Global Diversity, Host Associations, and New Insights into Aigialaceae, Astrosphaeriellaceae, and Pseudoastrosphaeriellaceae
Abstract
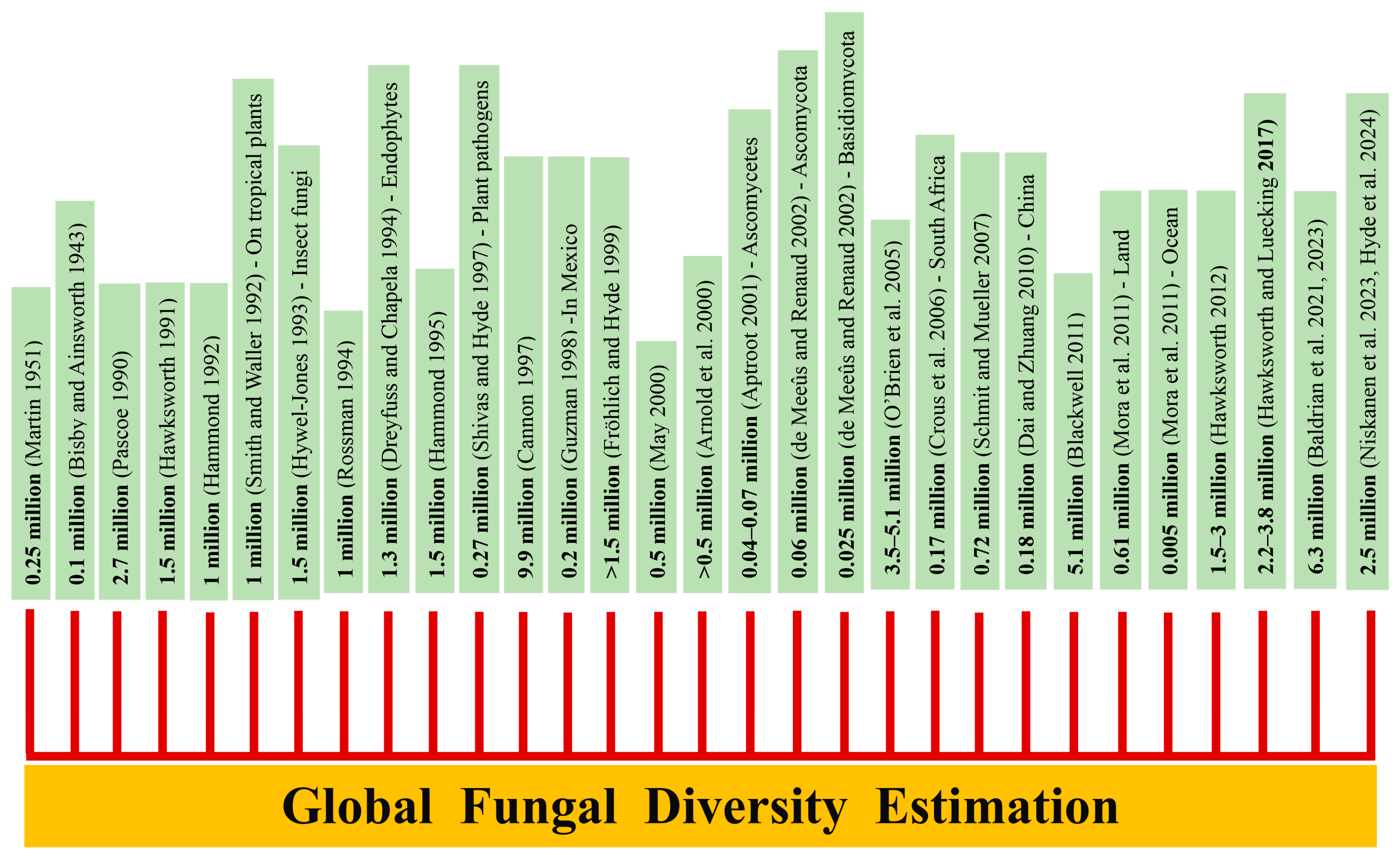
1. Introduction
2. Materials and Methods
2.1. Samples Collection, Microscopic Observations, and Fungal Isolation
2.2. DNA Extraction, PCR Amplification, and Sequencing
| Locus | Primers | Reference |
|---|---|---|
| LSU | Forward: LR0R GTACCCGCTGAACTTAAGC | [62] |
| Reverse: LR5 ATCCTGAGGGAAACTTC | ||
| SSU | Forward: NS1 GTAGTCATATGCTTGTCTC | [63] |
| Reverse: NS4 CTTCCGTCAATTCCTTTAAG | ||
| tef1-α | Forward: EF1-983F GCYCCYGGHCAYCGTGAYTTYAT | [64] |
| Reverse: EF1-2218R ATGACACCRACRGCRACRGTYTG |
2.3. Phylogenetic Analyses
2.4. Geographical Distribution and Host Associations
| Taxon | Strain/Voucher Number | GenBank Accession Numbers | Reference | ||
|---|---|---|---|---|---|
| LSU | SSU | tef1-α | |||
| Aigialus grandis | MFLUCC 15-1281 | MN420684 | MN420694 | - | [74] |
| A. parvus | BCC 32558 | GU479779 | GU479743 | GU479843 | [75] |
| Astrosphaeriella bakeriana | CBS 115556 | GU301801 | - | GU349015 | [76] |
| As. bambusae | MFLUCC 13-0230 | KT955461 | - | KT955424 | [58] |
| As. bambusae | MBSZU 25-061 | PX225037 | PX225048 | PX226738 | this study |
| As. fusispora | MFLUCC 10-0555 | KT955462 | KT955443 | KT955425 | [58] |
| As. lophiostomopsis | HKUCC 2984 | GU205215 | GU205232 | - | [77] |
| As. neofusispora | MFLUCC 11-0161 | KT955463 | KT955444 | KT955426 | [58] |
| As. neostellata | CX003 | MN629351 | MN629353 | MN635787 | [78] |
| As. neostellata | MFLUCC 11-0625 | KT955464 | - | - | [58] |
| As. stellata | MFLUCC10-0555 | JN846723 | JN846733 | - | [79] |
| As. thailandica | MFLUCC 11-0191 | KT955465 | KT955445 | KT955427 | [58] |
| As. thysanolaenae | MFLUCC 11-0186 | KT955466 | KT955446 | KT955428 | [58] |
| Astrosphaeriellopsis caryotae | MFLUCC 13-0830 | MF588990 | MF588980 | - | [59] |
| Carinispora nypae | BCC 36316 | - | GU479749 | GU479849 | [75] |
| C. aquatica | MFLUCC 11-0008 | MH057847 | MH057850 | - | [80] |
| C. aquatica | HKAS 112608 | OP377899 | OP377986 | - | [81] |
| C. aquatica | MFLUCC 18-1030 | MT627666 | MT864295 | MT954394 | [60] |
| C. minima | taxon:492516 | EU196550 | EU196551 | - | [82] |
| C. pruni | MBSZU 25-055 | PX225040 | PX225051 | PX226741 | this study |
| C. pruni | MBSZU 25-056 | PX225041 | PX225052 | PX226742 | this study |
| C. pruni | MBSZU 25-057 | PX225042 | PX225053 | PX226743 | this study |
| C. quercus | MFLU 18-2151 | MK347979 | MK347869 | - | [83] |
| C. quercus | MFLUCC 17-2342 | MN913681 | MT864311 | - | [60] |
| C. quercus | MFLUCC 17-2323 | MN913683 | MT864309 | - | [60] |
| C. quercus | SZU25-042 | PX225043 | PX225054 | - | this study |
| C. quercus | SZU25-043 | PX225044 | PX225055 | - | this study |
| C. submersa | MFLUCC 18-1283 | NG_073802 | MN913720 | - | [60] |
| C. submersa | MFLUCC 18-1409 | MN913719 | - | - | [60] |
| Delitschia chaetomioides | SMH 3253.2 | GU390656 | - | GU327753 | [84] |
| D. winteri | AFTOL-ID 1599 | DQ678077 | DQ678026 | DQ677922 | [85] |
| Fissuroma calami | MFLUCC 13-0836 | MF588993 | MF588983 | MF588975 | [59] |
| F. caryotae | MFLU 17-1253 | MF588996 | MF588986 | MF588979 | [59] |
| F. caryotae | SNT12 | MN712335 | MN699322 | MN744228 | [86] |
| F. caryotae | SZU25-040 | PX225038 | PX225049 | PX226739 | this study |
| F. maculans | MFLUCC 10-0886 | JN846724 | JN846734 | - | [79] |
| F. neoaggregatum | MFLUCC 10-0554 | KT955470 | KT955450 | KT955432 | [58] |
| F. palmae | MFLU 19-0820 | MN712336 | - | MN744229 | [86] |
| F. taiwanense | FU30861 | MG189605 | MG189607 | MG252072 | [87] |
| F. taiwanense | FU30862 | MG189606 | MG189608 | MG252073 | [87] |
| F. thailandicum | MFLUCC 11-0189 | KT955472 | KT955452 | KT955434 | [58] |
| F. wallichiae | MFLUCC 15-0315 | MN726235 | MN726247 | MN953045 | [88] |
| Neoastrosphaeriella aquatica | MFLUCC 18-0209 | MK138829 | MK138789 | MK132866 | [89] |
| N. aquatica | SNT240 | MN712338 | - | MN744231 | [86] |
| N. aquatica | SNT190 | MN712337 | MN699323 | MN744230 | [86] |
| N. aquatica | SZU25-047 | PX225039 | PX225050 | PX226740 | this study |
| N. krabiensis | MFLUCC 11-0025 | JN846729 | JN846739 | - | [79] |
| N. krabiensis | MFLUCC 11-0022 | JN846727 | JN846735 | - | [79] |
| N. phoenicis | MFLUCC 18-1477 | MN712339 | MN699324 | MN744232 | [86] |
| N. sribooniensis | MFLUCC 13-0834 | MF588997 | MF588987 | MF588977 | [59] |
| Pithomyces caryotae | MFLUCC 13-0828 | MF588999 | MF588989 | MF588978 | [59] |
| P. licualae | MFLUCC 17-2031 | MF588995 | MF588985 | [59] | |
| Pseudoastrosphaeriella africana | MFLUCC 11-0176 | KT955474 | KT955454 | KT955436 | [58] |
| P. aquatica | MFLUCC 18-0984 | MN913742 | MT864336 | MT954400 | [60] |
| P. aquatica | MFLUCC 18-0991 | MN325076 | - | MT954401 | [89] |
| P. aquatica | KUMCC 19-0096 | MN913746 | MT627731 | - | [60] |
| P. bambusae | MFLUCC 10-0885 | MN913695 | - | - | [60] |
| P. bambusae | KUMCC 19-0091 | MN913698 | - | MT954365 | [60] |
| P. bambusae | KUMCC 19-0093 | MN913699 | MT864301 | MT954363 | [60] |
| P. bambusae | KUMCC 19-0095 | MN913700 | - | MT954364 | [60] |
| P. bambusae | MFLUCC 11-0205 | KT955475 | KT955455 | KT955437 | [58] |
| P. longicolla | MFLUCC 11-0171 | KT955476 | - | KT955438 | [58] |
| P. thailandensis | MFLUCC 14-0038 | KT955479 | KT955458 | KT955441 | [58] |
| P. thailandensis | MFLUCC 11-0144 | KT955478 | KT955457 | KT955440 | [58] |
| P. thailandensis | MFLUCC 10-0553 | KT955477 | KT955456 | KT955439 | [58] |
| P. zingiberacearum | MBSZU 25-058 | PX225045 | PX225056 | PX226744 | this study |
| P. zingiberacearum | MBSZU 25-059 | PX225046 | PX225057 | PX226745 | this study |
| P. zingiberacearum | MBSZU 25-060 | PX225047 | PX225058 | PX226746 | this study |
| Pseudoastrosphaeriellopsis kaveriana | PUFD33 | MG947595 | MG947598 | MG968955 | [58] |
| Pteridiospora bambusae | MFLU 10-0071 | MG831565 | MG831566 | MG833012 | [90] |
| Pt. chiangraiensis | MFLUCC 11-0162 | KT955480 | KT955459 | KT955442 | [58] |
| Pt. javanica | MFLUCC 11-0195 | KJ742941 | KT955460 | KJ739606 | [58] |
| Pt. javanica | MFLUCC 11-0159 | KJ742940 | KJ739607 | KJ739605 | [91] |
