Assessment of Recombinant β-Propeller Phytase of the Bacillus Species Expressed Intracellularly in Yarrowia lipolityca
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. DNA Extraction
2.3. In Silico Bioinformatics Analysis
2.4. Amplification of Genes Encoding Phytases
2.5. Isolation of PCR Products from the Gel
2.6. Construction of an Integrative Expression Vector tphyD-BS-1 or tphyD-BS-2 Phytase Gene
2.7. Transformation of the Y. lipolytica Yeast Using a Plasmid Structure with the Phytase Gene
2.8. Genomic DNA Extraction
2.9. Selection of Transformants with PCR
2.10. RNA Isolation and Reverse Transcription Reaction
2.11. Quantitative Real-Time Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR)
2.12. Enzyme Activity Quantification
2.13. Phytase Expression (Cultivation)
2.14. Preparation of Cellular Homogenate
2.15. Assay of Phytase Activity in the Y. lipolytica Transformants
2.16. Protein aggregation
2.17. Phytase Refolding
2.18. Electrophoresis
2.19. Assay of the Protein Amount
2.20. Structural Characteristics of PhyC and PhyD Phytases
3. Results
3.1. In Silico Search for PhyD Phytase Genes (Comparative Genome Analysis)
- Clade I: B. subtilis, B. spizizenii.
- Clade II: B. amyloliquefaciens, B. siamensis.
- Clade III: B. licheniformis.
3.2. Identification of PhyD Class Phytase-Encoding Genes
3.3. Generation of Integrative Genetic Constructs Containing the Phytase Gene Using the VDAC Promoter
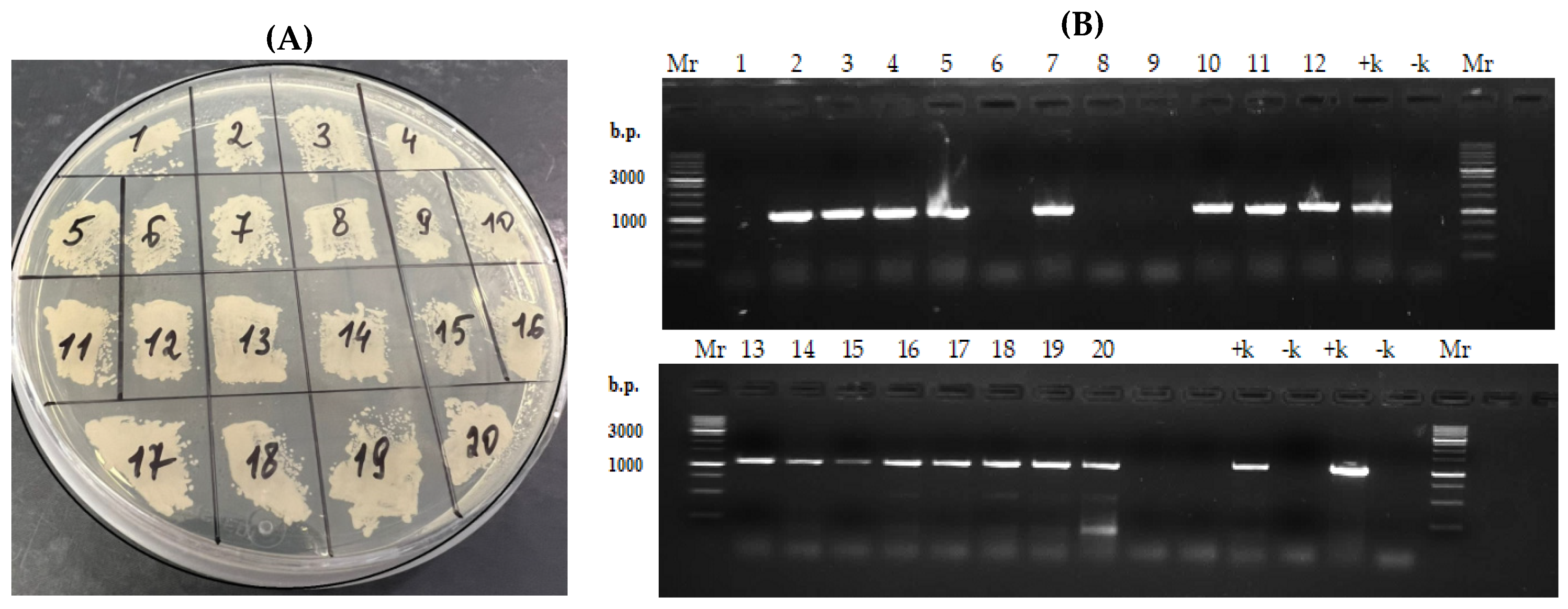
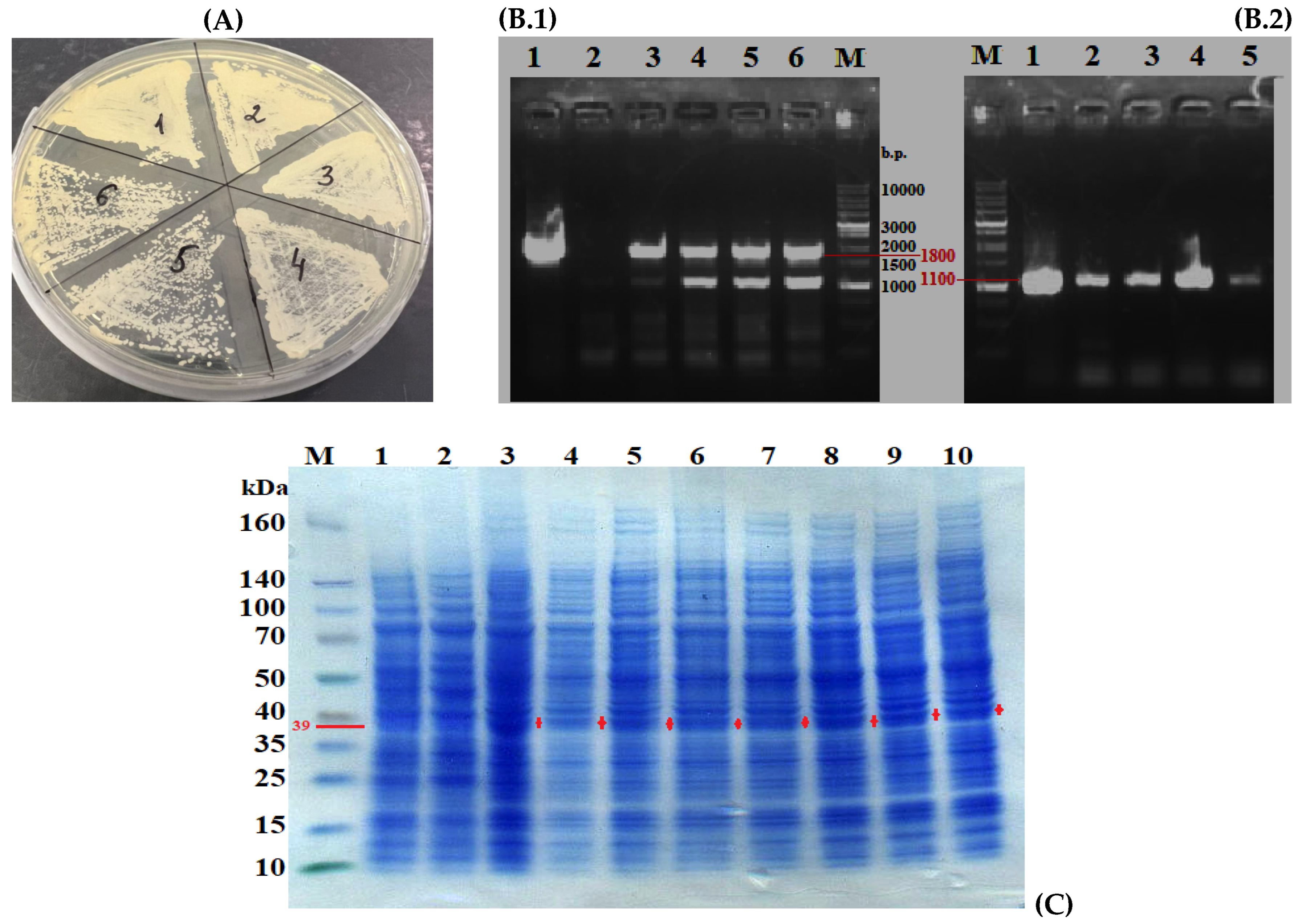
3.4. Selection of Y. lipolytica PO1f Transformants Carrying the Integrated Construct Expressing Phytase
3.5. Refolding of Phytase from Inclusion Bodies of Y. lipolytica Transformants
4. Discussion
4.1. Advantages of PhyD-Class Phytases and the Use of Encapsulated Enzyme Forms
4.2. Effect of Structural Features of PhyC and PhyD Phytases on Their Folding Process in Y. lipolytica
4.3. Protein Aggregation and Refolding of PhyD Phytase for Biotechnological Applications
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jeni, R.E.; Dittoe, D.K.; Olson, E.G.; Lourenco, J.; Corcionivoschi, N.; Ricke, S.C.; Callaway, T.R. Probiotics and Potential Applications for Alternative Poultry Production Systems. Poult. Sci. 2021, 100, 101156. [Google Scholar] [CrossRef] [PubMed]
- J Tejeda, O.; Kim, K.W. Role of Dietary Fiber in Poultry Nutrition. Animals 2021, 11, 461. [Google Scholar] [CrossRef] [PubMed]
- Selle, P.H.; Macelline, S.P.; Chrystal, P.V.; Liu, S.Y. The Contribution of Phytate-Degrading Enzymes to Chicken-Meat Production. Animals 2023, 13, 603. [Google Scholar] [CrossRef]
