Comparative Genomic Analysis of Two Monokaryons of Auricularia heimuer Hei29
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain and DNA Preparation
2.2. Genome Sequencing, Assembly, and Annotation
2.3. PSY Gene Family Analysis
2.4. Comparative Genome Analysis
3. Results
3.1. Acquisition and Morphological Description of Two Monokaryons
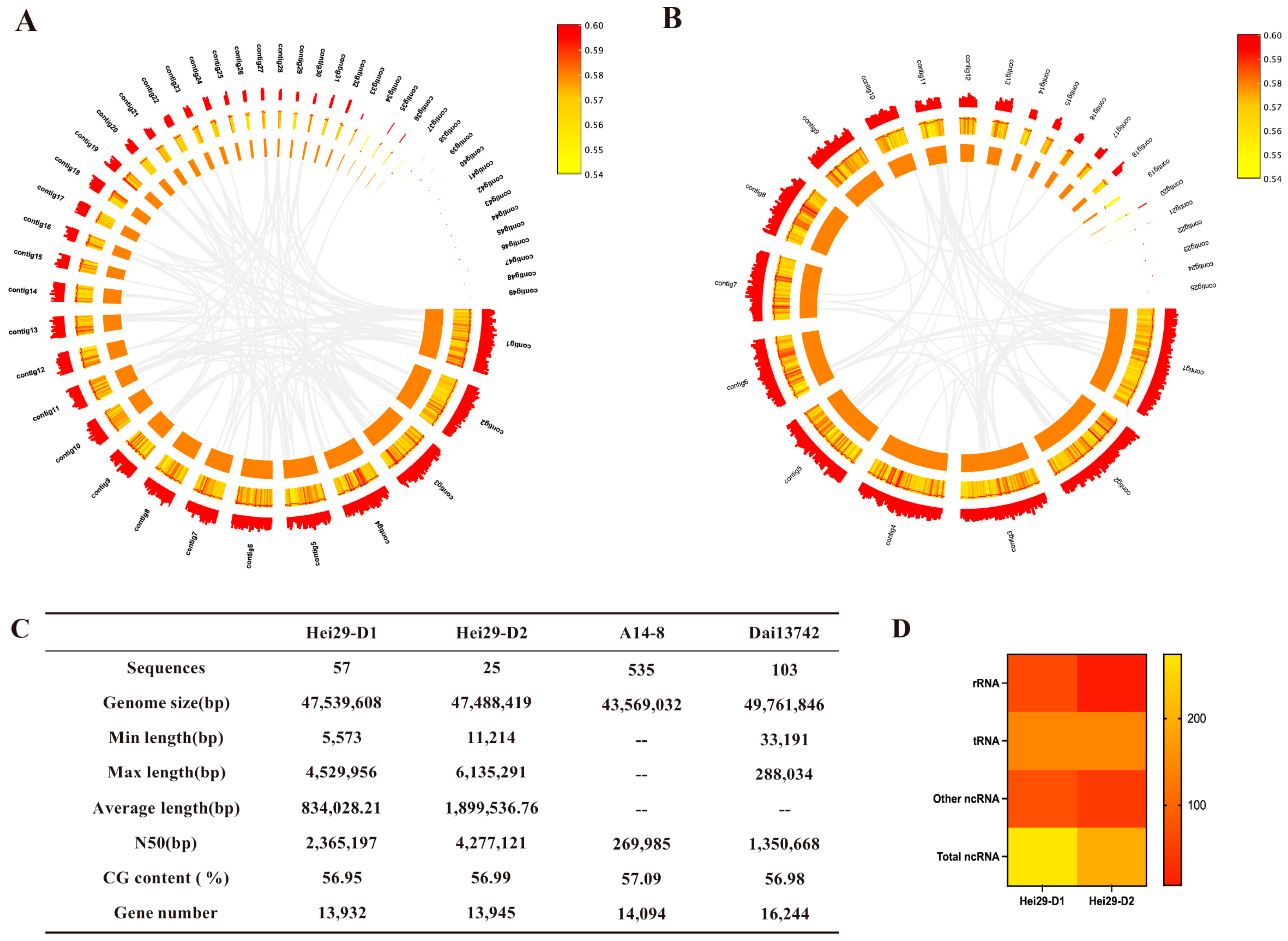
3.2. Genome Sequence Assembly
3.3. Genome Sequence Annotation