| Quercicola fusiformis | MFUCC 18-0479 | MK348009 | MK347898 | MK360085 | [83] |
| Q. guttulospora | MFUCC 18-0481 | MK348010 | MK347899 | MK360086 | [83] |
| Xenoastrosphaeriella aquatica | DLUCC 1525 | MZ420753 | MZ420755 | MZ442701 | [92] |
| X. tornata | MFLUCC 11-0196 | KT955467 | KT955447 | KT955429 | [58] |
| X. trochus | KUMCC 18-0194 | MT659668 | MT659669 | MT653597 | [93] |
3. Results
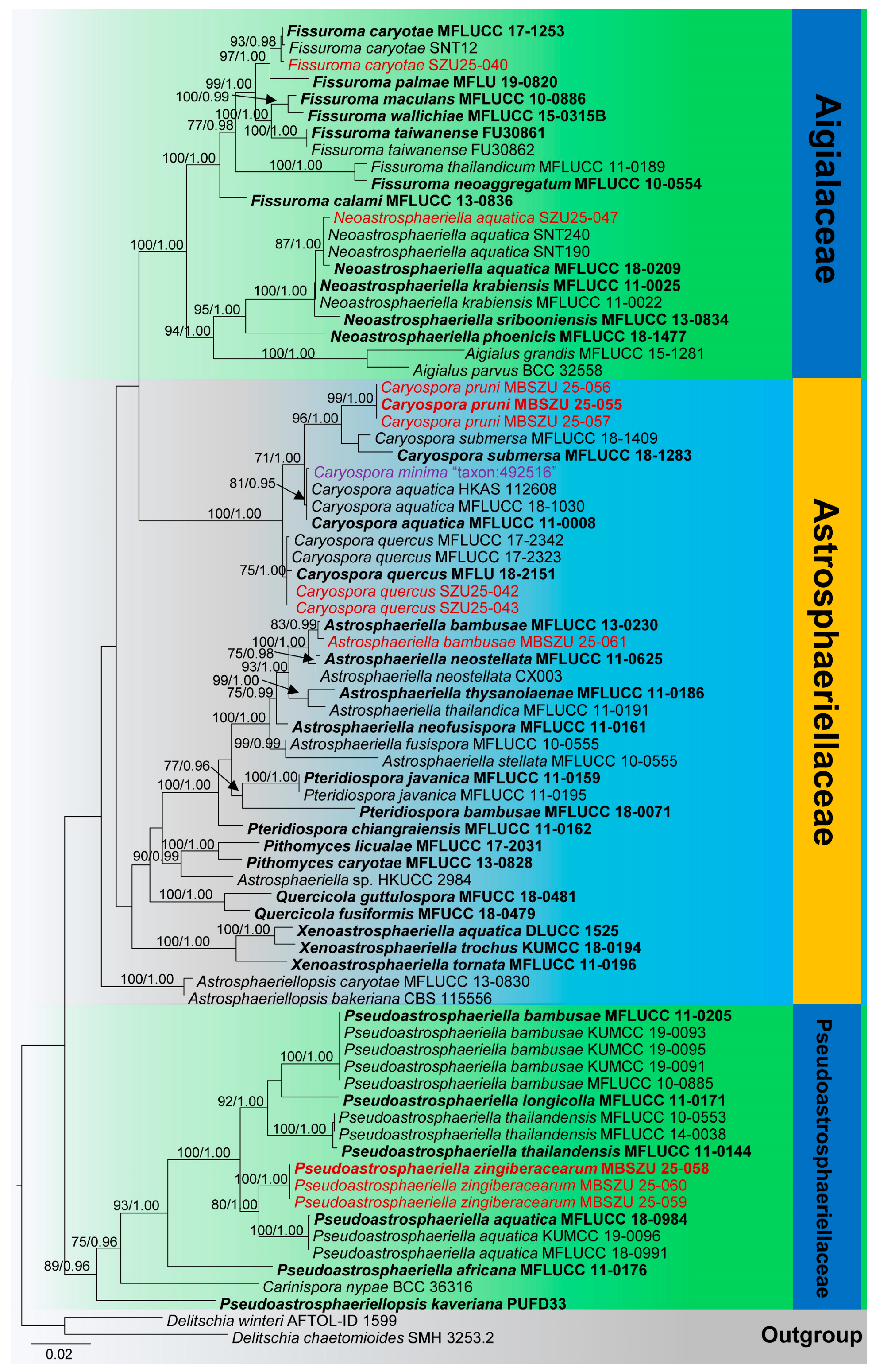
3.1. Phylogenetic Analyses
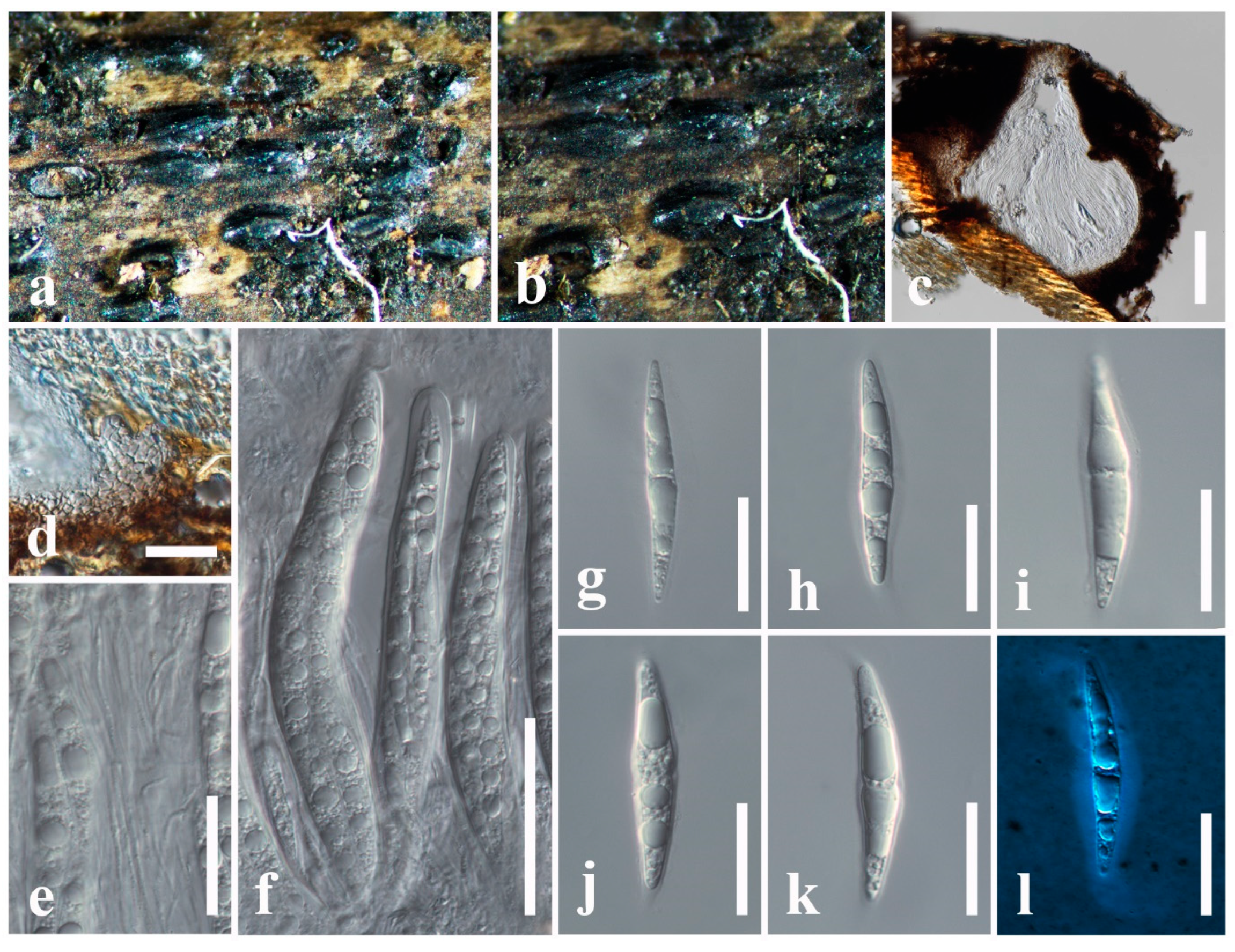
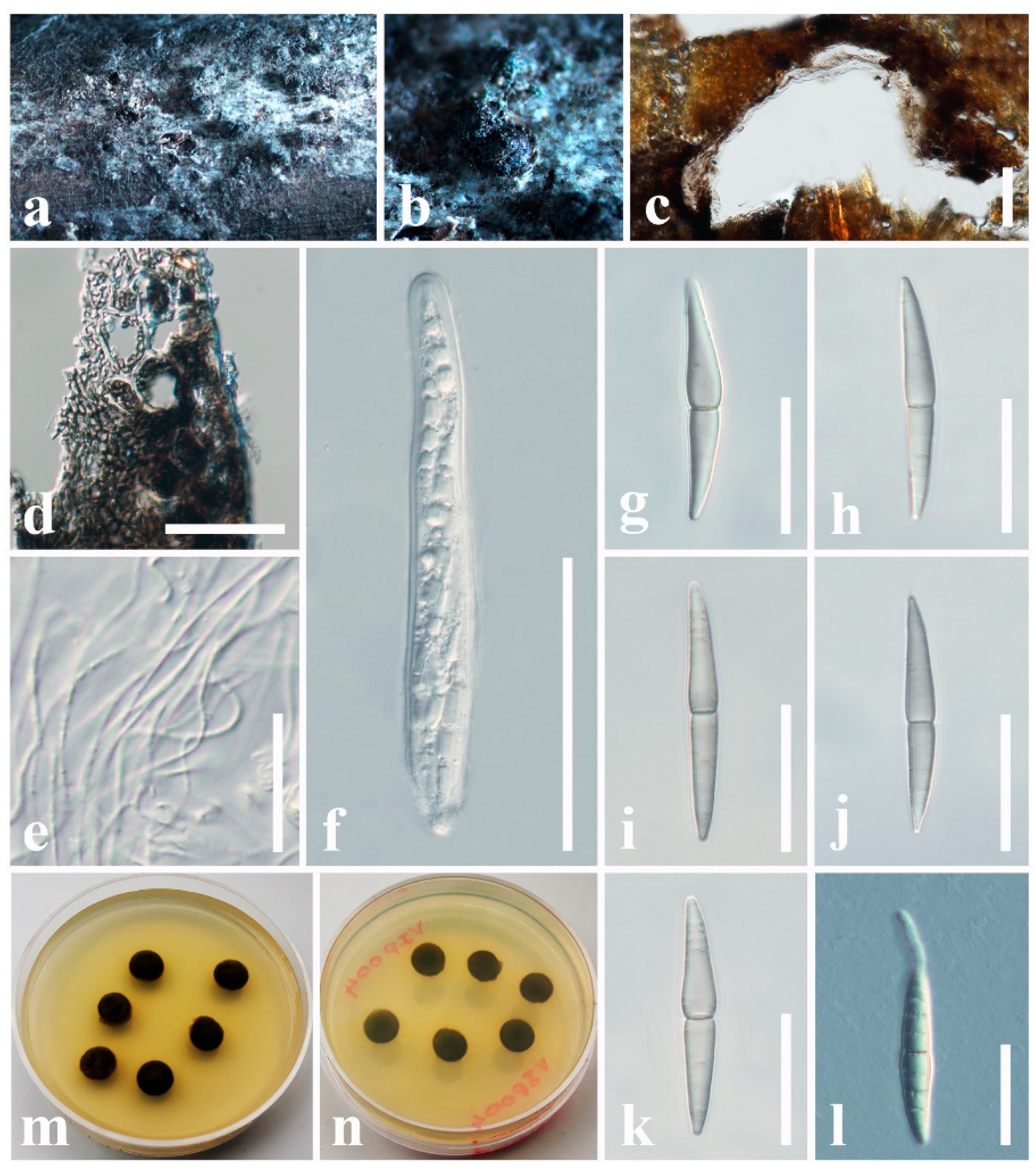
3.2. Taxonomy
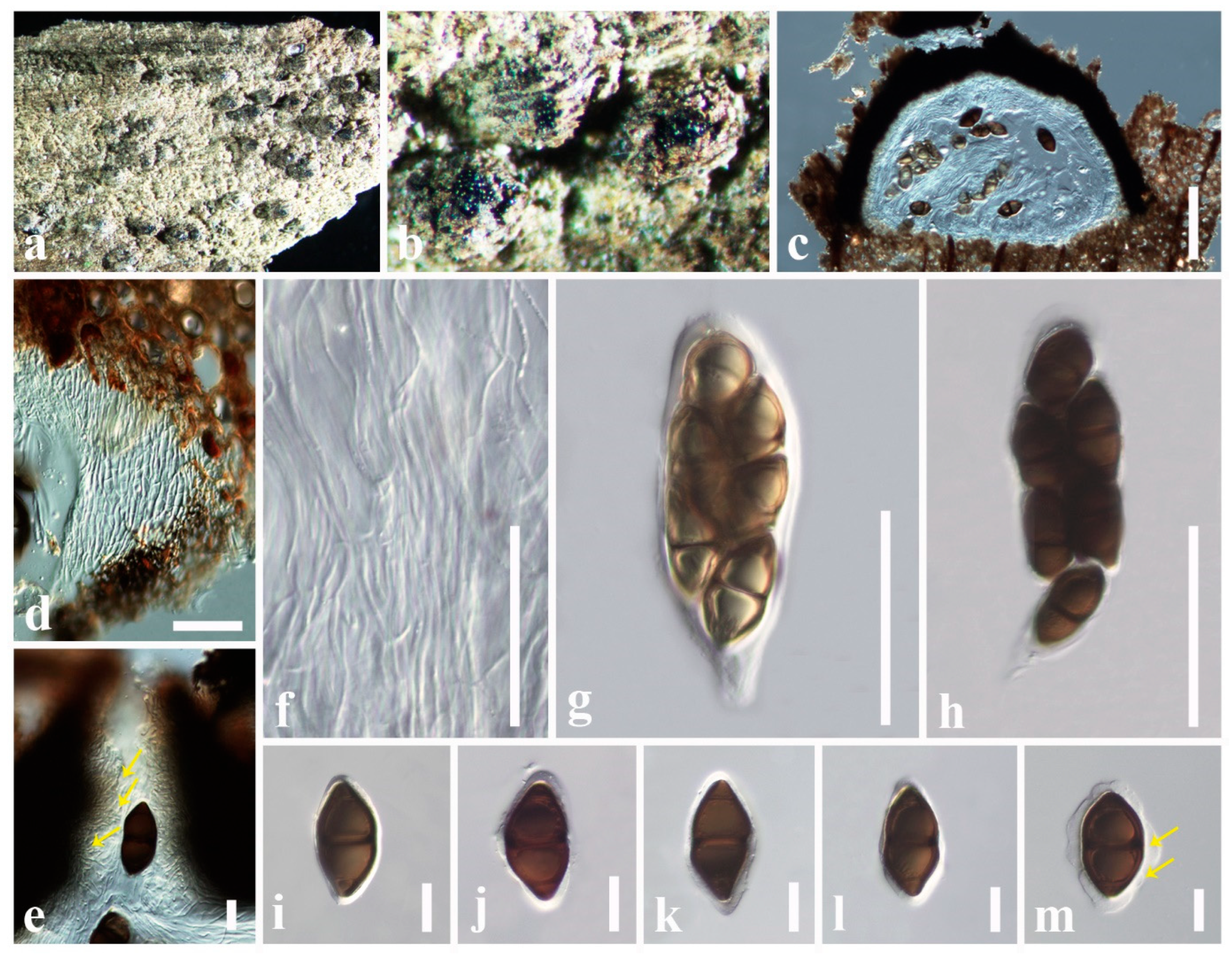
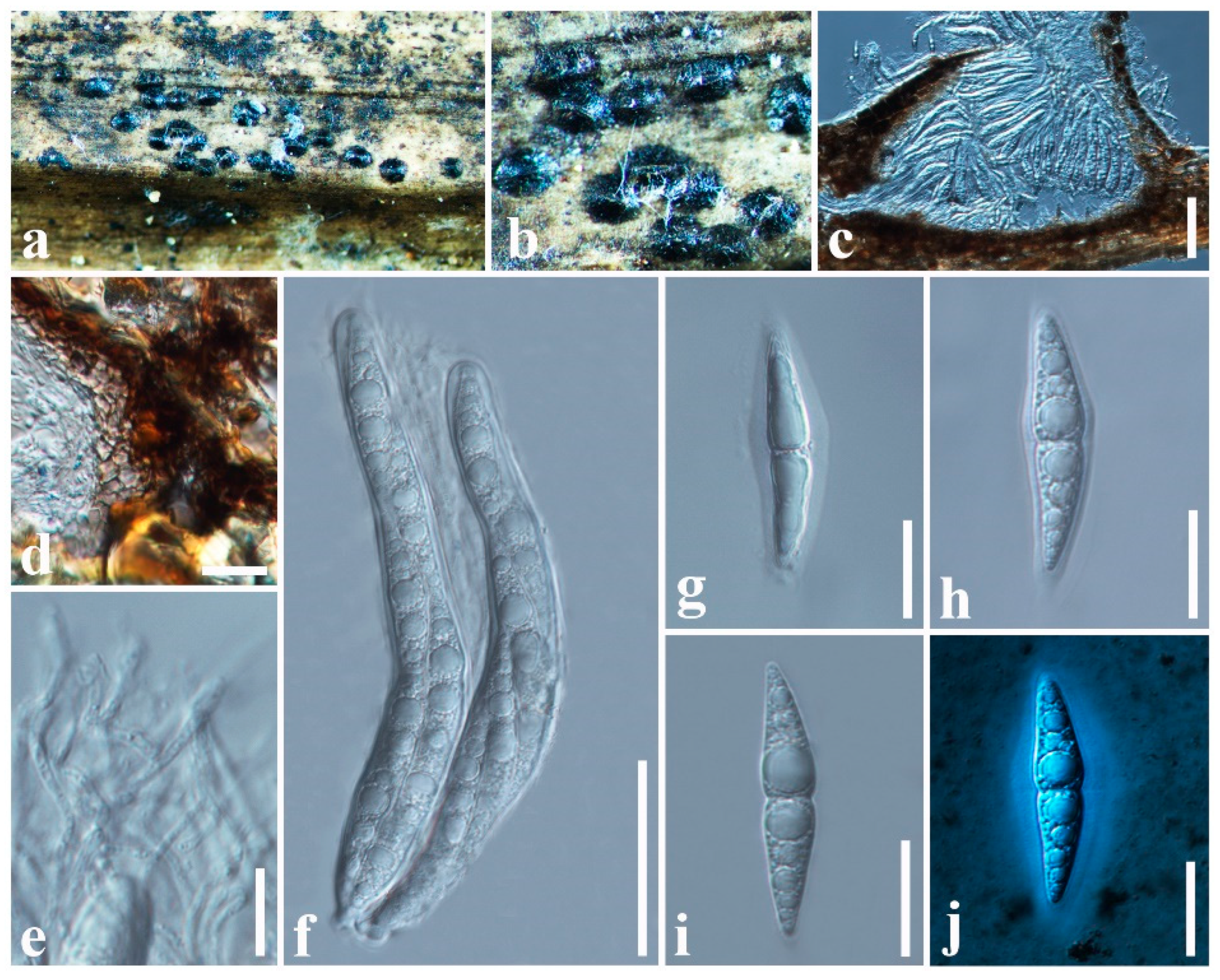
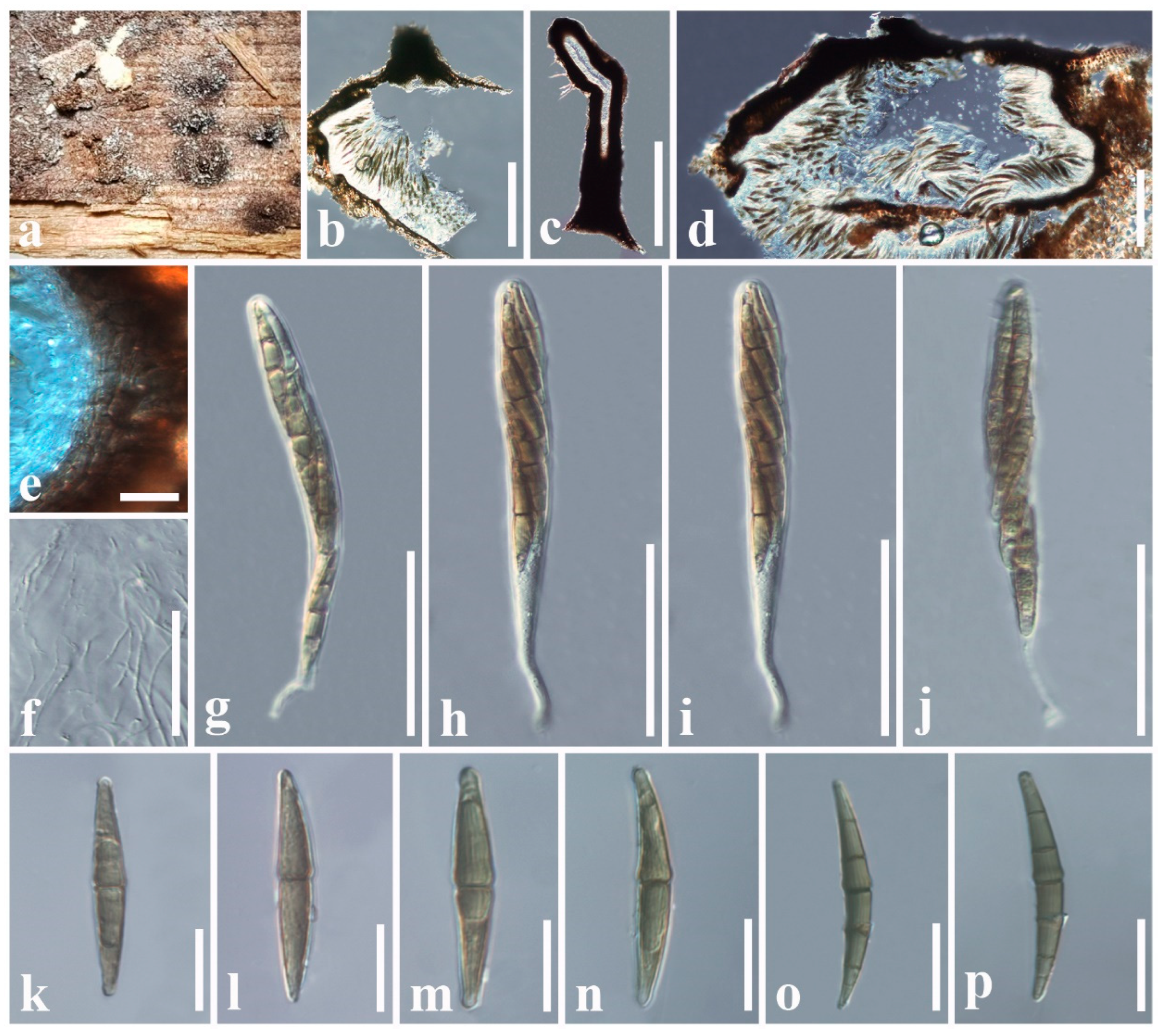
3.2.1. Aigialaceae Suetrong, Sakay., E.B.G. Jones, Kohlm., Volkm.-Kohlm. & C.L. Schoch, Stud. Mycol. 64: 166 (2009)
3.2.2. Astrosphaeriellaceae Phookamsak & K.D. Hyde, Fungal Diversity 74: 161 (2015)
3.2.3. Pseudoastrosphaeriellaceae Phookamsak & K.D. Hyde, Fungal Diversity: 181 (2015)
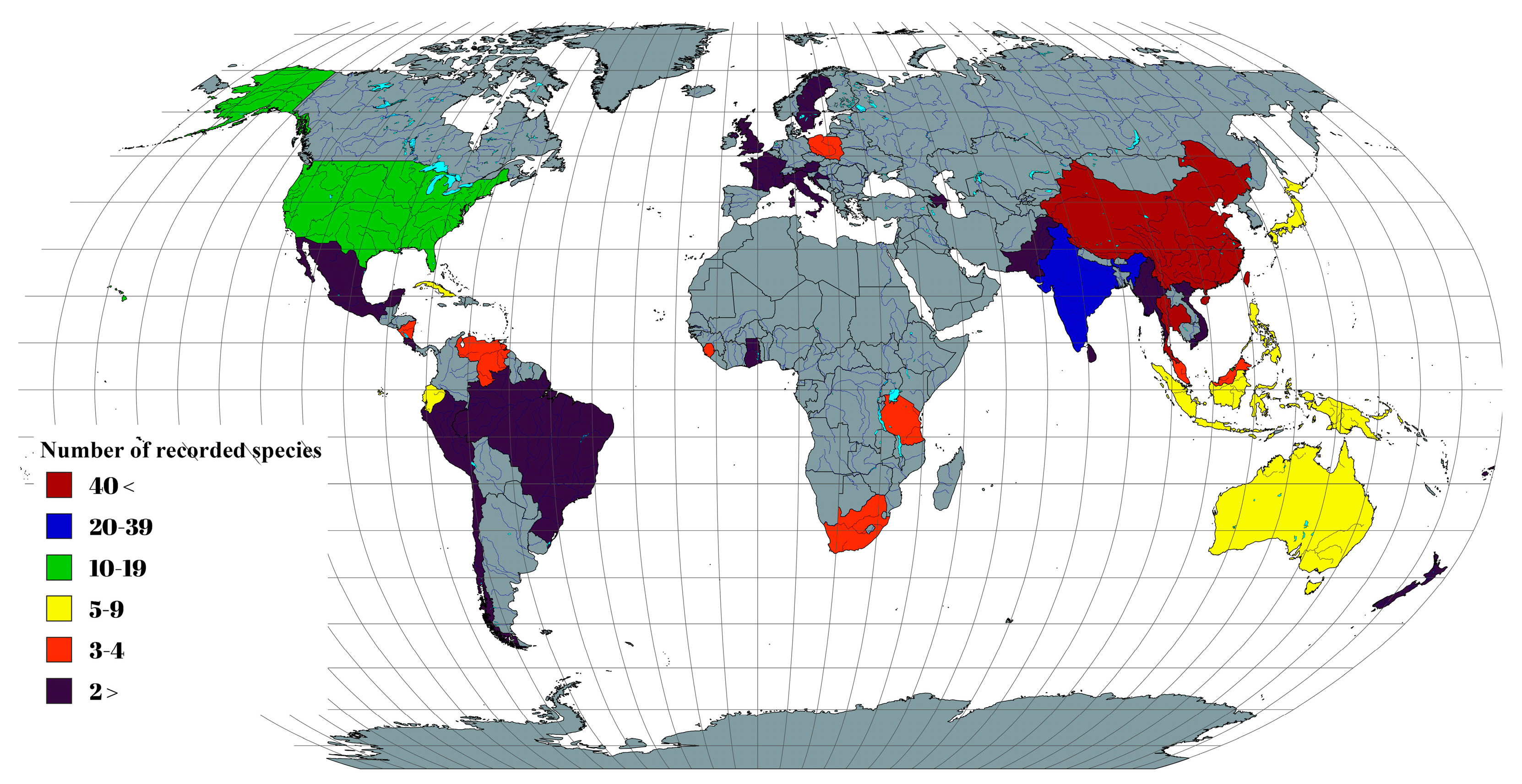
3.3. Geographical Distribution and Their Host Occurrences of Aigialaceae, Astrosphaeriellaceae, and Pseudoastrosphaeriellaceae Species
3.3.1. Aigialaceae
3.3.2. Astrosphaeriellaceae
3.3.3. Pseudoastrosphaeriellaceae
| Species | Host | Family | Locality | References |
| Aigialus grandis | Rhizophora mangle, Rhizophora mucronata, Sonneratia acida | Lythraceae, Rhizophoraceae | The United States, India | [120,121,122] |
| Aigialus mangrovis | Rhizophora mucronata | Rhizophoraceae | India | [121,123] |
| Aigialus parvus | Avicennia alba, Rhizophora mangle, Sonneratia acida | Lythraceae, Rhizophoraceae | The United States | [121,122] |
| Aigialus rhizophorae | Rhizophora mucronata | Rhizophoraceae | India | [121,123] |
| Aigialus striatisporus | Rhizophora apiculata | Rhizophoraceae | Thailand | [73] |
| Fissuroma aggregatum | Phyllostachys reticulata | Poaceae | Japan | [95,124,125,126,127] |
| Fissuroma arengae | Arenga pinnata | Arecaceae | Thailand | [88] |
| Fissuroma bambusae | Bambusa spp. | Poaceae | Thailand | [58] |
| Fissuroma bambusicola | Bambusa spp. | Poaceae | China | [128] |
| Fissuroma calami | Calamus rotang | Arecaceae | Thailand | [59] |
| Fissuroma caryotae | Arenga undulatifolia Caryota urens | Arecaceae | China | [59], this study |
| Fissuroma chinense | Bambusa spp. | Poaceae | China | [66] |
| Fissuroma fissuristomum | Bambusa spp. Calamus conirostris, Licuala sp., Mauritia flexuosa | Arecaceae, Poaceae | Australia, Brunei, China, Ecuador, Papua New Guinea | [73,107,129] |
| Fissuroma kavachabeejae | Calamus andamanicus | Arecaceae | India | [105] |
| Fissuroma maculans | Arenga spp. | Arecaceae | Philippines | [79,130] |
| Fissuroma microsporum | Borassus flabellifer | Arecaceae | India | [105] |
| Fissuroma neoaggregatum | Bambusa spp. | Poaceae | Thailand | [58] |
| Fissuroma palmae | Arenga pinnata | Arecaceae | Thailand | [86] |
| Fissuroma taiwanense | Hedychium coronarium | Zingiberaceae | China | [87]) |
| Fissuroma thailandicum | Bambusa spp. | Poaceae | Thailand | [58] |
| Fissuroma wallichiae | Wallichia sp. | Theaceae | Thailand | [88] |
| Neoastrosphaeriella alankrithabeejae | Calamus andamanicus | Arecaceae | India | [105] |
| Neoastrosphaeriella aquatica | Phoenix paludosa | Arecaceae | China, Thailand | [89], this study |
| Neoastrosphaeriella krabiensis | Metroxylon sagu | Arecaceae | Thailand | [79] |
| Neoastrosphaeriella phoenicis | Phoenix paludosa | Arecaceae | Thailand | [86] |
| Neoastrosphaeriella sribooniensis | Calamus rotang | Arecaceae | Thailand | [59] |
| Posidoniomyces atricolor | Posidonia oceanica | Posidoniaceae | Croatia | [131] |
| Rimora mangrovei | Avicennia officinalis, Rhizophora mangle | Rhizophoraceae, Verbenaceae | The United States, India | [73,121,122] |
| Aquatospora cylindrica | Unknown host | - | Thailand | [60] |
| Astrosphaeriella angustispora | Eleiodoxa conferta, Licuala spp. | Arecaceae | Brunei | [119,132,133] |
| Astrosphaeriella aosimensis | Livistona subglobosa | Arecaceae | Japan | [134,135] |
| Astrosphaeriella applanata | Alnus sp., Carpinus sp., Quercus spp. | - | Poland, Sweden, the United Kingdom, | [73,109,136] |
| Astrosphaeriella aquatica | Licuala longicalycata, Livistona spp. | Arecaceae | Ecuador, Papua New Guinea, Thailand | [73,119,133,137] |
| Astrosphaeriella asiana | Aegiceras corniculatum, Sonneratia alba | Lythraceae, Primulaceae | Thailand | [119,138] |
| Astrosphaeriella australiensis | Calamus spp. | Arecaceae | Australia | [119] |
| Astrosphaeriella bambusae | Bambusa spp., Metroxylon sagu | Arecaceae, Poaceae | China, Thailand | [58], this study |
| Astrosphaeriella bambusella | Bambusa spp. | Poaceae | Indonesia | [126,139] |
| Astrosphaeriella callicarpa | Unknown host | - | Indonesia | [140] |
| Astrosphaeriella daemonoropis | Daemonorops margaritae | Arecaceae | China | [115,129] |
| Astrosphaeriella erumpens | Unknown host | Arecaceae | Cuba | [141] |
| Astrosphaeriella exorrhiza | Iriartea sp. | Arecaceae | Venezuela | [119,142] |
| Astrosphaeriella floridana | Sabal palmetto | Arecaceae | The United States, Thailand | [133,143] |
| Astrosphaeriella frondicola | Calamus spp., Daemonorops spp., Laccospadix australasica, Oraniopsis appendiculata | Arecaceae | Australia, Brunei, China | [73,115,119,144] |
| Astrosphaeriella fuscomaculans | Phyllostachys nigra | Poaceae | Japan | [73,125,126] |
| Astrosphaeriella fusispora | Bambusa spinosa, Phyllostachys bambusoides, Pleioblastus pubescens | Poaceae | Japan, Philippines | [73,125,126,130,145] |
| Astrosphaeriella gaofengensis | Bambusa spp. | Poaceae | China | [66] |
| Astrosphaeriella immersa | Archontophoenix alexandrae | Arecaceae | China | [115,129] |
| Astrosphaeriella lageniformis | Cocos nucifera | Arecaceae | China | [73,146] |
| Astrosphaeriella lenticularis | Geonoma sp. | Arecaceae | Brunei, Ecuador | [73,119] |
| Astrosphaeriella linguiformis | Bambusa spp. | Poaceae | China | [147] |
| Astrosphaeriella livistonicola | Livistona chinensis | Arecaceae | China, Thailand | [73,115,129,133] |
| Astrosphaeriella longispora | Unknown host | - | Costa Rica | [148] |