- Filippovich, S.Y.; Isakova, E.P.; Gessler, N.N.; Deryabina, Y.I. Advances in Immobilization of Phytases and Their Application. Bioresour. Technol. 2023, 379, 129030. [Google Scholar] [CrossRef]
- Farhat-Khemakhem, A.; Ben Farhat, M.; Boukhris, I.; Bejar, W.; Bouchaala, K.; Kammoun, R.; Maguin, E.; Bejar, S.; Chouayekh, H. Heterologous Expression and Optimization Using Experimental Designs Allowed Highly Efficient Production of the PHY US417 Phytase in Bacillus Subtilis 168. AMB Express 2012, 2, 10. [Google Scholar] [CrossRef] [PubMed]
- Urgessa, O.E.; Koyamo, R.; Dinka, H.; Tefese, K.; Gemeda, M.T. Review on Desirable Microbial Phytases as a Poultry Feed Additive: Their Sources, Production, Enzymatic Evaluation, Market Size, and Regulation. Int. J. Microbiol. 2024, 2024, 9400374. [Google Scholar] [CrossRef]
- Kumar, V.; Yadav, A.N.; Verma, P.; Sangwan, P.; Saxena, A.; Kumar, K.; Singh, B. β-Propeller Phytases: Diversity, Catalytic Attributes, Current Developments and Potential Biotechnological Applications. Int. J. Biol. Macromol. 2017, 98, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.G.; Porres, J.M.; Mullaney, E.J.; Brinch-Pedersen, H. Phytase: Source, Structure and Application. In Industrial Enzymes: Structure, Function and Applications, 1st ed.; Polaina, J., MacCabe, A.P., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 505–529. ISBN 978-1-4020-5377-1. [Google Scholar]
- Greiner, R.; Lim, B.L.; Cheng, C.; Carlsson, N.-G. Pathway of Phytate Dephosphorylation by β-Propeller Phytases of Different Origins. Can. J. Microbiol. 2007, 53, 488–495. [Google Scholar] [CrossRef] [PubMed]
- Moita, V.H.C.; Kim, S.W. Nutritional and Functional Roles of Phytase and Xylanase Enhancing the Intestinal Health and Growth of Nursery Pigs and Broiler Chickens. Animals 2022, 12, 3322. [Google Scholar] [CrossRef] [PubMed]
- Gessler, N.N.; Serdyuk, E.G.; Isakova, E.P.; Deryabina, Y.I. Phytases and the Prospects for Their Application (Review). Appl. Biochem. Microbiol. 2018, 54, 352–360. [Google Scholar] [CrossRef]
- Lee, S.H.; Cho, J.; Bok, J.; Kang, S.; Choi, Y.; Lee, P.C.W. Characterization, Gene Cloning, and Sequencing of a Fungal Phytase, PhyA, from Penicillium Oxalicum PJ3. Prep. Biochem. Biotechnol. 2015, 45, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Han, N.; Miao, H.; Yu, T.; Xu, B.; Yang, Y.; Wu, Q.; Zhang, R.; Huang, Z. Enhancing Thermal Tolerance of Aspergillus niger PhyA Phytase Directed by Structural Comparison and Computational Simulation. BMC Biotechnol. 2018, 18, 36. [Google Scholar] [CrossRef]
