera | Chalastospora | CBS 121340 | Australia | Elymus scabrus | JN383482 | AY562398 | KC584699 | KC584441 | AY563278 |
| A. chartarum | Pseudoulocladium | CBS 200.67 | Canada | Populus sp. | AF229488 | KC584172 | KC584741 | KC584481 | AY563319 |
| A. cheiranthi | Cheiranthus | CBS 109384 | Italy | Cheiranthus cheiri | AF229457 | KC584107 | KC584646 | KC584387 | AY563290 |
| A. chlamydospora | Phragmosporae | CBS 491.72 | Egypt | Soil | KC584189 | KC584108 | KC584647 | KC584388 | - |
| A. chlamydosporigena | Embellisia | CBS 341.71 | USA | Air | KC584231 | KC584156 | KC584710 | KC584451 | - |
| A. cinerariae | Sonchi | CBS 116495 | USA | Ligularia sp. | KC584190 | KC584109 | KC584648 | KC584389 | - |
| A. conjuncta | Infectoriae | CBS 196.86 | Switzerland | Pastinaca sativa | FJ266475 | AY562401 | KC584649 | KC584390 | AY563281 |
| A. conoidea | Brassicicola | CBS 132.89 | Saudi Arabia | Ricinus communis | AF348226 | FJ348227 | KC584711 | KC584452 | FJ348228 |
| A. cucurbitae | Ulocladioides | CBS 483.81 | New Zealand | Cucumis sativus | FJ266483 | AY562418 | KC584743 | KC584483 | AY563315 |
| A. cumini | Eureka | CBS 121329 | India | Cuminum cyminum | KC584191 | KC584110 | KC584650 | KC584391 | - |
| A. cyperi | Nimbya | IRAN 4223C | Iran | Cyperus sp. | OM349106 | OM640676 | OM640745 | OM640712 | OM616918 |
| A. cyperi | Nimbya | FCCUU 1397 | Iran | Cyperus sp. | OM349107 | OM640677 | OM640746 | OM640713 | OM616919 |
| A. cypericola | Nimbya | IRAN 3423C | Iran | Cyperus sp. | MT176120 | MT187250 | MT187262 | MT187276 | MT187235 |
| A. cypericola | Nimbya | IRAN 3424C | Iran | Cyperus sp. | MT176122 | MT187251 | MT187263 | MT187277 | MT187236 |
| A. cypericola | Nimbya | FCCUU 1387 | Iran | Cyperus sp. | OM349101 | - | - | OM640716 | OM616926 |
| A. cypericola | Nimbya | FCCUU 1388 | Iran | Eleocharis sp. | OM349102 | - | - | OM640717 | OM616927 |
| A. cypericola | Nimbya | IRAN 3425C | Iran | Cyperus sp. | MT176121 | MT187252 | MT187264 | MT187278 | MT187237 |
| A. daucifolii | Alternata | CBS 118812 | USA | Daucus carota | KC584193 | KC584112 | KC584652 | KC584393 | KP123905 |
| A. dennisii | - | CBS 476.90 | Isle of Man | Senecio jacobaea | JN383488 | JN383469 | KC584713 | KC584454 | JN383505 |
| A. dianthicola | Dianthicola | CBS 116491 | New Zealand | Dianthus × allwoodii | KC584194 | KC584113 | KC584653 | KC584394 | - |
| A. elegans | Dianthicola | CBS 109159 | Burkina Faso | Lycopersicon esculentum | KC584195 | KC584114 | KC584654 | KC584395 | - |
| A. embellisia | Embellisia | CBS 339.71 | USA | Allium sativum | KC584230 | KC584155 | KC584708 | KC584449 | - |
| A. ershadii | Pseudoalternaria | IRAN 3275C | Iran | Triticum aestivum | MK829647 | MK829645 | - | - | - |
| A. eryngii | Panax | CBS 121339 | - | Eryngium sp. | JQ693661 | AY562416 | KC584656 | KC584397 | AY563313 |
| A. euphorbiicola | Euphorbiicola | CBS 133874 | USA | Euphorbia hyssopifolia | KJ718174 | KJ718019 | KJ718522 | KJ718347 | - |
| A. euphorbiicola | Euphorbiicola | CBS 119410 | USA | Euphorbia pulcherrima | KJ718173 | KJ718018 | KJ718521 | KJ718346 | - |