| Astrosphaeriella lophiostomopsis | Arenga undulatifolia | Arecaceae | Brunei, Thailand | [119,133] |
| Astrosphaeriella macrospora | Miscanthus spp. | Poaceae | China | [149] |
| Astrosphaeriella malayensis | Daemonorops sp., Licuala longicalycata | Arecaceae | Malaysia, Papua New Guinea, Thailand | [119,129,133] |
| Astrosphaeriella maquilingiana | Calamus spp., Iriartea sp. | Arecaceae | Australia, Ecuador, Philippines | [73,119] |
| Astrosphaeriella mauritiae | Mauritia flexuosa | Arecaceae | Ecuador | [119] |
| Astrosphaeriella minima | Bambusa spp. | Poaceae | China, Indonesia | [140,150] |
| Astrosphaeriella minoensis | Licuala ramsayi, Phyllostachys reticulata, Sasa kurilensis | Arecaceae, Poaceae | Australia, Japan | [119,142,150] |
| Astrosphaeriella neofusispora | Bambusa spp. | Poaceae | Thailand | [58] |
| Astrosphaeriella neostellata | Bambusa spp. | Poaceae | Thailand | [58] |
| Astrosphaeriella nipicola | Nipa sp. | Arecaceae | Indonesia | [119] |
| Astrosphaeriella nypae | Bambusa spp., Nypa fruticans, Phoenix hanceana | Arecaceae, Poaceae | Brunei, China | [73,115,119] |
| Astrosphaeriella pallidipolaris | Unknown host | - | China | [149] |
| Astrosphaeriella papuana | Bambusa spp. | Poaceae | Papua New Guinea | [126,150] |
| Astrosphaeriella picea | Unknown host | - | - | [151] |
| Astrosphaeriella pinicola | Pinus spp. | Pinaceae | Austria | [152] |
| Astrosphaeriella polymorpha | Ulmus spp. | Ulmaceae | The United States | [153] |
| Astrosphaeriella roseobrunnea | Bambusa spp. | Poaceae | China | [66] |
| Astrosphaeriella seychellensis | Unknown host | - | Seychelles | [154] |
| Astrosphaeriella splendida | Arundinaria hindsii, Astrocaryum sp., Iriartea sp., Jessenia bataua, Mauritia flexuosa | Arecaceae, Poaceae | China, Ecuador | [73,107,119] |
| Astrosphaeriella stellata | Bambusa spp., Calamus spp., Dendrocalamus spp., Phyllostachys heterocycla, Thysanolaena maxima | Arecaceae, Poaceae | Australia, China, India, Japan, Papua New Guinea, Philippines, Vietnam | [107,115,119,127,150,155,156,157] |
| Astrosphaeriella striaspora | Valota insularis | Poaceae | Venezuela | [119] |
| Astrosphaeriella sundarbanensis | Sonneratia apetala | Lythraceae | India | [73] |
| Astrosphaeriella thailandensis | Unknown host | - | Thailand | [158] |
| Astrosphaeriella thailandica | Bambusa spp. | Poaceae | Thailand | [58] |
| Astrosphaeriella thysanolaenae | Thysanolaena maxima | Poaceae | Thailand | [58] |
| Astrosphaeriella trochus | Bambusa spp., Phragmites spp., Phyllostachys spp. | Poaceae | China, Indonesia, South Africa | [73,107,119] |
| Astrosphaeriella uberina | Unknown host | Arecaceae | France, Nicaragua | [73,119] |
| Astrosphaeriella uniseptata | Miliusa tectona | Annonaceae | India | [159] |
| Astrosphaeriella vaginata | Bactris baculifera | Arecaceae | Mexico | [160] |
| Astrosphaeriella venezuelensis | Bambusa spp. | Poaceae | Venezuela | [126] |
| Astrosphaeriella vesuvius | Calamus spp., Daemonorops oxycarpus, Korthalsia sp., Licuala sp. | Arecaceae | Australia, Brunei, Indonesia, Thailand, Malaysia, Papua New Guinea, Sri Lanka | [73,119] |
| Astrosphaeriella yunnanensis | Bambusa spp. | Poaceae | China | [161] |
| Astrosphaeriellopsis bakeriana | Livistona sinensis | Arecaceae | Singapore | [162] |
| Astrosphaeriellopsis caryotae | Caryota sp. | Arecaceae | Thailand | [59] |
| Caryospora aquatica | Unknown host | - | Thailand | [80] |
| Caryospora australiensis | Unknown host | - | Australia | [163] |
| Caryospora coffeae | Coffea sp. | Rubiaceae | Venezuela | [164] |
| Caryospora daweiensis | Melocalamus compactiflorus | Poaceae | China | [165] |
| Caryospora langloisii | Arundinaria sp., Grewia asiatica | Malvaceae, Poaceae | India, The United States | [126,166,167,168] |
| Caryospora masonii | Eugenia caryophyllus | Myrtaceae | Tanzania | [169] |
| Caryospora minima | Amygdalus persica, Prunus persica | Rosaceae | The United States | [168] |
| Caryospora obclavata | Unknown host | - | [104] | |
| Caryospora phyllostachydis | Phyllostachys bambusoides | Poaceae | Japan | [135] |
| Caryospora pruni | Prunus persica | Rosaceae | China | this study |
| Caryospora quercus | Citus maxima, Quercus sp. | Fagaceae, Rutaceae | China, Thailand | [60,83], this study |
| Caryospora submersa | Unknown host | - | Thailand | [60] |
| Javaria samuelsii | Nothofagus sp. | Arecaceae, Nothofagaceae | Brazil, New Zealand | [170,171] |
| Javaria shimekii | Unknown host | - | Nicaragua | [143] |
| Mycopepon bambusae | Babmbusa spp. | Poaceae | China | [172] |
| Mycopepon fusoidisporus | Babmbusa spp. | Poaceae | China | [172] |
| Mycopepon smithii | Unknown host | - | Nicaragua | [170] |
| Pithomyces africanus | Borassus spp., Ficus spp., Hyphaene thebaica, Musa balbisiana, Ophiopogon japonicus, Trachelospermum jasminoides | Apocynaceae, Arecaceae, Asparagaceae, Moraceae, Musaceae | China, Ghana, Sierra Leone | [73,110,111] |
| Pithomyces alabamensis | Quercus spp. | Fagaceae | The United States | [173] |
| Pithomyces arecastri | Arecastrum romanzoffianum | Arecaceae | China | [174] |
| Pithomyces bulbilus | Eucalyptus spp. | Myrtaceae | India | [175] |
| Pithomyces caryotae | Caryota sp. | Arecaceae | Thailand | [59] |
| Pithomyces cateniformis | Wisteria sinensis | Fabaceae | China | [110] |
| Pithomyces cinnamomeus | Unknown host | - | Cuba | [176] |
| Pithomyces clavisporopsis | Lilium amoenum | Liliaceae | China | [110] |
| Pithomyces clavisporus | Unknown host | - | The United States | [177] |
| Pithomyces cupaniae | Actinodaphne angustifolia, Albizia ferruginea, Anisophyllea laurina, Carpodinus hirsuta, Clitandra sp., Cupania guatemalensis, Funtumia africana, Jasminum dichotomum, Millettia pallens, Sorindeia juglandifolia | Anacardiaceae, Anisophylleaceae, Apocynaceae, Fabaceae, Lauraceae, Oleaceae, Sapindaceae | Costa Rica, Myanmar, Sierra Leone | [73,111,178] |
| Pithomyces dimorphosporus | Unknown host | - | Brazil | [179] |
| Pithomyces divaricatus | Casearia tomentosa | Salicaceae | India | [180] |
| Pithomyces djbhatii | Unknown host | Arecaceae | India | [73] |