- Corrêa, T.L.R.; de Araújo, E.F. Fungal Phytases: From Genes to Applications. Braz. J. Microbiol. 2020, 51, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.O.; Lee, J.K.; Kim, H.K.; Yu, J.H.; Oh, T.K. Cloning of the Thermostable Phytase Gene (Phy) from Bacillus sp. DS11 and Its Overexpression in Escherichia coli. FEMS Microbiol. Lett. 1998, 162, 185–191. [Google Scholar] [CrossRef]
- Kim, Y.O.; Lee, J.K.; Oh, B.C.; Oh, T.K. High-Level Expression of a Recombinant Thermostable Phytase in Bacillus Subtilis. Biosci. Biotechnol. Biochem. 1999, 63, 2205–2207. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zou, L.-K.; Wang, H.-N.; Pan, X.; Xie, T.; Wu, Q.; Xie, Z.-W.; Zhou, W.-R. Design and Expression of a Synthetic phyC Gene Encoding the Neutral Phytase in Pichia Pastoris. Acta Biochim. Biophys. Sin. 2006, 38, 803–811. [Google Scholar] [CrossRef][Green Version]
- Saadi, M.I.; Doosti, A.; Jalali, H.; Abdolyousefi, E.N.; Hooshiyar, M.; Tabrizi, R.; Noshadi, E. Cloning of Bacillus Subtilis Phytase Gene Construct in Escherichia coli. Iran. J. Microbiol. 2021, 13, 664–670. [Google Scholar] [CrossRef]
- Christensen, T.; Dersjant-Li, Y.; Sewalt, V.; Mejldal, R.; Haaning, S.; Pricelius, S.; Nikolaev, I.; Sorg, R.A.; de Kreij, A. In Vitro Characterization of a Novel Consensus Bacterial 6-Phytase and One of Its Variants. Curr. Biochem. Eng. 2020, 6, 156–171. [Google Scholar] [CrossRef]
- Gulati, H.K.; Chadha, B.S.; Saini, H.S. Production and Characterization of Thermostable Alkaline Phytase from Bacillus Laevolacticus Isolated from Rhizosphere Soil. J. Ind. Microbiol. Biotechnol. 2007, 34, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Fang, C.; Jiang, Y.; Yan, R. Characterization of a Hemolysin Gene ytjA from Bacillus Subtilis. Curr. Microbiol. 2009, 58, 642–647. [Google Scholar] [CrossRef]
- Heinzmann, S.; Entian, K.-D.; Stein, T. Engineering Bacillus Subtilis ATCC 6633 for Improved Production of the Lantibiotic Subtilin. Appl. Microbiol. Biotechnol. 2006, 69, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Swain, M.R.; Ray, R.C. Alpha-Amylase Production by Bacillus Subtilis CM3 in Solid State Fermentation Using Cassava Fibrous Residue. J. Basic Microbiol. 2007, 47, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.T.; Fan, M.H.; Kuo, F.C.; Sung, H.Y. Potent Fibrinolytic Enzyme from a Mutant of Bacillus Subtilis IMR-NK1. J. Agric. Food Chem. 2000, 48, 3210–3216. [Google Scholar] [CrossRef]
- Tran, T.T.; Mamo, G.; Mattiasson, B.; Hatti-Kaul, R. A Thermostable Phytase from Bacillus Sp. MD2: Cloning, Expression, and High-Level Production in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2010, 37, 279–287. [Google Scholar] [CrossRef]