| A. eureka | Eureka | CBS 193.86 | Australia | Medicago rugosa | JN383490 | JN383471 | KC584715 | KC584456 | JN383507 |
| A. gypsophilae | Gypsophilae | CBS 107.41 | Netherlands | Gypsophila elegans | KC584199 | KC584118 | KC584660 | KC584401 | KJ718688 |
| A. helianthiinficiens | helianthiinficiens | CBS 208.86 | USA | Helianthus annuus | JX101649 | KC584120 | EU130548 | KC584403 | - |
| A. heterospora | Ulocladioides | CBS 123376 | China | Lycopersicon esculentum | KC584248 | KC584176 | KC584748 | KC584488 | EU855805 |
| A. heyranica | Nimbya | IRAN 3516C | Iran | Carex sp. | MT176114 | MT187244 | MT187256 | MT187270 | MT187238 |
| A. heyranica | Nimbya | FCCUU 1421 | Iran | Carex sp. | OM535905 | OM640671 | OM640758 | OM640726 | OM616930 |
| A. hyacinthi | Embellisioides | CBS 416.71 | Netherlands | Hyacinthus orientalis | KC584233 | KC584158 | KC584716 | KC584457 | - |
| A. indefessa | Cheiranthus | CBS 536.83 | USA | Soil | KC584234 | KC584159 | KC584717 | KC584458 | AY563323 |
| A. infectoria | Infectoriae | CBS 210.86 | UK | Triticum aestivum | DQ323697 | AY278793 | KC584662 | KC584404 | FJ266502 |
| A. japonica | Japonicae | CBS 118390 | USA | Brassica chinensis | KC584201 | KC584121 | KC584663 | KC584405 | - |
| A. junci-acuti | Nimbya | IRAN 3508C | Iran | Juncus acutus | MT176111 | MT187241 | MT187253 | MT187267 | MT187230 |
| A. junci-acuti | Nimbya | IRAN 3518C | Iran | Juncus acutus | MT176112 | MT187242 | MT187254 | MT187268 | MT187232 |
| A. junci-acuti | Nimbya | IRAN 3512C | Iran | Juncus acutus | MT176113 | MT187243 | MT187255 | MT187269 | MT187231 |
| A. junci-acuti | Nimbya | FCCUU 1389 | Iran | Carex sp. | OM349091 | OM640674 | - | OM640704 | OM616923 |
| A. junci-acuti | Nimbya | FCCUU 1390 | Iran | Carex sp. | OM349092 | OM640672 | - | OM640702 | OM616922 |
| A. junci-acuti | Nimbya | FCCUU 1391 | Iran | Carex sp. | OM349093 | OM640675 | - | OM640705 | OM616924 |
| A. junci-acuti | Nimbya | FCCUU 1392 | Iran | Carex sp. | OM349090 | OM640673 | - | OM640703 | OM616925 |
| A. juncigena | Nimbya | IRAN 4779C | Iran | Juncus sp. | OM349085 | OM640661 | OM640740 | OM640697 | OM616904 |
| A. juncigena | Nimbya | FCCUU 1409 | Iran | Juncus sp. | OM349086 | OM640662 | OM640741 | OM640698 | OM616905 |
| A. juncigena | Nimbya | FCCUU 1410 | Iran | Juncus sp. | OM349087 | OM640663 | OM640742 | OM640699 | OM616906 |
| A. juncigena | Nimbya | FCCUU 1411 | Iran | Juncus sp. | OM349088 | OM640664 | OM640743 | OM640700 | OM616907 |
| A. juncigena | Nimbya | FCCUU 1412 | Iran | Juncus sp. | OM349089 | OM640665 | OM640744 | OM640701 | OM616908 |
| A. junci-inflexi | Nimbya | IRAN 4227C | Iran | Juncus inflexus | OM349097 | OM640660 | OM640752 | OM640696 | OM616912 |
| A. junci-inflexi | Nimbya | FCCUU 1414 | Iran | Juncus inflexus | OM349098 | OM640659 | OM640751 | OM640695 | OM616914 |
| A. junci-inflexi | Nimbya | FCCUU 1415 | Iran | Juncus inflexus | OM349099 | OM640658 | OM640750 | OM640694 | OM616913 |