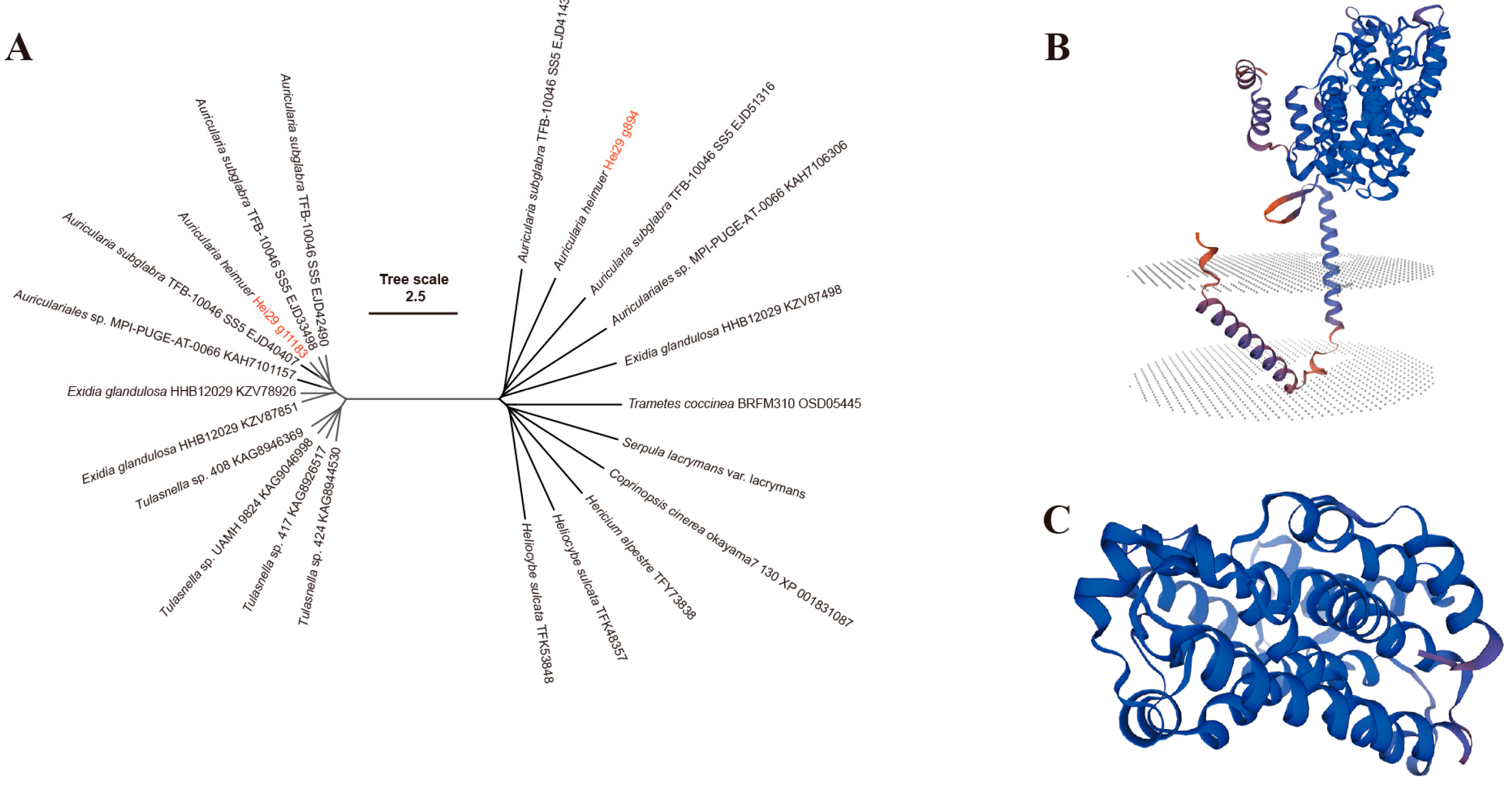
3.4. Identification of the PSY Gene
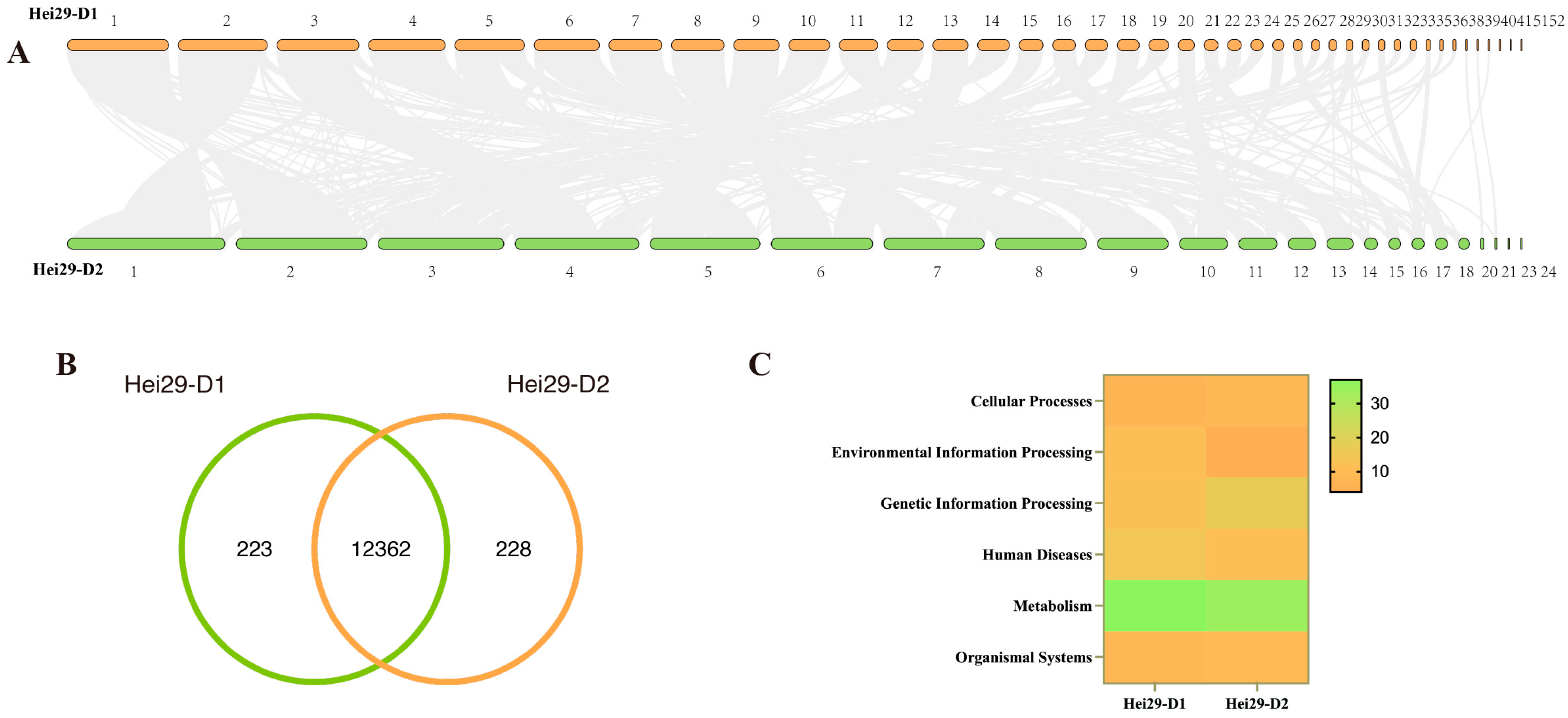
3.5. Comparative Genomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wu, F.; Yuan, Y.; Malysheva, V.F.; Du, P.; Dai, Y.C. Species clarification of the most important and cultivated Auricularia mushroom “Heimuer”: Evidence from morphological and molecular data. Phytotaxa 2014, 186, 241–253. [Google Scholar] [CrossRef]
- China Edible Fungi Association. Research on the Statistical Survey Results of Edible Fungi in China in 2023. Available online: https://mp.weixin.qq.com/s/aFSu1OVNjR8-Q-KVhR_iNw (accessed on 17 December 2024).
- China Edible Fungi Association. Market and Industry Analysis Report on Auricularia heimuer in China. Available online: https://mp.weixin.qq.com/s/ouvzCC6jLwZv-L-Z2WGuIg (accessed on 17 February 2023).