- Osman, A.A.; Babu, P.R.; Venu, K.; Rao, K.V.; Reddy, V.D. Prediction of substrate-binding site and elucidation of catalytic residue of a phytase from Bacillus sp. Enzyme Microb. Technol. 2012, 51, 35–39. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Varshavsky, A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011, 20, 1298–1345. [Google Scholar] [CrossRef] [PubMed]
- Epova, E.Y.; Balovneva, M.V.; Isakova, E.P.; Kudykina, Y.K.; Zylkova, M.V.; Deryabina, Y.I.; Shevelev, A.B. Expression System for Yarrowia Lipolytica Based on a Promoter of the Mitochondrial Potential-Dependent Porin VDAC Gene. Biotechnol. Bioproc. Eng. 2016, 21, 408–413. [Google Scholar] [CrossRef]
- Davidow, L.S.; Apostolakos, D.; O’Donnell, M.M.; Proctor, A.R.; Ogrydziak, D.M.; Wing, R.A.; Stasko, I.; DeZeeuw, J.R. Integrative Transformation of the Yeast Yarrowia Lipolytica. Curr. Genet. 1985, 10, 39–48. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; Volume 1. [Google Scholar]
- Borkowska, M.; Białas, W.; Celińska, E. A new set of reference genes for comparative gene expression analyses in Yarrowia lipolytica. FEMS Yeast Res. 2020, 20, foaa059. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(Delta Delta C (T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Divisi, D.; Di, L.G.; Zaccagna, G.; Crisci, R. Basic statistics with Microsoft Excel: A review. J. Thorac. Dis. 2017, 9, 1734–1740. [Google Scholar] [CrossRef] [PubMed]
- Serdyuk, E.; Issakova, E.; Gessler, N.; Antipov, A.; Deryabina, Y. Activity Phytases in Recombinant Strains Yarrowia lipolytica under Different Conditions of Cultivation. Bulletin Sci. Pract. 2018, 4, 18–30. [Google Scholar] [CrossRef]
- Seras-Franzoso, J.; Cano-Garrido, O.; Peternel, S.; Arís, A.; Garcia-Fruitós, E. Purification of Inclusion Bodies Produced in Bacteria and Yeast. Methods Mol. Biol. 2022, 2406, 401–416. [Google Scholar] [CrossRef]
- Deryabina, Y.; Isakova, E.; Sekova, V.; Antipov, A.; Saris, N.-E.L. Inhibition of Free Radical Scavenging Enzymes Affects Mitochondrial Membrane Permeability Transition during Growth and Aging of Yeast Cells. J. Bioenerg. Biomembr. 2014, 46, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Reddy, C.S.; Achary, V.M.M.; Manna, M.; Singh, J.; Kaul, T.; Reddy, M.K. Isolation and Molecular Characterization of Thermostable Phytase from Bacillus Subtilis (BSPhyARRMK33). Appl. Biochem. Biotechnol. 2015, 175, 3058–3067. [Google Scholar] [CrossRef] [PubMed]