| A. kordkuyana | Pseudoalternaria | IRAN 2764C | Iran | Triticum aestivum | MF033843 | MF033826 | - | - | - |
| A. kulundii | Soda | CBS 137525 | Russia | Soil | KJ443262 | KJ649618 | - | KJ443176 | - |
| A. leucanthemi | Teretispora | CBS 421.65 | Netherlands | Chrysanthemum maximum | KC584240 | KC584164 | KC584732 | KC584472 | - |
| A. leucanthemi | Teretispora | CBS 422.65 | USA | Chrysanthemum maximum | KC584241 | KC584165 | KC584733 | KC584473 | - |
| A. mimicula | Brassicicola | CBS 118696 | USA | Lycopersicon esculentum | FJ266477 | AY562415 | KC584669 | KC584411 | AY563310 |
| A. nepalensis | Japonicae | CBS 118700 | Nepal | Brassica sp. | KC584207 | KC584126 | KC584672 | KC584414 | - |
| A. nobilis | Gypsophilae | CBS 116490 | New Zealand | Dianthus caryophyllus | KC584208 | KC584127 | KC584673 | KC584415 | JQ646385 |
| A. omanensis | Omanensis | SQUCC 15560 | Oman | dead wood | MK878563 | MK880900 | MK880897 | MK880894 | - |
| A. omanensis | Omanensis | SQUCC 13580 | Oman | dead wood | MK878562 | MK880899 | MK880896 | MK880893 | - |
| A. oregonensis | Infectoriae | CBS 542.94 | USA | Triticum aestivum | FJ266478 | FJ266491 | KC584674 | KC584416 | FJ266503 |
| A. oudemansii | Ulocladium | CBS 114.07 | - | - | FJ266488 | KC584175 | KC584746 | KC584486 | FJ266514 |
| A. panax | Panax | CBS 482.81 | USA | Aralia racemosa | KC584209 | KC584128 | KC584675 | KC584417 | JQ646382 |
| A. penicillata | Crivellia | CBS 116607 | Austria | Papaver rhoeas | KC584229 | KC584153 | KC584706 | KC584447 | - |
| A. penicillata | Crivellia | CBS 116608 | Austria | Papaver rhoeas | FJ357311 | FJ357299 | KC584698 | KC584440 | JN383502 |
| A. perpunctulata | Althernantherae | CBS 115267 | USA | Alternanthera philoxeroides | KC584210 | KC584129 | KC584676 | KC584418 | JQ905111 |
| A. persica | Nimbya | IRAN 4229C | Iran | Juncus sp. | OM349082 | OM640655 | OM640737 | OM640691 | OM616901 |
| A. persica | Nimbya | IRAN 4262C | Iran | Juncus sp. | OM349083 | OM640656 | OM640738 | OM640692 | OM616902 |
| A. persica | Nimbya | FCCUU 1418 | Iran | Juncus sp. | OM349084 | OM640657 | OM640739 | OM640693 | OM616903 |
| A. petroselini | Radicina | CBS 112.41 | - | Petroselinum sativum | KC584211 | KC584130 | KC584677 | KC584419 | AY563288 |
| A. petuchovskii | Soda | CBS 137517 | Russia | Soil | KJ443254 | KJ649616 | - | KJ443170 | - |
| A. photistica | Panax | CBS 212.86 | UK | Digitalis purpurea | KC584212 | KC584131 | KC584678 | KC584420 | AY563282 |
| A. phragmospora | Phragmosporae | CBS 274.70 | Netherlands | Soil | JN383493 | JN383474 | KC584721 | KC584462 | JN383509 |
| A. porri | Porri | CBS 116699 | USA | Allium cepa | KJ718218 | KJ718053 | KJ718564 | KJ718391 | KJ718727 |
| A. proteae | Embellisioides | CBS 475.90 | Australia | Protea sp. | AY278842 | KC584161 | KC584723 | KC584464 | - |
| A. protenta | Porri | CBS 116651 | USA | Solanum tuberosum | KC584217 | KC584139 | KC584688 | KC584430 | GQ180097 |
| A. pseudorostrata | Porri | CBS 119411 | USA | Euphorbia pulcherrima | JN383483 | AY562406 | KC584680 | KC584422 | AY563295 |