- Sun, X.; Yang, C.; Ma, Y.; Zhang, J.; Wang, L. Research progress of Auricularia heimuer on cultivation physiology and molecular biology. Front. Microbiol. 2022, 3, 1048249. [Google Scholar] [CrossRef]
- Miao, J.; Regenstein, J.M.; Qiu, J.; Zhang, J.; Zhang, X.; Li, H.; Zhang, H.; Wang, Z. Isolation, structural characterization and bioactivities of polysaccharides and its derivatives from Auricularia—A review. Int. J. Biol. Macromol. 2020, 150, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, P.; Dai, X.; Yao, X.; Zhou, S.; Ma, Q.; Liu, J.; Tian, S.; Zhu, J.; Zhang, J.; et al. Extraction, physicochemical properties, and antioxidant activity of natural melanin from Auricularia heimuer fermentation. Front. Nutr. 2023, 10, 1131542. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhang, J.; Han, Z.; Kong, X.; Zhang, X. Characteristics of the Hei29 strain of Auricularia auricula-judae and key points for cultivation. Edible Fungi 2003, 1, 10–11. [Google Scholar]
- Yuan, Y.; Wu, F.; Si, J.; Zhao, Y.F.; Dai, Y.-C. Whole genome sequence of Auricularia heimuer (Basidiomycota, Fungi), the third most important cultivated mushroom worldwide. Genomics 2019, 111, 50–58. [Google Scholar] [CrossRef]
- Fang, M.; Wang, X.; Chen, Y.; Wang, P.; Lu, L.; Lu, J.; Yao, F.; Zhang, Y. Genome sequence analysis of Auricularia heimuer combined with genetic linkage map. J. Fungi 2020, 6, 37. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Feng, X.L.; Wang, Z.X.; Qi, J. The First Whole Genome Sequencing of Agaricus bitorquis and Its Metabolite Profiling. J. Fungi 2023, 9, 485. [Google Scholar] [CrossRef]
- Wei, J.; Cheng, M.; Zhu, J.F.; Zhang, Y.; Cui, K.; Wang, X.; Qi, J. Comparative Genomic Analysis and Metabolic Potential Profiling of a Novel Culinary-Medicinal Mushroom, Hericium rajendrae (Basidiomycota). J. Fungi 2023, 9, 1018. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Yan, D.; Song, S.; Fan, Y.; Wang, S.; Liu, Y.; Huang, Y.; Rong, C.; Guo, Y.; Zhao, S.; et al. Haplotype-resolved genome analyses reveal genetically distinct nuclei within a commercial cultivar of Lentinula edodes. J. Fungi 2022, 8, 496. [Google Scholar] [CrossRef]
- Chen, C.L.; Li, W.C.; Chuang, Y.C.; Liu, H.C.; Huang, C.H.; Lo, K.Y.; Chen, C.Y.; Chang, F.M.; Chang, G.A.; Lin, Y.L.; et al. Sexual Crossing, Chromosome-Level Genome Sequences, and Comparative Genomic Analyses for the Medicinal Mushroom Taiwanofungus Camphoratus (Syn Antrodia Cinnamomea, Antrodia Camphorata). Microbiol. Spectr. 2022, 10, e0203221. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Zhang, P.; Dai, X.; Kong, X.; Ma, Q.F.; Zhang, J.C. Preparation, Regeneration, and Fluorescence Identification of Mononuclear Bodies of Auricularia auricula-judae. J. Edible Fungi 2008, 3, 13–17. [Google Scholar] [CrossRef]
- McCarthy, A. Third generation DNA sequencing: Pacific biosciences’ single molecule real time technology. Chem. Biol. 2010, 17, 675–676. [Google Scholar] [CrossRef]
- Cheng, H.; Concepcion, G.T.; Feng, X.; Zhang, H.; Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 2021, 18, 170–175. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Walenz, B.P.; Rhie, A.; Vollger, M.R.; Logsdon, G.A.; Grothe, R.; Miga, K.H.; Eichler, E.E.; Phillippy, A.M.; Koren, S. HiCanu: Accurate assembly of segmental duplications, satellites, and allelic variants from high-fidelity long reads. Genome Res. 2020, 30, 1291–1305. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef]
- Stanke, M.; Schöffmann, O.; Morgenstern, B.; Waack, S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinform. 2006, 7, 62. [Google Scholar] [CrossRef]
- Lowe, T.M.; Eddy, S.R. TRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004, 32 (Suppl. S1), D258–D261. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant. 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Amalamol, D.; Ashwin, N.M.R.; Lakshana, K.V.; Nirmal Bharathi, M.; Ramesh Sundar, A.; Sukumaran, R.K.; Malathi, P.; Viswanathan, R. A highly efficient stratagem for protoplast isolation and genetic transformation in filamentous fungus Colletotrichum falcatum. Folia Microbiol. 2022, 67, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Li, S. Regulation of ribosomal proteins on viral infection. Cells 2019, 8, 508. [Google Scholar] [CrossRef]