| Pithomyces elaeidicola | Elaeis guineensis, Phoenix canariensis, Trachycarpus spp. | Arecaceae | China, Sierra Leone, Tanzania | [73,110,111,181] |
| Pithomyces ellipticus | Lagerstroemia speciosa, Trachycarpus fortunei | Arecaceae, Lythraceae | China | [73,110] |
| Pithomyces ellisii | Butea monosperma, Eucalyptus sp. | Fabaceae, Myrtaceae | India | [182] |
| Pithomyces flavus | Oncosperma sp. | Arecaceae | Sri Lanka | [73,111] |
| Pithomyces gladioli | Gladiolus communis | Iridaceae | China | [73,110] |
| Pithomyces graminicola | Arachis hypogaea, Canna spp., Dendrocalamus sp., Mangifera indica, Panicum spp., Persea americana, Phyllostachys spp., Ravenala spp., Saccharum spp., Sporobolus fertilis | Anacardiaceae, Cannaceae, Fabaceae, Lauraceae, Poaceae, Strelitziaceae | China, Fiji, India, South Africa | [73,110,111,112,113,114,115] |
| Pithomyces helminthosporioides | Ficus elastica | Moraceae | China | [73,110] |
| Pithomyces hyalosporus | Unknown host | - | India | [159] |
| Pithomyces leprosus | Faurea saligna | Proteaceae | Tanzania | [183] |
| Pithomyces licualae | Licuala sp. | Arecaceae | China | [59] |
| Pithomyces longiclavisporus | Soil | - | China | [117] |
| Pithomyces longipes | Bambusa ventricosa | Poaceae | China | [174] |
| Pithomyces musae | Musa wilsonii | Musaceae | China | [73] |
| Pithomyces niger | Bambusa spp. | Poaceae | Cuba | [184] |
| Pithomyces obpyriformis | Lagerstroemia speciosa | Lythraceae | China | [174] |
| Pithomyces obscuriseptatus | Acorus calamus, Butomus umbellatus, Carex spp., Cyperus fuscus, Eleocharis spp., Juncus spp., Sparganium spp. | Acoraceae, Arecaceae, Butomaceae, Cyperaceae, Juncaceae, Typhaceae | Peru, Poland | [73,116] |
| Pithomyces pallidus | Soil | - | China | [117] |
| Pithomyces prolatus | Pithecellobium cubense | Fabaceae | Cuba | [185] |
| Pithomyces pulvinatus | Ficus microcarpa, Phoenix sp., Setaria pumila | Arecaceae, Fabaceae, Moraceae, Poaceae | China, Indonesia, Pakistan, Poland | [73,109,110,186] |
| Pithomyces quadratus | Crataegus sp. | Rosaceae | The United States | [187] |
| Pithomyces saccharicola | Saccharum officinarum | Poaceae | China | [174] |
| Pithomyces sivaramaprasadii | Unknown host | - | India | [188] |
| Pithomyces subramanianii | Unknown host | - | India | [73] |
| Pithomyces sumiderensis | Unknown host | - | Cuba | [176] |
| Pithomyces taiwanensis | Arecastrum romanzoffianum | Arecaceae | China | [73,110,189] |
| Pithomyces trachelospermi | Trachelospermum jasminoides | Apocynaceae | China | [174] |
| Pithomyces valparadisiacus | Calopsis spp., Leucadendron sp., Puya chilensis, Puya coerulea, Rhodocoma capensis | Bromeliaceae, Proteaceae, Restionaceae | Chile, South Africa | [73,190,191] |
| Pithomyces variegatae | Bauhinia variegata | Fabaceae | China | [174] |
| Pteridiospora bambusae | Bambusa sp. | Poaceae | China | [90] |
| Pteridiospora chiangraiensis | Bambusa sp. | Poaceae | Thailand | [58] |
| Pteridiospora chochrjakovii | Quercus pedunculiflora | Fagaceae | Azerbaijan | [73] |
| Pteridiospora javanica | Bambusa sp. | Poaceae | Indonesia, Thailand | [91,126] |
| Pteridiospora munkii | Phoenix sylvestris | Arecaceae | India | [121] |
| Pteridiospora spinosispora | Fraxinus pennsylvanica, Liquidambar styraciflua | Altingiaceae, Oleaceae | The United States | [192] |
| Quercicola fusiformis | Quercus sp. | Fagaceae | Thailand | [83] |
| Quercicola guttulospora | - | Fagaceae | Thailand | [83] |
| Xenoastrosphaeriella aquatica | Unknown host | - | China | [193] |
| Xenoastrosphaeriella tornata | Bambusa sp. | Poaceae | Thailand | [58,194] |
| Carinispora nypae | Licuala longicalycata, Nypa fruticans | Arecaceae | Brunei, Thailand | [97,133,195] |
| Carinispora velatispora | Oncosperma tigillarium | Arecaceae | Brunei | [137] |
| Pseudoastrosphaeriella aequatoriensis | Phytelephas sp. | Arecaceae | Ecuador | [73,119] |
| Pseudoastrosphaeriella africana | Arenga undulatifolia, Calamus spp., Daemonorops sp., Phragmites australis | Arecaceae, Poaceae | Australia, Brunei, Malaysia, Philippines, Tanzania | [119,142] |
| Pseudoastrosphaeriella aquatica | Unknown host | - | Thailand | [60] |
| Pseudoastrosphaeriella bambusae | Bambusa spp. | Poaceae | Thailand | [58] |
| Pseudoastrosphaeriella longicolla | Bambusa spp. | Poaceae | Thailand | [58] |
| Pseudoastrosphaeriella papillata | Bambusa spp. | Poaceae | Brunei | [119] |
| Pseudoastrosphaeriella thailandensis | Bambusa spp. | Poaceae | Thailand | [119] |
| Pseudoastrosphaeriellopsis kaveriana | Avicennia marina | Arecaceae | India | [58] |
| Pseudoastrosphaeriella zingiberacearum | Hedychium coronarium | Zingiberaceae | China | this study |
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hawksworth, D.L. The fungal dimension of biodiversity—Magnitude, significance, and conservation. Mycol. Res. 1991, 95, 641–655. [Google Scholar] [CrossRef]
- Hammond, P.M. Species inventory. In Global Biodiversity: Status of the Earth’s Living Resources; Groombridge, B., Ed.; Chapman and Hall: London, UK, 1992; pp. 17–39. [Google Scholar]
- Rossman, A.Y. Strategy for an all-taxa inventory of fungal biodiversity. In Biodiversity and Terrestrial Ecosystems; Peng, C.I., Chou, C.H., Eds.; Academia Sinica Monograph Series No. 14; Institute of Botany: Taipei, Taiwan, 1994; pp. 169–194. [Google Scholar]
- May, R.M. The dimensions of life on earth. In Nature and Human Society: The Quest for a Sustainable World; Raven, P.H., Williams, T., Eds.; National Academy Press: Washington, DC, USA, 2000; pp. 30–45. [Google Scholar]
- Blackwell, M. The Fungi: 1, 2, 3…5.1 million species? Am. J. Bot. 2011, 98, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Hawksworth, D.L.; Lücking, R. Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol. Spectr. 2017, 5, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Hussain, M.; Zhang, W.; Stadler, M.; Liu, X.; Xiang, M. Current insights into fungal species diversity and perspective on naming the environmental DNA sequences of fungi. Mycology 2019, 10, 127–140. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Saleh, A.; Aumentado, H.D.R.; Boekhout, T.; Bera, I.; Khyaju, S.; Bhunjun, C.S.; Chethana, K.T.; Phukhamsakda, C.; Doilom, M.; et al. Fungal numbers: Global needs for a realistic assessment. Fungal Divers. 2024, 128, 191–225. [Google Scholar] [CrossRef]