| A. radicina | Radicina | CBS 245.67 | USA | Daucus carota | KC584213 | KC584133 | KC584681 | KC584423 | FN689405 |
| A. rosae | Pseudoalternaria | CBS 121341 | New Zealand | Rosa rubiginosa | JQ693639 | JQ646279 | - | - | JQ646370 |
| A. salkadehensis | Nimbya | IRAN 4226C | Iran | Carex sp. | OM349094 | OM640690 | OM640747 | OM640706 | OM616911 |
| A. salkadehensis | Nimbya | FCCUU 1399 | Iran | Cyperus sp. | OM349095 | OM640688 | OM640732 | OM640707 | OM616909 |
| A. salkadehensis | Nimbya | IRAN 4225C | Iran | Cyperus sp. | OM349096 | OM640689 | OM640733 | OM640708 | OM616910 |
| A. schoenoplecti | Nimbya | IRAN 4263C | Iran | Schoenoplectu sp. | OM349103 | OM640678 | OM640729 | OM640709 | OM616915 |
| A. schoenoplecti | Nimbya | FCCUU 1394 | Iran | Schoenoplectu sp. | OM349104 | OM640679 | OM640730 | OM640710 | OM616916 |
| A. schoenoplecti | Nimbya | FCCUU 1395 | Iran | Schoenoplectu sp. | OM349105 | OM640680 | OM640731 | OM640711 | OM616917 |
| A. scirpicola | Nimbya | CBS 481.90 | UK | Scirpus sp. | KC584237 | KC584163 | KC584728 | KC584469 | - |
| A. scirpinfestans | Nimbya | EGS 49-185 | USA | Scirpus acutus | JN383499 | JN383480 | - | - | JN383514 |
| A. scirpivora | Nimbya | EGS 50-021 | USA | Scirpus acutus | JN383500 | JN383481 | - | - | JN383515 |
| A. scirpivora | Nimbya | FCCUU 1419 | Iran | Scirpus acutus | OM535903 | OM640681 | OM640759 | OM640727 | OM616928 |
| A. scirpivora | Nimbya | FCCUU 1420 | Iran | Scirpus acutus | OM535904 | OM640682 | OM640760 | OM640728 | OM616929 |
| A. scirpivora | Nimbya | IRAN 3421C | Iran | Scirpus acutus | MT176118 | MT187248 | MT187260 | MT187274 | MT187240 |
| A. scirpivora | Nimbya | IRAN 3419C | Iran | Scirpus acutus | MT176119 | MT187249 | MT187261 | MT187275 | - |
| A. septospora | Pseudoulocladium | CBS 109.38 | Italy | Wood | FJ266489 | FJ266500 | KC584747 | KC584487 | FJ266515 |
| A. shukurtuzii | Soda | CBS 137520 | Russia | Soil | KJ443257 | KJ649620 | - | KJ443172 | - |
| A. simsimi | Dianthicola | CBS 115265 | Argentina | Sesamum indicum | JF780937 | KC584137 | KC584686 | KC584428 | - |
| A. smyrnii | Radicina | CBS 109380 | UK | Smyrnium olusatrum | AF229456 | KC584138 | KC584687 | KC584429 | AY563289 |
| A. soliaridae | - | CBS 118387 | USA | Soil | KC584218 | KC584140 | KC584689 | KC584431 | - |
| A. sonchi | Sonchi | CBS 119675 | Canada | Sonchus asper | KC584220 | KC584142 | KC584691 | KC584433 | - |
| A. tellustris | Embellisia | CBS 538.83 | USA | Soil | FJ357316 | AY562419 | KC584724 | KC584465 | AY563325 |
| A. thalictrigena | - | CBS 121712 | Germany | Thalictrum sp. | EU040211 | KC584144 | KC584694 | KC584436 | - |
| A. triglochinicola | Eureka | CBS 119676 | Australia | Triglochin procera | KC584222 | KC584145 | KC584695 | KC584437 | - |
| A. urmiana | Nimbya | IRAN 4228C | Iran | Juncus inflexus | OM349108 | OM640683 | OM640754 | OM640721 | OM616896 |
| A. urmiana | Nimbya | FCCUU 1404 | Iran | Juncus inflexus | OM349109 | OM640684 | OM640755 | OM640722 | OM616897 |
| A. urmiana | Nimbya | IRAN 4224C | Iran | Juncus acutus | OM349110 | OM640685 | OM640753 | OM640723 | OM616898 |