- Rao, D.E.C.S.; Rao, K.V.; Reddy, V.D. Cloning and Expression of Bacillus Phytase Gene (Phy) in Escherichia coli and Recovery of Active Enzyme from the Inclusion Bodies. J. Appl. Microbiol. 2008, 105, 1128–1137. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Davis, B.J. Disc Electrophoresis-II Method and Application to Human Serum Proteins. Ann. N. Y. Acad. Sci. 1964, 121, 404–427. [Google Scholar] [CrossRef]
- Kumar, V.; Sinha, A.K. Chapter 3-General aspects of phytases. In Enzymes in Human and Animal Nutrition, 1st ed.; Kumar, V., Nunes, C.S., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 53–72. ISBN 978-0-12-805419-2. [Google Scholar]
- Khurana, H.; Sharma, M.; Verma, H.; Lopes, B.S.; Lal, R.; Negi, R.K. Genomic insights into the phylogeny of Bacillus strains and elucidation of their secondary metabolic potential. Genomics 2020, 112, 3191–3200. [Google Scholar] [CrossRef]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Blom, J.; Klenk, H.P.; Borriss, R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an “Operational Group B. amyloliquefaciens” within the B. subtilis Species Complex. Front. Microbiol. 2017, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, R.; Cheng, Y.; Middaugh, C.R.; Volkin, D.B. High-throughput biophysical analysis of protein therapeutics to examine interrelationships between aggregate formation and conformational stability. AAPS J. 2014, 16, 48–64. [Google Scholar] [CrossRef]
- Ha, N.C.; Oh, B.C.; Shin, S.; Kim, H.J.; Oh, T.K.; Kim, Y.O.; Choi, K.Y.; Oh, B.H. Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states. Nat. Struct. Biol. 2000, 7, 147–153. [Google Scholar] [CrossRef]
- Kerovuo, J.; Lauraeus, M.; Nurminen, P.; Kalkkinen, N.; Apajalahti, J. Isolation, characterization, molecular gene cloning, and sequencing of a novel phytase from Bacillus subtilis. Appl. Environ. Microbiol. 1998, 64, 2079–2085. [Google Scholar] [CrossRef]
- Kim, Y.O.; Kim, H.K.; Bae, K.S.; Yu, J.H.; Oh, T.K. Purification and properties of a thermostable phytase from Bacillus sp. DS11. Enzyme Microb. Technol. 1998, 22, 2–7. [Google Scholar] [CrossRef]
- Oh, B.C.; Choi, W.C.; Park, S.; Kim, Y.O.; Oh, T.K. Biochemical properties and substrate specificities of alkaline and histidine acid phytases. Appl. Microbiol. Biotechnol. 2004, 63, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Olazarán, M.; Rodríguez-Blanco, L.; Carreon-Treviño, J.G.; Gallegos-López, J.A.; Viader-Salvadó, J.M. Expression of a Bacillus phytase C gene in Pichia pastoris and properties of the recombinant enzyme. Appl. Environ. Microbiol. 2010, 76, 5601–5608. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.G.; Porres, J.M. Phytase enzymology, applications, and biotechnology. Biotechnol. Lett. 2003, 25, 1787–1794. [Google Scholar] [CrossRef] [PubMed]