- Hammond, P.M. The current magnitude of biodiversity. In Global Biodiversity Assessment; Heywood, V., Ed.; Cambridge University Press: Cambridge, UK, 1995; pp. 113–138. [Google Scholar]
- Cannon, P.F. Diversity of the Phyllachoraceae with special reference to the tropics. In Biodiversity of Tropical Microfungi; Hyde, K.D., Ed.; Hong Kong University Press: Hong Kong, 1997; pp. 255–278. [Google Scholar]
- Arnold, A.E.; Maynard, Z.; Gilbert, G.S.; Coley, P.D.; Kursar, T.A. Are tropical fungal endophytes hyperdiverse? Ecol. Lett. 2000, 3, 267–274. [Google Scholar] [CrossRef]
- Hawksworth, D.L. The magnitude of fungal diversity: The 1.5 million species estimate revisited. Mycol. Res. 2001, 105, 1422–1432. [Google Scholar] [CrossRef]
- Brien, H.E.; Parrent, J.L.; Jackson, J.A.; Moncalvo, J.M.; Vilgalys, R. Fungal community analysis by large-scale sequencing of environmental samples. Appl. Environ. Microb. 2005, 71, 5544–5550. [Google Scholar] [CrossRef]
- Schmit, J.P.; Mueller, G.M. An estimate of the lower limit of global fungal diversity. Biodivers. Conserv. 2007, 16, 99–111. [Google Scholar] [CrossRef]
- Hawksworth, D.L. Global species numbers of fungi: Are tropical studies and molecular approaches contributing to a more robust estimate? Biodivers. Conserv. 2012, 21, 2425–2433. [Google Scholar] [CrossRef]
- Baldrian, P.; Větrovský, T.; Lepinay, C.; Kohout, P. High-throughput sequencing view on the magnitude of global fungal diversity. Fungal Divers. 2022, 114, 539–547. [Google Scholar] [CrossRef]
- Baldrian, P.; Kohout, P.; Větrovský, T. Global fungal diversity estimated from high-throughput sequencing. In Evolution of Fungi and Fungal-Like Organisms; Springer International Publishing: Cham, Switzerland, 2023; pp. 227–238. [Google Scholar] [CrossRef]
- Djemiel, C.; Dequiedt, S.; Horrigue, W.; Bailly, A.; Lelièvre, M.; Tripied, J.; Guilland, C.; Perrin, S.; Comment, G.; Saby, N.P.; et al. Unraveling biogeographical patterns and environmental drivers of soil fungal diversity at the French national scale. Soil 2024, 10, 251–273. [Google Scholar] [CrossRef]
- Martin, G.W. The numbers of fungi. Proc. Iowa Acad. Sci. 1951, 58, 175–178. [Google Scholar]
- Bisby, G.R.; Ainsworth, G.C. The numbers of fungi. Br. Mycol. Soc. 1943, 26, 16–19. [Google Scholar] [CrossRef]
- Pascoe, I. History of Systematic Mycology in Australia; Short, P.S., Ed.; Australian Systematic Botany Society: South Yarra, Australia, 1990; pp. 259–264. [Google Scholar]
- Smith, D.; Waller, J.M. Culture collections of microorganisms: Their importance in tropical plant pathology. Fitopatol. Bras. 1992, 17, 1–8. [Google Scholar]
- Hywel-Jones, N. A systematic survey of insect fungi from natural tropical forest in Thailand. In Aspects of Tropical Mycology; Isaac, S., Frankland, J.C., Watling, R., Whalley, A.J.S., Eds.; Cambridge University Press: Cambridge, UK, 1993; pp. 300–301. [Google Scholar]
- Dreyfuss, M.M.; Chapela, I.H. Potential of fungi in the discovery of novel, low-molecular weight pharmaceuticals. In The Discovery of Natural Products with Therapeutic Potential; Gullo, V., Ed.; Butterworth Heinemann: London, UK, 1994; pp. 49–80. [Google Scholar]
- Shivas, R.G.; Hyde, K.D. Biodiversity of plant pathogenic fungi in the tropics. In Biodiversity of Tropical Microfungi; Hyde, K.D., Ed.; Hong Kong University Press: Hong Kong, 1997; pp. 47–56. [Google Scholar]
- Guzman, G. Inventorying the fungi of Mexico. Biodivers. Conserv. 1998, 7, 369–384. [Google Scholar] [CrossRef]
- Fröhlich, J.; Hyde, K.D.; Guest, D.I. Fungi associated with leaf spots of palms in north Queensland, Australia. Mycol. Res. 1997, 101, 721–732. [Google Scholar] [CrossRef]
- Aptroot, A. Lichenized and saprobic fungal biodiversity of a single Elaeocarpus tree in Papua New Guinea, with the report of 200 species of ascomycetes associated with one tree. Fungal Divers. 2001, 6, 1–11. [Google Scholar]
- de Meeûs, T.; Renaud, F. Parasites within the new phylogeny of eukaryotes. Trends Parasitol. 2002, 18, 247–251. [Google Scholar] [CrossRef]
- Crous, P.W.; Rong, I.H.; Wood, A.; Lee, S.; Glen, H.; Botha, W.; Slippers, B.; de Beer, W.Z.; Wingfield, M.J.; Hawksworth, D.L. How many species of fungi are there at the tip of Africa? Stud. Mycol. 2006, 55, 13–33. [Google Scholar] [CrossRef]
- Dai, Y.C.; Zhuang, J.Y. Numbers of fungal species hitherto known in China. Mycosystema 2010, 29, 625–628. [Google Scholar]
- Mora, C.; Tittensor, D.P.; Adl, S.; Simpson, A.G.B.; Worm, B. How many species are there on Earth and in the ocean? PLoS Biol. 2011, 9, e1001127. [Google Scholar] [CrossRef]
- Niskanen, T.; Lücking, R.; Dahlberg, A.; Gaya, E.; Laura, M.S.; Mikryukov, V.; Liimatainen, K.; Druzhinina, I.; Westrip, J.R.S.M.; Mueller, G.M.; et al. Pushing the frontiers of biodiversity research: Unveiling the global diversity, distribution, and conservation of fungi. Annu. Rev. Environ. 2023, 48, 49–176. [Google Scholar] [CrossRef]
- Dai, Y.C.; Cui, B.K.; Si, J.; He, S.H.; Hyde, K.D.; Yuan, H.S.; Liu, X.Y.; Zhou, L.W. Dynamics of the worldwide number of fungi with emphasis on fungal diversity in China. Mycol. Prog. 2015, 14, 62. [Google Scholar] [CrossRef]
- Feng, B.; Yang, Z. Studies on diversity of higher fungi in Yunnan, southwestern China: A review. Plant Divers. 2018, 40, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ma, L.; Zuo, X.; Ye, X.; Wang, R.; Huang, Z.; Liu, G.; Cornelissen, J.H. Plant diversity has stronger linkage with soil fungal diversity than with bacterial diversity across grasslands of northern China. Glob. Ecol. Biogeogr. 2022, 31, 886–900. [Google Scholar] [CrossRef]
- Cai, J.; Yu, W.B.; Zhang, T.; Wang, H.; Li, D.Z. China’s biodiversity hotspots revisited: A treasure chest for plants. PhytoKeys 2019, 130, 1–24. [Google Scholar] [CrossRef]
- Liu, S.L.; Zhao, P.; Cai, L.; Shen, S.; Wei, H.W.; Na, Q.; Han, M.; Wei, R.; Ge, Y.; Ma, H.; et al. Catalogue of fungi in China 1. New taxa of plant-inhabiting fungi. Mycology 2025, 16, 1–58. [Google Scholar] [CrossRef]