| A. urmiana | Nimbya | FCCUU 1406 | Iran | Juncus acutus | OM349111 | OM640686 | OM640756 | OM640724 | OM616899 |
| A. urmiana | Nimbya | FCCUU 1407 | Iran | Juncus acutus | OM349112 | OM640687 | OM640757 | OM640725 | OM616900 |
| A. vaccariicola | Gypsophilae | CBS 118714 | USA | Vaccaria hispanica | KC584224 | KC584147 | KC584697 | KC584439 | JQ646384 |
| Stemphylium botryosum | - | CBS 714.68 | Canada | Medicago sativa | KC584238 | AF443881 | KC584729 | AF107804 | - |
| S. vesicarium | - | CBS 191.86 | India | Medicago sativa | KC584239 | AF443884 | KC584731 | KC584471 | - |
| Gene | Parameter | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Number of Taxa | Total Characters | Constant Sites | Variable Sites | Parsimony Informative Sites | Parsimony Uninformative Sites | AIC Substitution Model * | Lset Nst, Rates | −lnL | ||
| Section Nimbya | ITS | 55 | 469 | 388 | 81 | 76 | 5 | SYM + I + G | 6, invgamma | 1393.2854 |
| GAPDH | 53 | 514 | 368 | 146 | 138 | 8 | GTR + I | 6, propinv | 2019.9746 | |
| TEF1 | 46 | 177 | 113 | 64 | 60 | 4 | K80 + G | 2, gamma | 758.6035 | |
| RPB2 | 53 | 716 | 526 | 190 | 178 | 12 | GTR + G | 6, gamma | 2521.9597 | |
| Alt a 1 | 50 | 431 | 215 | 216 | 193 | 23 | GTR + I + G | 6, invgamma | 2506.1685 | |
| Combined | 55 | 2307 | 1610 | 697 | 645 | 52 | – | – | 10,258.9258 | |
| All Alternaria sections | ITS | 132 | 462 | 333 | 129 | 111 | 18 | GTR + I + G | 6, invgamma | 2978.5835 |
| GAPDH | 130 | 524 | 311 | 213 | 192 | 21 | GTR + I + G | 6, invgamma | 5840.7612 | |
| TEF1 | 107 | 187 | 59 | 127 | 109 | 18 | GTR + G | 6, gamma | 2732.1633 | |
| RPB2 | 125 | 690 | 437 | 253 | 247 | 6 | SYM + I + G | 6, invgamma | 7530.5337 | |
| Alt a 1 | 87 | 436 | 117 | 320 | 277 | 43 | HKY + I + G | 6, invgamma | 7424.5391 | |
| Combined | 132 | 2299 | 1257 | 1042 | 936 | 106 | – | – | 28,937.1276 | |
| Species | Host Family (Genus or Species) | Conidial Characteristics | Colony Characteristics (on PCA) | Sexual Morph | Distribution in Iran (province) |
|---|---|---|---|---|---|
| A. caricicola | Cyperaceae (Cyperus sp. and Carex sp.) | 87–205 × 10–15 μm; 7–14 transverse septa; smooth; Beaks 30–100 | Flat, entire, olivaceous green to grey olivaceous; 60–70 mm/7d | Not observed | East Azarbaijan; West Azarbaijan |
| A. caricifolia | Cyperaceae (Carex sp.) | 35–100 × 5–9 μm; 3–11 transverse septa; smooth, apical cell extension up to 45 μm | Flat, floccose, white to rosy buff center, hazel margins; 58 mm/7d | Present; ascomata 120–240 × 90–220 μm | West Azarbaijan |
| A. cyperi | Cyperaceae (Cyperus sp.) | 55–100 × 10–12 μm; 5–12 transverse septa; smooth; apical cell extension up to 15 μm | Fawn with salmon tints; velvety; 59 mm/7d | Not observed | East Azarbaijan |
| A. cypericola | Cyperaceae (Cyperus sp. and Eleocharis sp.) | 87–155 × 10–15 μm; 6–12 transverse septa; smooth; Beaks 37.5–137 | Flat, entire, dark green to olivaceous brown; 60–70 mm/7d | Not observed | West Azarbaijan |