- Akira, S.; Maeda, K. Control of RNA stability in immunity. Annu. Rev. Immunol. 2021, 39, 481–509. [Google Scholar] [CrossRef]
- Colonna Romano, N.; Fanti, L. Transposable elements: Major players in shaping genomic and evolutionary patterns. Cells 2022, 11, 1048. [Google Scholar] [CrossRef]
- Baduel, P.; Quadrana, L. Jumpstarting evolution: How transposition can facilitate adaptation to rapid environmental changes. Curr. Opin. Plant Biol. 2021, 61, 102043. [Google Scholar] [CrossRef] [PubMed]
- Merenciano, M.; González, J. The interplay between developmental stage and environment underlies the adaptive effect of a natural transposable element insertion. Mol. Biol. Evol. 2023, 40, msad044. [Google Scholar] [CrossRef]
- Baduel, P.; Leduque, B.; Ignace, A.; Gy, I.; Gil, J., Jr.; Loudet, O.; Colot, V.; Quadrana, L. Genetic and environmental modulation of transposition shapes the evolutionary potential of Arabidopsis thaliana. Genome Biol. 2021, 22, 138. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y. KEGG Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020, 29, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Sun, X.; Qi, Q.; Wang, X.T.; Sun, T.T. Identification of key enzymes involved in the biosynthesis of triterpenes in Auricularia auricula. North. Hortic. 2020, 21, 123–130. [Google Scholar]
- Kristensen, A.T.; Olsen, L.I.; Axelsen, K.B.; Fuglsang, T. The PSY Peptide Family—Expression, Modification and Physiological Implications. Genes 2021, 12, 218. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, L.; Dong, C.; Guo, J.; Jin, L.; Wei, P.; Li, F.; Zhang, X.; Wang, R. Characterization and functional analysis of phytoene synthase gene family in tobacco. BMC Plant Biol. 2021, 21, 32. [Google Scholar] [CrossRef]
- Chettry, U.K.; Chrungoo, N.K. A multifocal approach towards understanding the complexities of carotenoid biosynthesis and accumulation in rice grains. Brief. Funct. Genom. 2020, 19, 324–335. [Google Scholar] [CrossRef]
- Li, F.; Vallabhaneni, R.; Yu, J.; Rochord, T.; Wurtzel, E.T. The maize phytoene synthase gene family: Overlapping roles for carotenogenesis in endosperm, photomorphogenesis and thermal stress tolerance. Plant Physiol. 2008, 147, 1334–1346. [Google Scholar] [CrossRef]
- Ezquerro, M.; Burbano-Erazo, E.; Rodriguez-Concepcion, M. Overlapping and specialized roles of tomato phytoene synthases in carotenoid and abscisic acid production. Plant Physiol. 2023, 193, 2021–2036. [Google Scholar] [CrossRef] [PubMed]
- Parada, R.; Royo, C.; Gadaleta, A.; Colasuonno, P.; Marcotuli, I.; Matus, I.; Castillo, D.; de Camargo, A.C.; Araya-Flores, J.; Villegas, D.; et al. Phytoene synthase 1 (Psy-1) and lipoxygenase 1 (Lpx-1) Genes Influence on Semolina Yellowness in Wheat Mediterranean Germplasm. Int. J. Mol. Sci. 2020, 21, 4669. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Qiao, S.; Ge, C.; Wei, Z.; Xu, C. Identification and expression analysis of the PSY gene family in spinach. Biotechnol. Bull. 2023, 9, 169–178. [Google Scholar]
- Floudas, D.; Binder, M.; Riley, R.; Barry, K.; Blanchette, R.A.; Henrissat, B.; Martínez, A.T.; Otillar, R.; Spatafora, J.W.; Yadav, J.S.; et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 2012, 336, 1715–1719. [Google Scholar] [CrossRef]
- Liu, D.; Tao, X.; Wei, H. cDNA cloning and bioinformatics analysis of the phytoene synthase encoding gene psy in Chlorella vulgaris. J. Shanghai Ocean. Univ. 2017, 26, 171–182. [Google Scholar]
- Rogers, R.L.; Grizzard, S.L.; Titus-McQuillan, J.E.; Bockrath, K.; Patel, S.; Wares, J.P.; Garner, J.T.; Moore, C.C. Gene family amplification facilitates adaptation in freshwater unionid bivalve Megalonaias nervosa. Mol. Ecol. 2021, 30, 1155–1173. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Xiao, Y.; Xu, D.; Xue, H.; Song, H.; Li, P. Detoxification effect of edible fungi on aflatoxin B1. Chin. J. Microecol. 2017, 29, 37–41. [Google Scholar]