- Shen, D. Microbial diversity and application of microbial products for agricultural purposes in China. Agric. Ecosyst. Environ. 1997, 62, 237–245. [Google Scholar] [CrossRef]
- Ping, Z.; Zhiling, D.; Huijun, G.; Biyun, L. Influence of Land Use on Numbers and Diversity of Soil Microorganisms in Gaoligong Mountains. Acta Bot. Yunnan. 1999, 2, 84–89. [Google Scholar]
- Dequn, Z.; Hyde, K.D.; Vrijmoed, L.L.P. Resources and diversity of bambusicolous fungi in China. Guizhou Sci. 2000, 18, 62–70. [Google Scholar]
- Cai, L.; Tsui, C.K.M.; Zhang, K.Q.; Hyde, K.D. Aquatic fungi from lake Fuxian, Yunnan, China. Fungal Divers. 2002, 9, 57–70. [Google Scholar]
- Li, L.F.; Li, T.; Zhao, Z.W. Differences of arbuscular mycorrhizal fungal diversity and community between a cultivated land, an old field, and a never-cultivated field in a hot and arid ecosystem of southwest China. Mycorrhiza 2007, 17, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.Y.; Cai, Y.Z.; Hyde, K.D.; Corke, H.; Sun, M. Biodiversity of endophytic fungi associated with 29 traditional Chinese medicinal plants. Fungal Divers. 2008, 33, 61–75. [Google Scholar]
- Zhang, Z.F.; Liu, F.; Zhou, X.; Liu, X.Z.; Liu, S.J.; Cai, L. Culturable mycobiota from Karst caves in China, with descriptions of 20 new species. Persoonia Mol. Phylogeny Evol. Fungi 2017, 39, 1–31. [Google Scholar] [CrossRef]
- Redhead, S.A.; Bo, L. New species and new records of Tricholomataceae (Agaricales) from China. Can. J. Bot. 1982, 60, 1479–1486. [Google Scholar] [CrossRef]
- Shiao, M.S.; Lin, L.J.; Yeh, S.F.; Chou, C.S. Two new triterpenes of the fungus Ganoderma lucidum. J. Nat. Prod. 1987, 50, 886–890. [Google Scholar] [CrossRef]
- Wei, S.X.; Zhuang, J.Y. Additions to the graminicolous rust fungi of China. Mycosystema 1989, 2, 217–221. [Google Scholar]
- Zang, M.; Petersen, R.H. Endophallus, a new genus in the Phallaceae from China. Mycologia 1989, 81, 486–489. [Google Scholar] [CrossRef]
- Dai, Y.C.; Yang, Z.L.; Cui, B.K.; Yu, C.J.; Zhou, L.W. Species diversity and utilization of medicinal mushrooms and fungi in China. Int. J. Med. Mushrooms 2009, 11, 287–302. [Google Scholar] [CrossRef]
- Cui, B.K.; Li, H.J.; Ji, X.; Zhou, J.L.; Song, J.; Si, J.; Yang, Z.L.; Dai, Y.C. Species diversity, taxonomy and phylogeny of Polyporaceae (Basidiomycota) in China. Fungal Divers. 2019, 97, 137–392. [Google Scholar] [CrossRef]
- Zhao, R.; Chen, K.Y.; Mao, L.J.; Zhang, C.L. Eleven new species of Trichoderma (Hypocreaceae, Hypocreales) from China. Mycology 2025, 16, 180–209. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Chen, J.; Liao, X.; Chen, X.; Huang, L.; Huang, J.; Huang, R.; Zhong, S.; Zhang, X. Biodiversity, distribution and functional differences of fungi in four species of corals from the South China sea, elucidated by high-throughput sequencing Technology. J. Fungi 2024, 10, 452. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bai, Q.; Meng, F.; Dong, W.; Fan, H.; Zhu, X.; Duan, Y.; Chen, L. High-Throughput Sequence Analysis of Microbial Communities of Soybean in Northeast China. Agronomy 2025, 15, 436. [Google Scholar] [CrossRef]
- Yan, Q.; Wang, P.; Liu, Z.; Yu, Y.; Tan, X.; Huang, X.; Wen, J.; Zhang, W. High-Throughput sequencing uncovers fungal community succession during Morchella sextelata development. J. Fungi 2025, 11, 364. [Google Scholar] [CrossRef]
- Tennakoon, D.S.; Thambugala, K.M.; de Silva, N.I.; Song, H.Y.; Suwannarach, N.; Chen, F.S.; Hu, D.M. An overview of Melanommataceae (Pleosporales, Dothideomycetes): Current insight into the host associations and geographical distribution with some interesting novel additions from plant litter. MycoKeys 2024, 106, 43–96. [Google Scholar] [CrossRef]
- Hongsanan, S.; Khuna, S.; Manawasinghe, I.S.; Tibpromma, S.; Chethana, K.W.T.; Xie, N.; Bagacay, J.F.E.; Calabon, M.S.; Chen, C.; Doilom, M. Mycosphere Notes 521-571: A special edition of fungal biodiversity to celebrate Kevin, D. Hyde’s 70th birthday and his exceptional contributions to Mycology. Mycosphere 2025, 16, 2002–2180. [Google Scholar] [CrossRef]
- Phookamsak, R.; Norphanphoun, C.; Tanaka, K.; Dai, D.Q.; Luo, Z.L.; Liu, J.K.; Su, H.Y.; Bhat, D.J.; Bahkali, A.H.; Mortimer, P.E.; et al. Towards a natural classification of Astrosphaeriella-like species; introducing Astrosphaeriellaceae and Pseudoastrosphaeriellaceae fam. nov. and Astrosphaeriellopsis, gen. nov. Fungal Divers. 2015, 74, 143–197. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Jeewon, R.; Jones, E.B.G.; Boonmee, S.; Kaewchai, S.; Manawasinghe, I.S.; Lumyong, S.; Hyde, K.D. Novel palmicolous taxa within Pleosporales: Multigene phylogeny and taxonomic circumscription. Mycol. Prog. 2018, 17, 571–590. [Google Scholar] [CrossRef]
- Dong, W.; Wang, B.; Hyde, K.D.; McKenzie, E.H.C.; Raja, H.A.; Tanaka, K.; Abdel-Wahab, M.A.; Abdel-Aziz, F.A.; Doilom, M.; Phookamsak, R.; et al. Freshwater Dothideomycetes. Fungal Divers. 2020, 105, 319–575. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Rathnayaka, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dissanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. Res. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Habib, K.; Li, W.H.; Ren, Y.L.; Liu, L.L.; Lu, C.T.; Zhang, Q.F.; Yao, Z.Q.; Luo, X.Y.; Zhou, X.; Zeng, W.Y.; et al. Exploration of ascomycetous fungi revealing novel taxa in Southwestern China. Mycosphere 2025, 16, 1412–1529. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the SC10 Workshop on Gateway Computing Environments (GCE10), New Orleans, LA, USA, 14 November 2010. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nylander, J.A.A. MrModeltest2 v. 2.3 (Program for Selecting DNA Substitution Models Using PAUP*); Evolutionary Biology Centre: Uppsala, Sweden, 2008. [Google Scholar]
- Rambaut, A. FigTree, version 1.4.0; Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2012. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 10 August 2025).