- Golovan, S.; Wang, G.; Zhang, J.; Forsberg, C.W. Characterization and overproduction of the Escherichia coli appA encoded bifunctional enzyme that exhibits both phytase and acid phosphatase activities. Can. J. Microbiol. 2000, 46, 59–71. [Google Scholar] [CrossRef]
- Singh, S.; Singh, S.; Sharma, P.K.; Sharma, D. Isolation, identification and molecular characterization of phytase producing bacteria, Pseudomonas sp. aazad. J. Pure Appl. Microbiol. 2017, 11, 1845–1850. [Google Scholar] [CrossRef]
- Suleimanova, A.D.; Bulmakova, D.S.; Itkina, D.L.; Sharipova, M.R. Expression of Pantoea sp. 3.5.1 AgpP Phytase in Three Expression Systems. BioNanoScience 2021, 11, 648–652. [Google Scholar]
- Kostrewa, D.; Wyss, M.; D’Arcy, A.; van Loon, A.P. Crystal structure of Aspergillus niger pH 2.5 acid phosphatase at 2.4 A resolution. J. Mol. Biol. 1999, 288, 965–974. [Google Scholar] [CrossRef]
- Ullah, A.H.; Mullaney, E.J. Disulfide bonds are necessary for structure and activity in Aspergillus ficuum phytase. Biochem. Biophys. Res. Commun. 1996, 227, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Meng, F.G.; Zhou, H.M. The role of disulfide bonds in the conformational stability and catalytic activity of phytase. Biochem. Cell Biol. 2004, 82, 329–334. [Google Scholar] [CrossRef]
- Cheng, C.; Wong, K.B.; Lim, B.L. The effect of disulfide bond on the conformational stability and catalytic activity of beta-propeller phytase. Protein Pept. Lett. 2007, 14, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Kumar, T.K.; Samuel, D.; Jayaraman, G.; Srimathi, T.; Yu, C. The role of proline in the prevention of aggregation during protein folding in vitro. Biotechnol. Mol. Biol. Int. 1998, 46, 509–517. [Google Scholar] [CrossRef]

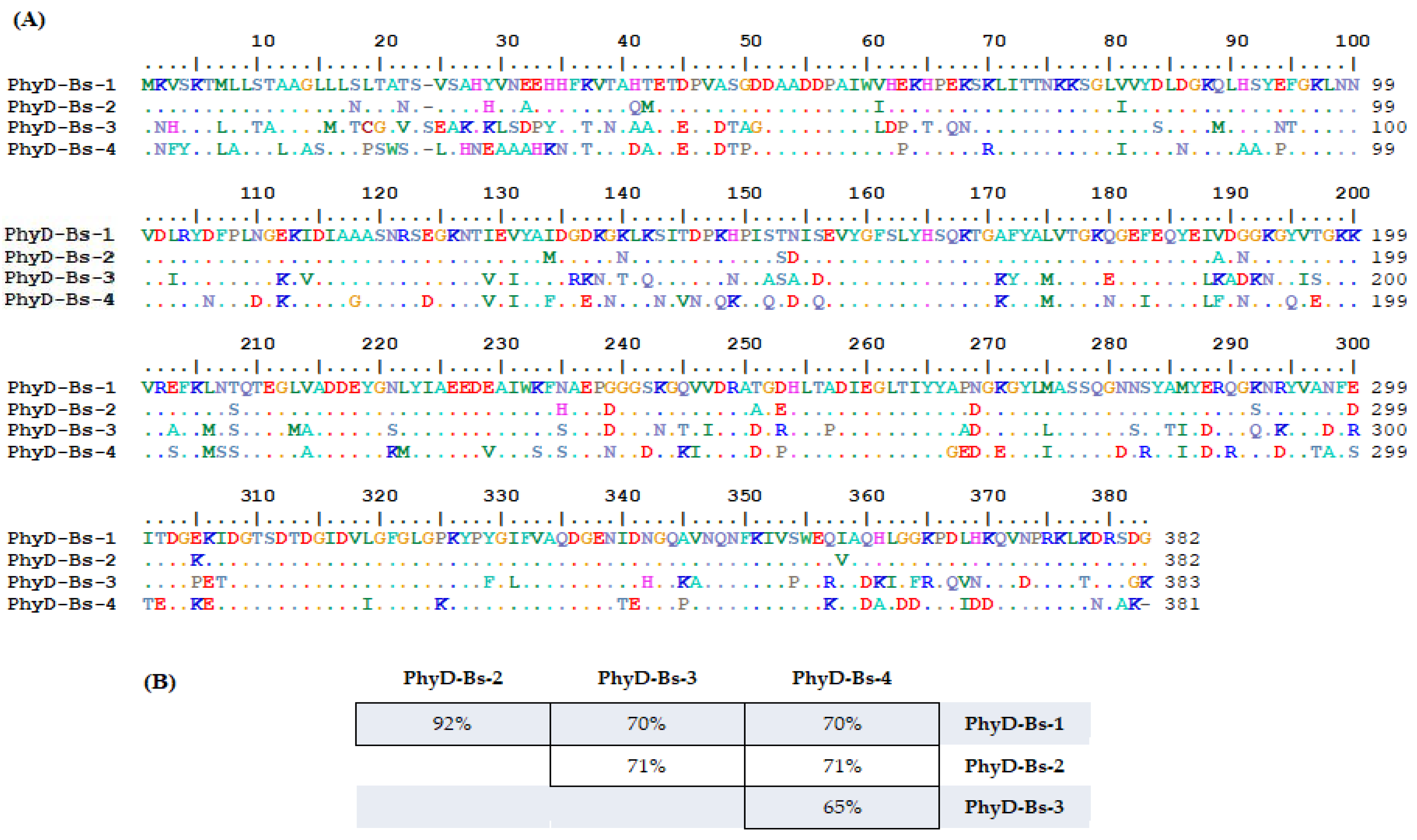
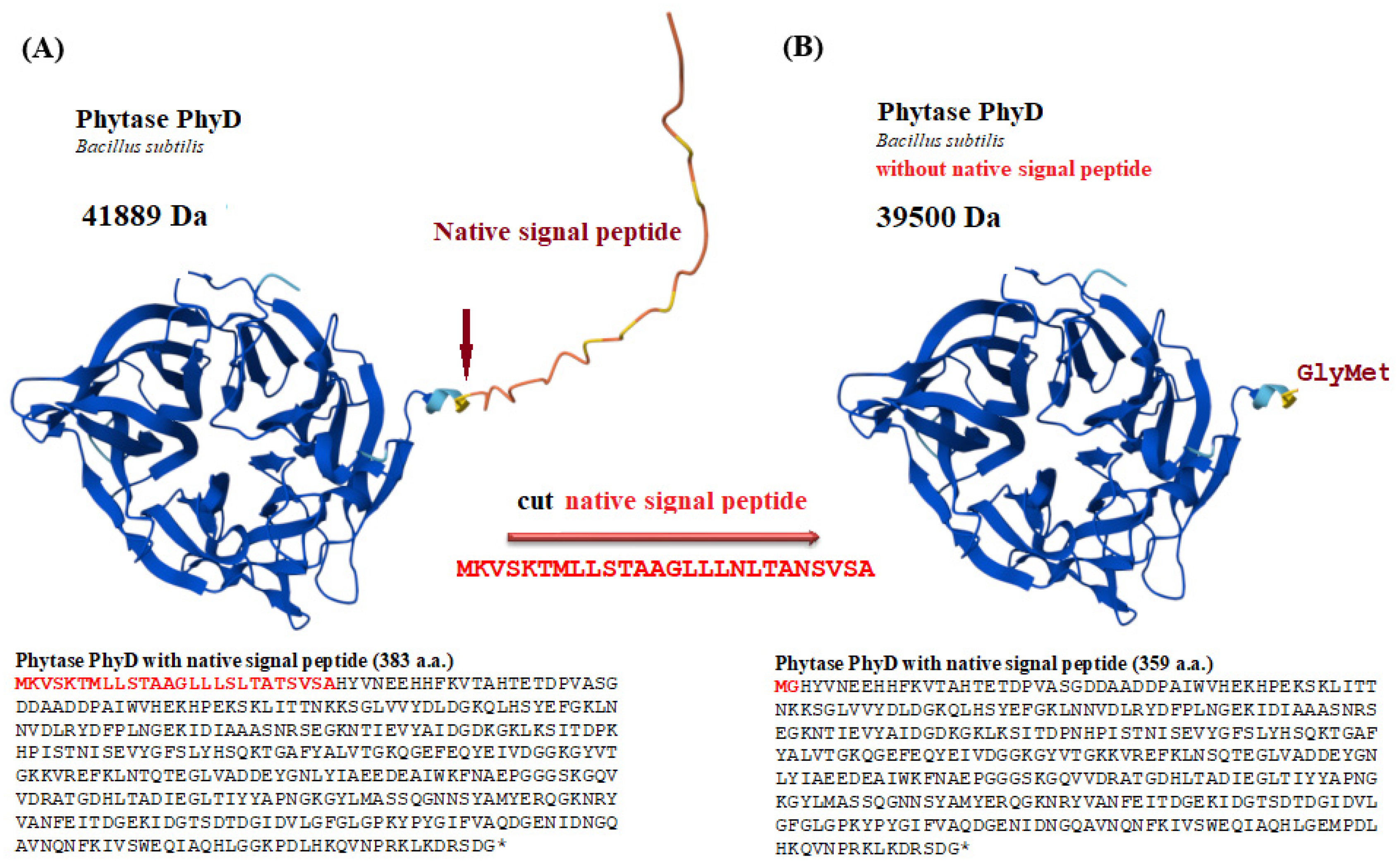
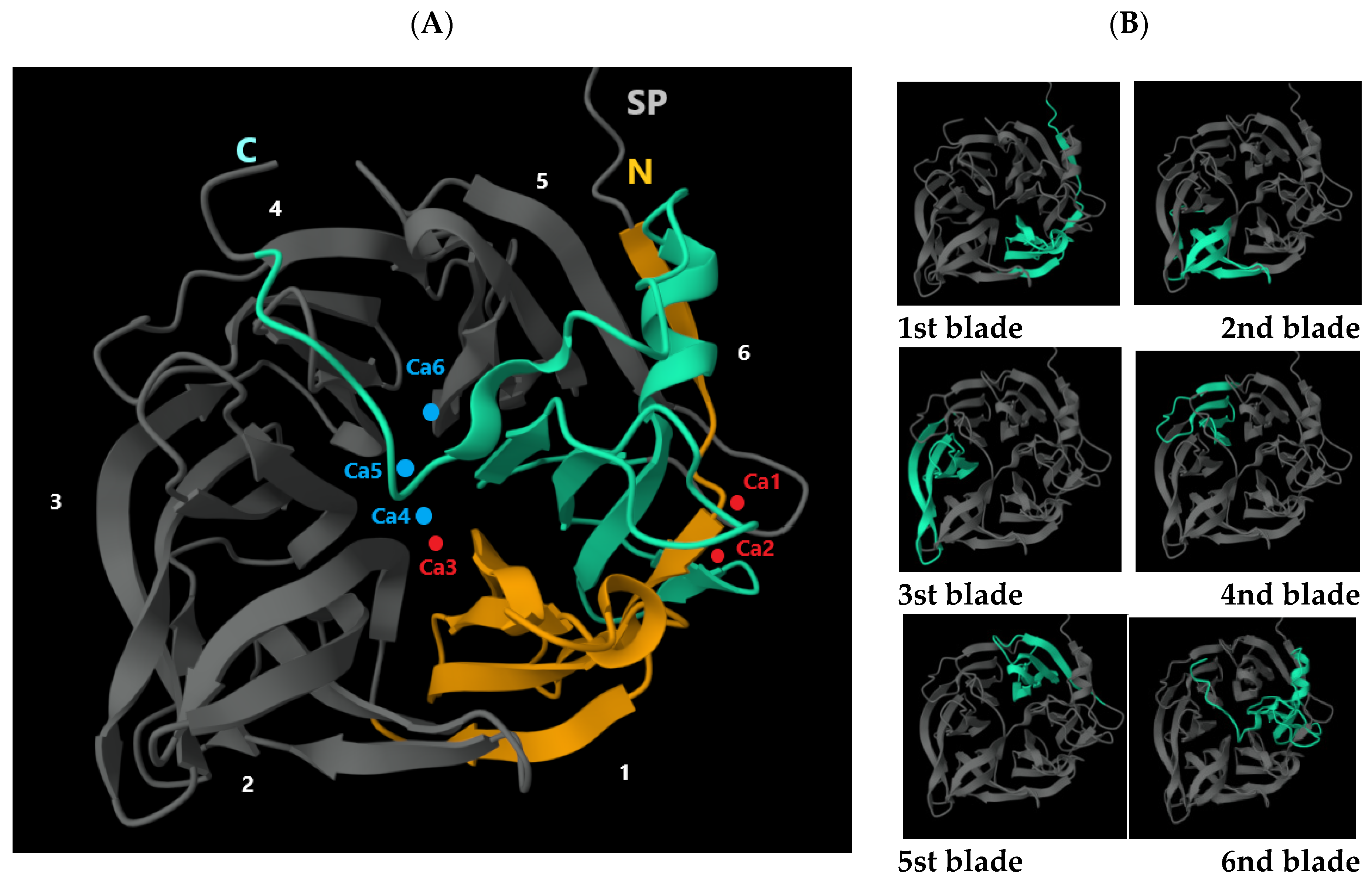
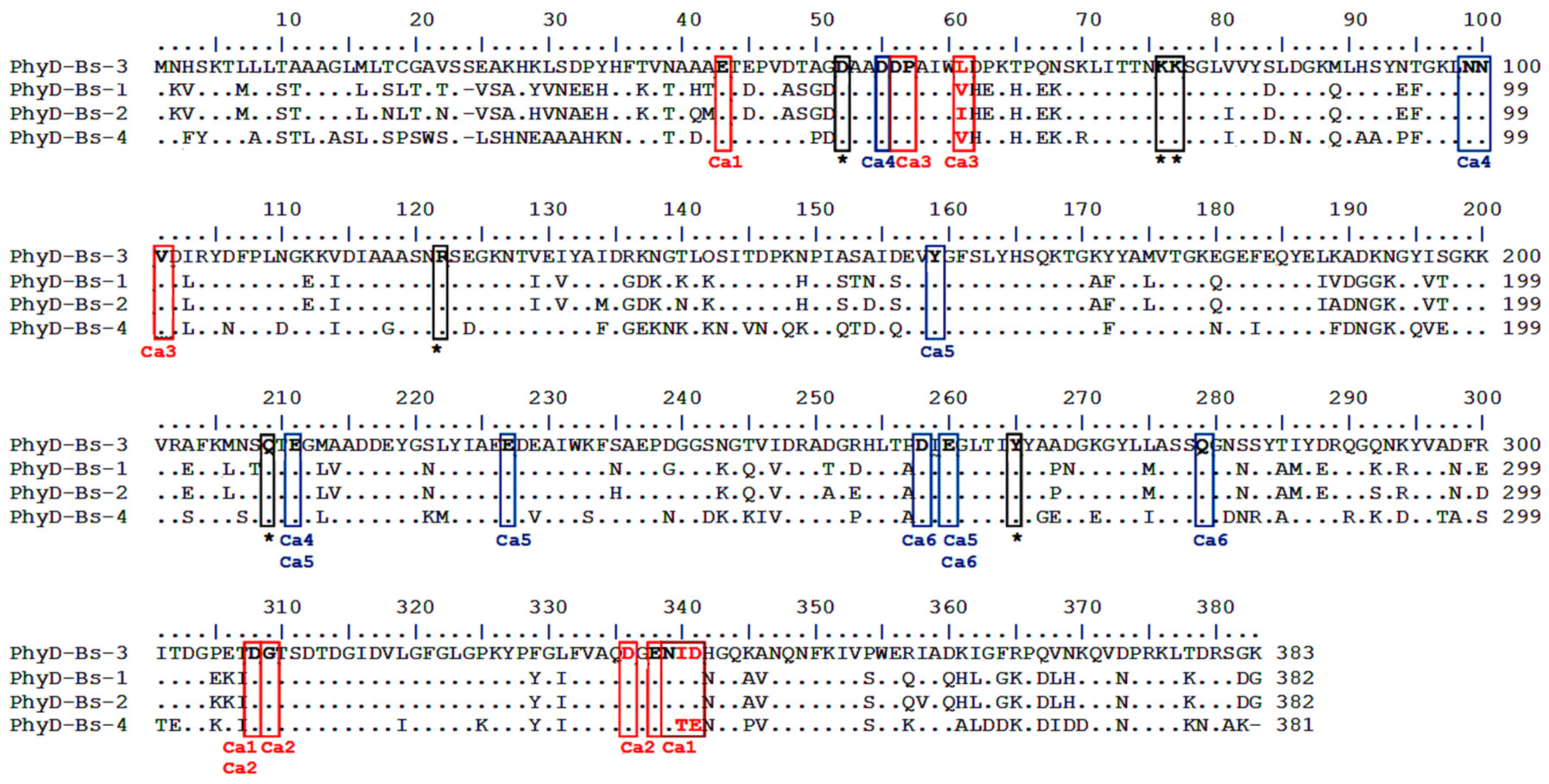
| Isolated Isoform | Organism Scientific Name | Sequence (a.a) of Phytase (PhyD), *—Stop Codon | Length (a.a.) | Identical Proteins, Number in GenBank | Percent Identity, % | Isoform In Silico |
|---|---|---|---|---|---|---|
| PhyD-Bs-1 |
| MKVSKTMLLSTAAGLLLSLTATSVSAHYVNEEHHFKVTAHTETDPVASGDDAADDPAIWVHEKHPEKSKLITTNKKSGLVVYDLDGKQLHSYEFGKLNNVDLRYDFPLNGEKIDIAAASNRSEGKNTIEVYAIDGDKGKLKSITDPKHPISTNISEVYGFSLYHSQKTGAFYALVTGKQGEFEQYEIVDGGKGYVTGKKVREFKLNTQTEGLVADDEYGNLYIAEEDEAIWKFNAEPGGGSKGQVVDRATGDHLTADIEGLTIYYAPNGKGYLMASSQGNNSYAMYERQGKNRYVANFEITDGEKIDGTSDTDGIDVLGFGLGPKYPYGIFVAQDGENIDNGQAVNQNFKIVSWEQIAQHLGGKPDLHKQVNPRKLKDRSDG* | 383 |
| 100% 100% 98% | PhyD-isoform-1 |