- Gordon, G.C.; Cameron, J.C.; Gupta, S.T.P.; Engstrom, M.D.; Reed, J.L.; Pfleger, B.F. Genome-wide analysis of RNA decay in the Cyanobacterium Synechococcus sp. Strain PCC 7002. mSystems 2020, 5, e00224-20. [Google Scholar] [CrossRef] [PubMed]
- Bao, D.; Xu, J.; Pan, Y.; Tan, Q. Analysis of the concepts related to heterothallic edible fungi: Diploid and monokaryotic strains. Acta Edulis Fungi 2024, 31, 1–11. [Google Scholar]
- Zhang, X.O.; Pratt, H.; Weng, Z. Investigating the potential roles of SINEs in the human genome. Annu. Rev. Genom. Hum. Genet. 2021, 22, 199–218. [Google Scholar] [CrossRef]
- Morrish, T.A.; Gilbert, N.; Myers, J.S.; Vincent, B.J.; Stamato, T.D.; Taccioli, G.E.; Batzer, M.A.; Moran, J.V. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 2002, 31, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Li, P.; Ma, L.; Lai, S.; Sun, S.; Hu, K.; Zhang, L. Analysis and modification of central carbon metabolism in Hypsizygus marmoreus for improving mycelial growth performance and fruiting body yield. Front. Microbiol. 2023, 14, 1233512. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Yang, G.; Ma, Y.; Zhang, J. Extraction and Component Determination of Auricularia auricula-judae. China New Technol. New Prod. 2019, 1, 6–7. [Google Scholar] [CrossRef]
- Xiang, H.; Sun-Waterhouse, D.; Cui, C. Hypoglycemic polysaccharides from Auricularia auricula and Auricularia polytricha inhibit oxidative stress, NF-κB signaling, and proinflammatory cytokine production in streptozotocin-induced diabetic mice. Food Sci. Hum. Wellness 2020, 9, 104–115. [Google Scholar] [CrossRef]
- Kong, X.; Duan, W.; Li, D.; Tang, X.; Duan, Z. Effects of polysaccharides from Auricularia auricula on the immuno-stimulatory activity and gut microbiota in immunosuppressed mice induced by cyclophosphamide. Front. Immunol. 2020, 11, 595700. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.Z.; Yao, F.; Yin, C.M.; Shi, D.F.; Gao, H. Optimization of Fermentation Process and Its Impact on Gene Transcription of Intracellular Polysaccharide Synthesis in the Wood Ear Medicinal Mushroom Auricularia auricula-judae (Agaricomycetes). Int. J. Med. Mushrooms 2020, 22, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Su, S.; Wu, Y.; Liu, C.; Jiang, S.; Liu, Q.; Chen, M.; Long, C.; Wang, T.; Xu, Y.; Tian, F.; et al. Genome-wide identification of the UGP gene family in Auricularia auricula-judae and its correlation with mycelial growth and polysaccharide synthesis. Seed 2024, 43, 69–80. [Google Scholar]
- Zan, X.Y.; Wu, X.H.; Cui, F.J.; Zhu, H.A.; Sun, W.J.; Jiang, L.H.; Tao, T.L.; Zhao, X. UDP-glucose pyrophosphorylase gene affects mycelia growth and polysaccharide synthesis of Grifola frondosa. Int. J. Biol. Macromol. 2020, 161, 1161–1170. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Bai, Y.; Dai, R.; Guo, X.; Liu, Z.H.; Yuan, S. Improved Polysaccharide Production by Homologous Co-overexpression of Phosphoglucomutase and UDP Glucose Pyrophosphorylase Genes in the Mushroom Coprinopsis cinerea. J. Agric. Food Chem. 2018, 66, 4702–4709. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.L.; Yuan, H.; Li, N.; Xiao, J.H.; Xu, J.W. Increased production and anti-senescence activity of exopolysaccharides in Ganoderma lingzhi by co-overexpression of β-1,3-glucan synthase and UDP-glucose pyrophosphorylase. Int. J. Biol. Macromol. 2023, 253 (Pt 2), 126778. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, L.; Zhang, D.; Li, R.; Cheng, T.; Zhang, Y.; Liu, X.; Wong, G.; Tang, Y.; Wang, H.; et al. Comparative transcriptome analysis reveals relationship of three major domesticated varieties of Auricularia auricula-judae. Sci. Rep. 2019, 9, 78. [Google Scholar] [CrossRef]
- Shahar, O.; Pereman, I.; Khamisie, H.; Ezov, N.; Danay, O.; Khattib, A.; Schweitzer, R.; Khatib, S.; Mahajna, J. Compounds originating from the edible mushroom Auricularia auricula-judae inhibit tropomyosin receptor kinase B activity. Heliyon 2023, 9, e13756. [Google Scholar] [CrossRef] [PubMed]