- Farr, D.F.; Rossman, A.Y. Fungal Databases, Systematic Mycology and Microbiology Laboratory. ARS, USDA. 2025. Available online: https://fungi.ars.usda.gov/ (accessed on 1 August 2025).
- Dayarathne, M.C.; Jones, E.B.G.; Maharachchikumbura, S.S.N.; Devadatha, B.; Sarma, V.V.; Khongphinitbunjong, K.; Chomnunti, P.; Hyde, K.D. Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 2020, 11, 1–188. [Google Scholar] [CrossRef]
- Suetrong, S.; Schoch, C.L.; Spatafora, J.W.; Kohlmeyer, J.; Volkmann-Kohlmeyer, B.; Sakayaroj, J.; Phongpaichit, S.; Tanaka, K.; Hirayama, K.; Jones, E.B.G. Molecular systematics of the marine Dothideomycetes. Stud. Mycol. 2009, 64, 155–173. [Google Scholar] [CrossRef]
- Schoch, C.L.; Crous, P.W.; Groenewald, J.Z.; Boehm, E.W.A.; Burgess, T.I.; De Gruyter, J.; De Hoog, G.S.; Dixon, L.J.; Grube, M.; Gueidan, C.; et al. A class-wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 2009, 64, 1–15. [Google Scholar] [CrossRef]
- Hu, F.J.; Fournier, J.; Jeewon, R.; Hyde, K.D. Relationships among Astrosphaeriella, Caryospora and Trematosphaeria. Ph.D. Thesis, The University of Hong Kong, Hong Kong, 2009. [Google Scholar]
- Hyde, K.D.; de Silva, N.I.; Jeewon, R.; Bhat, D.J.; Phookamsak, R.; Doilom, M.; Boonmee, S.; Jayawardena, R.S.; Maharachchikumbura, S.S.N.; Senanayake, I.C.; et al. AJOM new records and collections of fungi: 1–100. Asian J. Mycol. 2020, 3, 22–294. [Google Scholar] [CrossRef]
- Liu, J.K.; Phookamsak, R.; Jones, E.B.G.; Zhang, Y.; Ko-Ko, T.W.; Hu, H.L.; Boonmee, S.; Doilom, M.; Chukeatirote, E.; Bahkali, A.H.; et al. Astrosphaeriella is polyphyletic, with species in Fissuroma gen. nov., and Neoastrosphaeriella gen. nov. Fungal Divers. 2011, 51, 135–154. [Google Scholar] [CrossRef]
- Ariyawansa, H.A.; Hyde, K.D.; Jayasiri, S.C.; Buyck, B.; Chethana, K.W.T.; Dai, D.Q.; Dai, Y.C.; Daranagama, D.A.; Jayawardena, R.S.; Lücking, R. Fungal diversity notes 111–252 taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2015, 75, 27–274. [Google Scholar] [CrossRef]
- Yang, J.; Liu, L.L.; Jones, E.G.; Hyde, K.D.; Liu, Z.Y.; Bao, D.F.; Liu, N.G.; Li, W.L.; Shen, H.W.; Yu, X.D.; et al. Freshwater fungi from karst landscapes in China and Thailand. Fungal Divers. 2023, 119, 1–212. [Google Scholar] [CrossRef]
- Cai, L.; Hyde, K.D. Ascorhombispora aquatica gen. et sp. nov. from a freshwater habitat in China, and its phylogenetic placement based on molecular data. Cryptogam. Mycol. 2007, 28, 291–300. [Google Scholar]
- Jayasiri, S.C.; Hyde, K.D.; Jones, E.B.G.; McKenzie, E.H.C.; Jeewon, R.; Phillips, A.J.L.; Bhat, D.J.; Wanasinghe, D.N.; Liu, J.K.; Lu, Y.Z.; et al. Diversity, morphology and molecular phylogeny of Dothideomycetes on decaying wild seed pods and fruits. Mycosphere 2019, 10, 1–186. [Google Scholar] [CrossRef]
- Mugambi, G.K.; Huhndorf, S.M. Molecular phylogenetics of Pleosporales: Melanommataceae and Lophiostomataceae re-circumscribed (Pleosporomycetidae, Dothideomycetes, Ascomycota). Stud. Mycol. 2009, 64, 103–121. [Google Scholar] [CrossRef]
- Schoch, C.L.; Shoemaker, R.A.; Seifert, K.A.; Hambleton, S.; Spatafora, J.W.; Crous, P.W. A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 2006, 98, 1041–1052. [Google Scholar] [CrossRef]
- Zhang, S.N.; Hyde, K.D.; Jones, E.B.G.; Cheewangkoon, R.; Liu, J.K. Additions to Fissuroma and Neoastrosphaeriella (Aigialaceae, Pleosporales) from palms. Mycosphere 2020, 11, 269–284. [Google Scholar] [CrossRef]
- Tennakoon, D.S.; Phookamsak, R.; Kuo, C.H.; Goh, T.K.; Jeewon, R.; Hyde, K.D. Morphological and phylogenetic evidence reveal Fissuroma taiwanense sp. nov. (Aigialaceae, Pleosporales) from Hedychium coronarium. Phytotaxa 2018, 338, 265–275. [Google Scholar] [CrossRef]
- Konta, S.; Hyde, K.D.; Eungwanichayapant, P.D.; Doilom, M.; Tennakoon, D.S.; Senwanna, C.; Boonmee, S. Fissuroma (Aigialaceae: Pleosporales) appears to be hyperdiverse on Arecaceae: Evidence from two new species from southern Thailand. Acta Bot. Bras. 2020, 34, 384–393. [Google Scholar] [CrossRef]
- Bao, D.F.; Luo, Z.L.; Jeewon, R.; Nalumpang, S.; Su, H.Y.; Hyde, K.D. Neoastrosphaeriella aquatica sp. nov. (Aigialaceae), a new species from freshwater habitat in Southern Thailand. Phytotaxa 2019, 391, 197. [Google Scholar] [CrossRef]
- Hyde, K.D.; Chaiwan, N.; Norphanphoun, C.; Boonmee, S.; Camporesi, E.; Chethana, K.W.T.; Dayarathne, M.C.; de Silva, N.I.; Dissanayake, A.J.; Ekanayaka, A.H.; et al. Mycosphere notes 169–224. Mycosphere 2018, 9, 271–430. [Google Scholar] [CrossRef]
- Phookamsak, R.; Liu, J.K.; Manamgoda, D.S.; Wanasinghe, D.N.; Ariyawansa, H.A.; Mortimer, P.E.; Chukeatirote, E.; McKenzie, E.H.C.; Hyde, K.D. Epitypification of two bambusicolous fungi from Thailand. Cryptogam. Mycol. 2014, 35, 239–256. [Google Scholar] [CrossRef]
- Bao, D.F.; Hyde, K.D.; McKenzie, E.H.; Jeewon, R.; Su, H.Y.; Nalumpang, S.; Luo, Z.L. Biodiversity of lignicolous freshwater hyphomycetes from China and Thailand and description of sixteen species. J. Fungi 2021, 7, 669. [Google Scholar] [CrossRef] [PubMed]
- Hongsanan, S.; Hyde, K.D.; Phookamsak, R.; Wanasinghe, D.N.; McKenzie, E.H.C.; Sarma, V.V.; Lücking, R.; Boonmee, S.; Bhat, J.D.; Liu, N.-G.; et al. Refined families of Dothideomycetes: Orders and families incertae sedis in Dothideomycetes. Fungal Divers. 2020, 105, 17–318. [Google Scholar] [CrossRef]
- Rehm, H. Ascomycetes philippinenses—III. Philipp. J. Sci. Sect. C 1913, 8, 391–406. [Google Scholar]
- Tanaka, K.; Harada, Y. Bambusicolous fungi in Japan (4): A new combination, Astrosphaeriella aggregata. Mycoscience 2005, 46, 114–118. [Google Scholar] [CrossRef]
- Hongsanan, S.; Phookamsak, R.; Bhat, D.J.; Wanasinghe, D.N.; Promputtha, I.; Suwannarach, N.; Sandamali, D.; Lumyong, S.; Xu, J.; Xie, N. Exploring ascomycete diversity in Yunnan, China I: Resolving ambiguous taxa in Phaeothecoidiellaceae and investigating conservation implications of fungi. Front. Cell. Infect. Microbiol. 2023, 13, 1252387. [Google Scholar] [CrossRef]
- Zhang, Y.; Crous, P.W.; Schoch, C.L.; Hyde, K.D. Pleosporales. Fungal Divers. 2012, 53, 1–221. [Google Scholar] [CrossRef] [PubMed]
- Species Fungorum. 2025. Available online: http://www.indexfungorum.org (accessed on 15 September 2025).