| A. heyranica | Cyperaceae (Carex sp.) | 50–75 × 10–12 μm; 4–9 transverse septa; smooth, Beaks 50–175 | Flat, entire, olivaceous brown; 25–30 mm/7d | Not observed | Guilan |
| A. junci–acuti | Cyperaceae (Carex sp.); Juncaceae (Juncus acutus) | 50–87 × 7–9 μm; 5–14 transverse septa; smooth, Beaks 20–200 | Flat, entire, dark green to olivaceous brown; 70–80 mm/7d | Not observed | Golestan; West Azarbaijan |
| A. juncigena sp. nov. | Juncaceae (Juncus sp.) | 40–110 × 10–16 μm; 5–15 transverse septa; smooth; apical cell extension up to 50 μm | Flat, entire, velvety, pale vinaceous to vinaceous buff; 50 mm/7d | Not observed | West Azarbaijan |
| A. junci–inflexi | Juncaceae (Juncus inflexus) | 21–80 × 8–12 μm; 2–9 transverse septa; smooth to verrucose; apical cell extension up to 15 μm | Flat, entire, velvety, fawn; 65 mm/7d | Not observed | East Azarbaijan; West Azarbaijan |
| A. persica | Juncaceae (Juncus sp.) | 40–85 × 8–11 μm; 4–15 transverse septa; smooth to verrucose; apical cell extension up to 50 μm | Floccose, vinaceous buff; 60 mm/7d | Not observed | West Azarbaijan |
| A. salkadehensis | Cyperaceae (Cyperus sp. and Carex sp.) | 35–100 × 10–13 μm; 4–13 transverse septa, smooth; apical cell extension up to 45 μm | Flat, entire, velvety, hazel; 63 mm/7d | Not observed | West Azarbaijan |
| A. schoenoplecti | Cyperaceae (Schoenoplectus sp.) | 40–88 × 12–18 μm; 3–11 transverse septa; smooth; apical cell extension up to 22 μm | Flat, entire, velvety, sepia to cinnamon; 49 mm/7d | Not observed | West Azarbaijan |
| A. scirpivora | Cyperaceae (Scirpus acutus) | 30–60 × 5–8 μm; 3–10 transverse septa; smooth; apical cell extension up to 50 μm | Flat, entire, dark green to olivaceous brown; 60–70 mm/7d | Present; ascomata 280–500 × 250–450 μm | Multiple provinces |
| A. urmiana | Juncaceae (Juncus acutus and Juncus inflexus) | 35–55 × 12–15 μm; 3–8 transverse septa; smooth to verrucose; apical cell extension up to 10 μm | Flat, entire, velvety, rosy buff; 51 mm/7d | Not observed | West Azarbaijan |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmadpour, A.; Ghosta, Y.; Alavi, Z.; Alavi, F.; Poursafar, A.; Rampelotto, P.H. Diversity of Alternaria Section Nimbya in Iran, with the Description of Eight New Species. J. Fungi 2025, 11, 225. https://doi.org/10.3390/jof11030225
Ahmadpour A, Ghosta Y, Alavi Z, Alavi F, Poursafar A, Rampelotto PH. Diversity of Alternaria Section Nimbya in Iran, with the Description of Eight New Species. Journal of Fungi. 2025; 11(3):225. https://doi.org/10.3390/jof11030225
Chicago/Turabian StyleAhmadpour, Abdollah, Youbert Ghosta, Zahra Alavi, Fatemeh Alavi, Alireza Poursafar, and Pabulo Henrique Rampelotto. 2025. "Diversity of Alternaria Section Nimbya in Iran, with the Description of Eight New Species" Journal of Fungi 11, no. 3: 225. https://doi.org/10.3390/jof11030225
APA StyleAhmadpour, A., Ghosta, Y., Alavi, Z., Alavi, F., Poursafar, A., & Rampelotto, P. H. (2025). Diversity of Alternaria Section Nimbya in Iran, with the Description of Eight New Species. Journal of Fungi, 11(3), 225. https://doi.org/10.3390/jof11030225