- Reza, M.A.; Hossain, M.A.; Lee, S.J.; Yohannes, S.B.; Damte, D.; Rhee, M.H.; Jo, W.S.; Suh, J.W.; Park, S.C. Dichloromethane extract of the jelly ear mushroom Auricularia auricula-judae (higher Basidiomycetes) inhibits tumor cell growth in vitro. Int. J. Med. Mushrooms 2014, 16, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Chis, A.A.; Arseniu, A.M.; Morgovan, C.; Dobrea, C.M.; Frum, A.; Juncan, A.M.; Butuca, A.; Ghibu, S.; Gligor, F.G.; Rus, L.L. Biopolymeric prodrug systems as potential antineoplastic therapy. Pharmaceutics 2022, 14, 1773. [Google Scholar] [CrossRef]
- Bao, Z.; Yao, L.; Zhang, X.; Lin, S. Isolation, purification, characterization, and immunomodulatory effects of polysaccharide from Auricularia auricula on RAW264.7 macrophages. J. Food Biochem. 2020, 44, e13516. [Google Scholar] [CrossRef] [PubMed]
- Vanherle, L.; Matuskova, H.; Don-Doncow, N.; Uhl, F.E.; Meissner, A. Improving cerebrovascular function to increase neuronal recovery in neurodegeneration associated to Cardiovascular Disease. Front. Cell Dev. Biol. 2020, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Dailah, H.G. Potential of therapeutic small molecules in apoptosis regulation in the treatment of Neurodegenerative Diseases: An Updated Review. Molecules 2022, 27, 7207. [Google Scholar] [CrossRef]
- Dowaraka-Persad, B.; Neergheen, V.S. Mushroom-derived compounds as metabolic modulators in cancer. Molecules 2023, 28, 1441. [Google Scholar] [CrossRef]
- Wong, J.H.; Ng, T.B.; Chan, H.H.L.; Liu, Q.; Man, G.C.W.; Zhang, C.Z.; Guan, S.; Ng, C.C.W.; Fang, E.F.; Wang, H.; et al. Mushroom extracts and compounds with suppressive action on breast cancer: Evidence from studies using cultured cancer cells, tumor-bearing animals, and clinical trials. Appl. Microbiol. Biotechnol. 2020, 104, 4675–4703. [Google Scholar] [CrossRef]
- Chai, Y.; Wang, G.; Fan, L.; Zhao, M. A proteomic analysis of mushroom polysaccharide-treated HepG2 cells. Sci. Rep. 2016, 6, 23565. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Zhang, H.; Wang, Z.; Liu, D.; Liu, S.; Han, W.; Regenstein, J.M.; Geng, L. The antitumor effect of folic acid conjugated-Auricularia auricular polysaccharide-cisplatin complex on cervical carcinoma cells in nude mice. Int. J. Biol. Macromol. 2018, 107 (Pt B), 2180–2189. [Google Scholar] [CrossRef]
- Tong, Z.; Chu, G.; Wan, C.; Wang, Q.; Yang, J.; Meng, Z.; Du, L.; Yang, J.; Ma, H. Multiple metabolites derived from mushrooms and their beneficial effect on Alzheimer’s Diseases. Nutrients 2023, 15, 2758. [Google Scholar] [CrossRef] [PubMed]
- Phan, C.W.; David, P.; Naidu, M.; Wong, K.H.; Sabaratnam, V. Therapeutic potential of culinary-medicinal mushrooms for the management of neurodegenerative diseases: Diversity, metabolite, and mechanism. Crit. Rev. Biotechnol. 2015, 35, 355–368. [Google Scholar] [CrossRef] [PubMed]
- Abitbol, A.; Mallard, B.; Tiralongo, E.; Tiralongo, J. Mushroom natural products in Neurodegenerative Disease drug discovery. Cells 2022, 11, 3938. [Google Scholar] [CrossRef]
- Paisansak, S.; Sangtanoo, P.; Srimongkol, P.; Saisavoey, T.; Reamtong, O.; Choowongkomon, K.; Karnchanatat, A. Angiotensin-I converting enzyme inhibitory peptide derived from the shiitake mushroom (Lentinula edodes). J. Food Sci. Technol. 2021, 58, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Li, Y.; Peng, K.; Wang, X.L.; Ding, Z.; Liu, L.; Xu, P.; Liu, G.Q. Isolation and characterization of three antihypertension peptides from the mycelia of Ganoderma Lucidum (Agaricomycetes). J. Agric. Food Chem. 2019, 67, 8149–8159. [Google Scholar] [CrossRef] [PubMed]
- Diana, D.; Ismaya, W.T.; Meidianto, V.F.; Tandrasasmita, O.M.; Tjandrawinata, R.R.; Rachmawati, H. Bioconjugation of captopril-light subunit of Agaricus bisporus mushroom tyrosinase: Characterization and potential use as a drug carrier for oral delivery. Biol. Pharm. Bull. 2018, 41, 1837–1842. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Chen, X.; Mao, X.; Kong, X. Optimization of the enzymatic hydrolysis process for the preparation of ACE-inhibiting peptides from Auricularia auricular-judae. Food Nutr. China 2024. Available online: https://xueshu.baidu.com/usercenter/paper/show?paperid=1v1e0a60ay020m20027v0080m8544055&site=xueshu_se (accessed on 17 December 2024).