| PhyD-Bs-2 |
| MKVSKTMLLSTAAGLLLNLTANSVSAHHVNAEHHFKVTAQMETDPVASGDDAADDPAIWIHEKHPEKSKLITTNKKSGLIVYDLDGKQLHSYEFGKLNNVDLRYDFPLNGEKIDIAAASNRSEGKNTIEVYAMDGDKGNLKSITDPKHPISSDISEVYGFSLYHSQKTGAFYALVTGKQGEFEQYEIADNGKGYVTGKKVREFKLNSQTEGLVADDEYGNLYIAEEDEAIWKFHAEPDGGSKGQVVDRAAGEHLTADIEGLTIYYAPDGKGYLMASSQGNNSYAMYERQGSNRYVANFDITDGKKIDGTSDTDGIDVLGFGLGPKYPYGIFVAQDGENIDNGQAVNQNFKIVSWEQVAQHLGGKPDLHKQVNPRKLKDRSDG* | 383 |
| 100% 95% 94% 93% 93% | PhyD-isoform-1 |
| PhyD-Bs-3 |
| MNHSKTLLLTAAAGLMLTCGAVSSEAKHKLSDPYHFTVNAAAETEPVDTAGDAADDPAIWLDPKTPQNSKLITTNKKSGLVVYSLDGKMLHSYNTGKLNNVDIRYDFPLNGKKVDIAAASNRSEGKNTVEIYAIDRKNGTLQSITDPKNPIASAIDEVYGFSLYHSQKTGKYYAMVTGKEGEFEQYELKADKNGYISGKKVRAFKMNSQTEGMAADDEYGSLYIAEEDEAIWKFSAEPDGGSNGTVIDRADGRHLTPDIEGLTIYYAADGKGYLLASSQGNSSYTIYDRQGQNKYVADFRITDGPETDGTSDTDGIDVLGFGLGPKYPFGLFVAQDGENIDHGQKANQNFKIVPWERIADKIGFRPQVNKQVDPRKLTDRSGK* | 383 |
| 100% 96% 96% | PhyD-isoform-2 |
| PhyD-Bs-4 |
| MNFYKTLALSTLAASLLSPSWSSLSHNEAAAHKNFTVTADAETEPVDTPDDAADDPAIWVHPKHPEKSRLITTNKKSGLIVYDLNGKQLAAYPFGKLNNVDLRYNFPLDGKKIDIAGASNRSDGKNTVEIYAFDGEKNKLKNIVNPQKPIQTDIQEVYGFSLYHSQKTGKFYAMVTGKNGEIEQYELFDNGKGQVEGKKVRSFKMSSQTEGLAADDEYGKMYIAEEDVAIWSFSAEPNGGDKGKIVDRADGPHLTADIEGLTIYYGEDGEGYLIASSQGDNRYAIYDRRGKNDYVTAFSTEDGKEIDGTSDTDGIDVIGFGLGKKYPYGIFVAQDGENTENGQPVNQNFKIVSWEKIADALDDKPDIDDQVNPRKLKNRAK* | 382 |
| 100% 96% 97% | PhyD-isoform-3 |
| Non-identified |
| MLKEIATVALMTSVLFSTVNSTPDLVEKFPNSSVQEKQKHSKLVKVEAKEETDAVANAGDAADDPAIWVHPHDSEKSKIIGTNKEGGISIYNLKGKQLYSYNFGKLNNVDVRYGFPIEGKKIDIAAASNRSTNTIDIFAINPKTGELENIMGSPIQSSMKEVYGFSLYHSQKTGIFYALVVGKDGTFEQYELFDNGKGKIEGKKVRELKLSSQSEGIVADDEYGTIYIGEEDVAIWKFNAEPDGGNQPIAKVDGADGNHLTADIEGLTIYYGKNGTGYLIASSQGNNSYAVYDRKGNNEYIGNFAIVDGKNTDGTSDTDGIDVMSFGLGEKYPNGIFLAQDGENMDHGKIVNQNFKMVDWKKIAKGFDSRLQAENNVNPRKLKYREILSK* | 390 |
| 100% 95% 93% | PhyD-isoform-4 |
| Non-identified |
| MQVLPKLAVSHYQAPVIAQVQAIAQTLPADRIGDAADDPAIWVNHQHPEQSRVLGTDKRGALEVYDLNGQRLQRLAVGRVNNVDVRQGFRLGGKTVDIATASHRDHNAISVFAIAPDSGEVSLLGEVPTPLKDIYGLCMYQPQGQIQVFVNDKNGRVLQYRLDDNHGAIKGTLVRDFRVNTQPEGCVADDKRGRFFLGEEDVGIWAFNADDTQAPAGTLIAKVGPMLHADVEGLALWQGERPILVASSQGNDSYVAYSALPPYQLLGRFRIGLNSEAGIDGTSXTDGIDITSLALGKAYPQGLLAVQDGRKRLPEQGQNFKLVPFDAVLKLLQQ* | 334 |
AHM26864.1
| 100% 97% 98% | PhyD-isoform-5 |
| Sample | Isolation Stage and Incubation Conditions | Phytase Activity, U/mg of Protein |
|---|---|---|
| 1 | After urea treatment on ice for 20 min, followed by incubation for 40 min with proline | 0.5 ± 0.01 * |
| 2 | After urea treatment for 30 min at room temperature and incubation with proline for 40 min. | 0.94 ± 0.04 * |
| 3 | 3 days of incubation with proline. | 0.05 ± 0.001 |