- Yu, N.; Huang, Y.; Jiang, Y.; Zou, L.; Liu, X.; Liu, S.; Chen, F.; Luo, J.; Zhu, Y. Ganoderma lucidum triterpenoids (GLTs) reduce neuronal apoptosis via inhibition of ROCK signal pathway in APP/PS1 transgenic Alzheimer’s Disease mice. Oxidative Med. Cell. Longev. 2020, 2020, 9894037. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zhao, L.; Song, Y.; Qiao, Y.; Wang, Z.X.; Qi, J. Genome-wide characterization and metabolite profiling of Cyathus olla: Insights into the biosynthesis of medicinal compounds. BMC Genom. 2024, 25, 618. [Google Scholar] [CrossRef] [PubMed]
- Cordaro, M.; Salinaro, A.T.; Siracusa, R.; D’Amico, R.; Impellizzeri, D.; Scuto, M.; Ontario, M.L.; Cuzzocrea, S.; Di Paola, R.; Fusco, R.; et al. Key Mechanisms and potential implications of Hericium erinaceus in NLRP3 inflammasome activation by reactive oxygen species during Alzheimer’s Disease. Antioxidants 2021, 10, 1664. [Google Scholar] [CrossRef]
- Naguib, A.M.; Apparoo, Y.; Xiong, C.; Phan, C.W. Maitake medicinal mushroom, Grifola frondosa (Agaricomycetes), and its neurotrophic properties: A Mini-Review. Int. J. Med. Mushrooms 2023, 25, 11–22. [Google Scholar] [CrossRef]
- Liu, Y.L.; Cao, Y.G.; Hao, F.X.; Zeng, M.N.; Niu, Y.; Chen, L.; Chen, X.; Zheng, X.K.; Feng, W.S. Chemical constituents from stipes of Lentinus edodes and their protective effects against Aβ25-35-induced N9 microglia cells injury. Phytochemistry 2024, 222, 114098. [Google Scholar] [CrossRef] [PubMed]
- Chu, J.Z.; Zheng, N.; Cui, Q.; Li, M.X.; Li, Y.; Wu, G.C.; Cheng, X.H.; Zhang, R. Analyses of potential mechanism of “sanghuang” in treatment of diseases based on network pharmacology and molecular docking. Mycosystema 2023, 41, 365–382. [Google Scholar] [CrossRef]



| Sample | Type | Number of Elements | Length Occupied | Percentage of Sequence |
|---|---|---|---|---|
| Hei29-D1 | SINEs | 22 | 7987 | 0.02 |
| LINEs | 612 | 786,037 | 1.65 | |
| LTR elements | 1031 | 1,777,874 | 3.74 | |
| DNA elements | 67 | 20,390 | 0.04 | |
| Unclassified | 1843 | 1,409,131 | 2.96 | |
| Hei29-D2 | SINEs | 16 | 7460 | 0.02 |
| LINEs | 397 | 494,891 | 1.04 | |
| LTR elements | 918 | 1,798,866 | 3.97 | |
| DNA elements | 217 | 166,481 | 0.35 | |
| Unclassified | 1667 | 1,268,699 | 2.67 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, F.; Han, C.; Zhang, J.; Zhang, P.; Zhang, X.; Yue, X.; Zhao, Y.; Dai, X. Comparative Genomic Analysis of Two Monokaryons of Auricularia heimuer Hei29. J. Fungi 2025, 11, 122. https://doi.org/10.3390/jof11020122
Wang F, Han C, Zhang J, Zhang P, Zhang X, Yue X, Zhao Y, Dai X. Comparative Genomic Analysis of Two Monokaryons of Auricularia heimuer Hei29. Journal of Fungi. 2025; 11(2):122. https://doi.org/10.3390/jof11020122
Chicago/Turabian StyleWang, Fengli, Chuang Han, Jiechi Zhang, Piqi Zhang, Xiaojia Zhang, Xin Yue, Yanshu Zhao, and Xiaodong Dai. 2025. "Comparative Genomic Analysis of Two Monokaryons of Auricularia heimuer Hei29" Journal of Fungi 11, no. 2: 122. https://doi.org/10.3390/jof11020122
APA StyleWang, F., Han, C., Zhang, J., Zhang, P., Zhang, X., Yue, X., Zhao, Y., & Dai, X. (2025). Comparative Genomic Analysis of Two Monokaryons of Auricularia heimuer Hei29. Journal of Fungi, 11(2), 122. https://doi.org/10.3390/jof11020122