| 4 | 4 days of incubation with proline at +10 °C. | 0 |
| Sample | Isolation Stage and Incubation Conditions | Phytase Activity, U/mg of Protein |
|---|---|---|
| 1 | Urea treatment on ice for 20 min followed by incubation with proline for 40 min | 0.26 ± 0.01 * |
| 2 | After urea treatment for 30 min at room temperature and incubation with proline for 40 min. | 0.47 ± 0.01 * |
| 3 | 3 days of incubation with proline. | 0.06 ± 0.001 |
| 4 | 4 days of incubation with proline at +10 °C. | 0 |
| Samples | PO1f (MatA, leu2-270, ura3-302, xpr2-322, asp-2) | PO1f Transformant Biomass (pUV3-tPhyD-Bs-1)_5 | PO1f Transformant Biomass (pUV3-tPhyD-Bs-1)_6 |
|---|---|---|---|
| Turbidity OD350 urea free | 0.12 ± 0.02 | 0.35 ± 0.07 | 0.24 ± 0.05 |
| Turbidity OD350 8 M urea | 0.10± 0.03 | 0.12 ± 0.03 | 0.11 ± 0.04 |
| Phytase | Predicted N-Glycosylation Sites * | Predicted O-Glycosylation Sites ** | GRAVY # | Number of Cys $ | Total Different Combinations of S |
|---|---|---|---|---|---|
| PhyD-Bs-1 | 2 | 2 | −0.597 | no | 0 |
| PhyD-Bs-2 | 1 | 1 | −0.618 | no | 0 |
| PhyD-Bs-3 | 2 | 0 | −0.620 | no | 1 |
| PhyD-Bs-4 | 2 | 1 | −0.699 | no | 0 |
| phy-OP | 0 | 4 | −0.364 | 8 | 764 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maloshenok, L.G.; Panina, Y.S.; Bruskin, S.A.; Zherdeva, V.V.; Gessler, N.N.; Rozumiy, A.V.; Antonov, E.V.; Deryabina, Y.I.; Isakova, E.P. Assessment of Recombinant β-Propeller Phytase of the Bacillus Species Expressed Intracellularly in Yarrowia lipolityca. J. Fungi 2025, 11, 186. https://doi.org/10.3390/jof11030186
Maloshenok LG, Panina YS, Bruskin SA, Zherdeva VV, Gessler NN, Rozumiy AV, Antonov EV, Deryabina YI, Isakova EP. Assessment of Recombinant β-Propeller Phytase of the Bacillus Species Expressed Intracellularly in Yarrowia lipolityca. Journal of Fungi. 2025; 11(3):186. https://doi.org/10.3390/jof11030186
Chicago/Turabian StyleMaloshenok, Liliya G., Yulia S. Panina, Sergey A. Bruskin, Victoria V. Zherdeva, Natalya N. Gessler, Alena V. Rozumiy, Egor V. Antonov, Yulia I. Deryabina, and Elena P. Isakova. 2025. "Assessment of Recombinant β-Propeller Phytase of the Bacillus Species Expressed Intracellularly in Yarrowia lipolityca" Journal of Fungi 11, no. 3: 186. https://doi.org/10.3390/jof11030186
APA StyleMaloshenok, L. G., Panina, Y. S., Bruskin, S. A., Zherdeva, V. V., Gessler, N. N., Rozumiy, A. V., Antonov, E. V., Deryabina, Y. I., & Isakova, E. P. (2025). Assessment of Recombinant β-Propeller Phytase of the Bacillus Species Expressed Intracellularly in Yarrowia lipolityca. Journal of Fungi, 11(3), 186. https://doi.org/10.3390/jof11030186