- De Notaris. Micromyc Ital. Novi 1855, 9, 7. [Google Scholar]
- Jeffers, W.F. Studies on Caryospora putaminum. Mycologia 1940, 32, 550–566. [Google Scholar] [CrossRef]
- Lumbsch, H.T.; Huhndorf, S.M. Myconet Volume 14. Part One. Outline of Ascomycota–2009. Fieldiana Life Earth Sci. 2010, 1, 1–60. [Google Scholar] [CrossRef]
- Hyde, K.D.; Jones, E.B.G.; Liu, J.K.; Ariyawansa, H.A.; Boehm, E.; Boonmee, S.; Braun, U.; Chomnunti, P.; Crous, P.W.; Dai, D.Q.; et al. Families of Dothideomycetes. Fungal Divers. 2013, 63, 1–313. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Dai, D.Q.; Sánchez-García, M.; Goto, B.T.; Saxena, R.K.; Erdoğdu, M.; Selçuk, F.; Rajeshkumar, K.C.; Aptroot, A.; et al. Outline of Fungi and fungus-like taxa—2021. Mycosphere 2022, 13, 53–453. [Google Scholar] [CrossRef]
- Raja, H.A.; Shearer, C.A. Freshwater ascomycetes: New and noteworthy species from aquatic habitats in Florida. Mycologia 2008, 100, 467–489. [Google Scholar] [CrossRef]
- Niranjan, M.; Sarma, V.V. New Ascomycetous fungi in the family Aigialaceae from Andaman Islands, India. Curr. Res. Environm. Appl. Mycol. 2018, 8, 351–359. [Google Scholar] [CrossRef]
- Vitória, N.S.; Cavalcanti, M.A.D.; Bezerra, J.L. Species of Astrosphaeriella and Fissuroma from palms: New records for South America and Brazil. Nova Hedwig. 2016, 102, 129–140. [Google Scholar] [CrossRef]
- Zhou, D.Q.; Cai, L.; Hyde, K.D. Astrosphaeriella and Roussoella species on bamboo from Hong Kong and Yunnan, China including a new species of Roussoella. Cryptogam. Mycol. 2003, 24, 191–197. [Google Scholar]
- He, S.C.; Hyde, K.D.; Jayawardena, R.S.; Thiyagaraja, V.; Wanasinghe, D.N.; Zhao, Y.W.; Wang, Z.Y.; Cai, T.; Yang, Y.Y.; Al-Otibi, F.; et al. Taxonomic contributions to Pleosporales and Kirschsteiniotheliales from the Xizang Autonomous Region, China. Mycology 2025, 1–38. [Google Scholar] [CrossRef]
- Mulenko, W.; Majewski, T.; Ruszkiewicz-Michalska, M. A Preliminary Checklist of Micromycetes in Poland; The Władysław Institute of Botany, Polish Academy of Sciences: Krakow, Poland, 2008; Volume 9, pp. 1–752. [Google Scholar]
- Tianyu, Z. Flora Fungorum Sincorum Vol. 31: 26 Genera of Dematiaceous Dictyosporous Hyphomycetes Excluding Alternaria; Science Press: Beijing, China, 2009; Volume 31, pp. 1–231. [Google Scholar]
- Ellis, M.B. Dematiaceous Hyphomycetes. I. Mycol. Pap. 1960, 76, 1–36. [Google Scholar]
- Thaung, M.M. A list of Hypomycetes (and Agonomycetes) in Burma. Australas. Mycol. 2008, 27, 149–172. [Google Scholar]
- Matsushima, T. Matsushima Mycological Memoirs No. 1. Saprophytic microfungi from Taiwan, Part 1. Hyphomycetes; Matsushima Fungus Collection: Kobe, Japan, 1980; p. 82. [Google Scholar]
- Thomas, T.V.; Eicker, A.; Robbertse, P.J. Possible role of fungi in negatively affecting fruit-set in avocados. S. Afr. J. Bot. 1994, 60, 251–256. [Google Scholar] [CrossRef]
- Lu, B.; Hyde, K.D.; Ho, W.H.; Tsui, K.M.; Taylor, J.E.; Wong, K.M.; Yanna, D.; Zhou, D. Checklist of Hong Kong Fungi. Fungal Divers. 2000, 207. Available online: https://www.fungaldiversity.org/fdp/researchseriesbooks/checklist.html (accessed on 1 August 2025).
- Czeczuga, B.; Muszynska, E.; Godlewska, A.; Mazalska, B. Aquatic fungi and straminipilous organisms on decomposing fragments of wetland plants. Mycol. Balcan. 2007, 4, 31–44. [Google Scholar]
- Liu, H.M.; Zhang, T.Y. Species of Pithomyces (Hyphomycetes) from soil in the warm temperate zone of eastern China. Mycosystema 2007, 26, 484–489. [Google Scholar]
- Phookamsak, R.; Hyde, K.D.; Jeewon, R.; Bhat, D.J.; Jones, E.B.G.; Maharachchikumbura, S.S.N.; Raspé, O.; Karunarathna, S.C.; Wanasinghe, D.N.; Hongsanan, S.; et al. Fungal Diversity Notes 929–1035: Taxonomic and Phylogenetic Contributions on Genera and Species of Fungi. Fungal Divers. 2019, 95, 1–273. [Google Scholar] [CrossRef]
- Hyde, K.D.; Fröhlich, J. Fungi from palms. XXXVII. The genus Astrosphaeriella, including ten new species. Sydowia 1997, 50, 81–132. [Google Scholar]
- Sarma, V.V.; Raghukumar, S. Manglicolous fungi from Chorao mangroves, Goa, West coast of India: Diversity and frequency of occurrence. Nova Hedwig. 2013, 97, 533–542. [Google Scholar] [CrossRef]
- Pande, A. Ascomycetes of Peninsular India; Scientific Publishers: Jodhpur, India, 2008; p. 584. [Google Scholar]
- Kohlmeyer, J.; Schatz, S. Aigialus gen. nov. (Ascomycetes) with two new marine species from Mangroves. Trans. Brit. Mycol. Soc. 1985, 85, 699–707. [Google Scholar] [CrossRef]
- Borse, B.D. New species of Aigialus from India. Trans. Brit. Mycol. Soc. 1987, 88, 424–426. [Google Scholar] [CrossRef]
- Hino, I.; Katumoto, K. Illustrationes fungorum bambusicolorum III. Bull. Fac. Agric. Yamaguchi Univ. 1955, 6, 29–68. [Google Scholar]
- Hino, I. Species of fungi parasitic on bamboos in Japan. Icon. Fungorum Bambusi 1961, 1–333. [Google Scholar]
- Eriksson, O.E.; Yue, J.Z. Bambusicolous pyrenomycetes, an annotated check-list. Myconet 1998, 1, 25–78. [Google Scholar]
- Hosoya, T.; Tanaka, K. Ascomycetes and Anamorphic Fungi collected from Yakushima Island, Southern Japan. Bull. Natl. Mus. Nat. Sci. 2007, 33, 47–54. [Google Scholar]
- Feng, Y.; Zhang, S.N.; Chen, Y.Y.; Liu, Z.Y. Fissuroma bambucicola sp. nov. (Aigialaceae, Pleosporales) from Bamboo in Guizhou, China. Phytotaxa 2022, 543, 64–72. [Google Scholar] [CrossRef]
- Hyde, K.D.; Aptroot, A.; Frohlich, J.; Taylor, J.E. Fungi from palms. XLIII. Lophiostoma and Astrosphaeriella species with slit-like ostioles. Nova Hedwig. 2000, 70, 143–160. [Google Scholar] [CrossRef]
- Teodoro, N.G. An Enumeration of Philippine Fungi. Techn. Bull. Dept. Agric. Comm. Manila 1937, 4, 1–585. [Google Scholar]
- Vohník, M. Are Lulworthioid fungi dark septate endophytes of the dominant Mediterranean seagrass Posidonia oceanica? Plant Biol. 2022, 24, 127–133. [Google Scholar] [CrossRef]
- Pinnoi, A.; Lumyong, S.; Hyde, K.D.; Jones, E.B.G. Biodiversity of fungi on the palm Eleiodoxa conferta in Sirindhorn peat swamp forest, Narathiwat, Thailand. Fungal Divers. 2006, 22, 205–218. [Google Scholar]
- Pinruan, U.; Hyde, K.D.; Lumyong, S.; McKenzie, E.H.C.; Jones, E.B.G. Occurrence of fungi on tissues of the peat swamp palm Licuala longicalycata. Fung. Divers. 2007, 25, 157–173. [Google Scholar]
- Hino, I.; Katumoto, K. Notes on fungi from western Japan (1). Bull. Fac. Agric. Yamaguchi Univ. 1956, 7, 257–266. [Google Scholar]
- Kobayashi, T. Index of Fungi Inhabiting Woody Plants in Japan, Host, Distribution and Literature; Zenkoku-Noson-Kyoiku Kyokai Publishing Co., Ltd.: Tokyo, Japan, 2007; p. 1227. [Google Scholar]
- Dennis, R.W.G. British Ascomycetes. J. Cramer Vaduz 1978, 585. [Google Scholar]
- Hyde, K.D. Fungi from palms. XII.* Three new intertidal ascomycetes from submerged palm fronds. Sydowia 1994, 46, 257–264. [Google Scholar]
- Hyde, K.D. Lophiostoma asiana sp. nov. from Thailand mangroves. Mycotaxon 1995, 55, 283–288. [Google Scholar] [CrossRef]
- von Höhnel, F. Fragmente zur Mykologie. XXIV. Mitteilung (Nr. 1189 bis 1214). Sitzungsberichte Kais. Akad. Wiss. Math.-Naturw. Kl. Abt. I 1920, 129, 137–184. [Google Scholar]
- Penzig, A.J.O.; Saccardo, P.A. Diagnoses fungorum novorum in insula Java collectorum. Series I. Malpighia 1897, 11, 387–409. [Google Scholar]
- Theissen, F. Beiträge zur Systematik der Ascomyzeten. Annales Mycologici. 1916, 14, 401–439. [Google Scholar]
- Hawksworth, D.L.; Boise, J.R. Some additional species of Astrosphaeriella, with a key to the members of the genus. Sydowia 1985, 38, 114–124. [Google Scholar]
- Barr, M.E. Melanommatales (Loculoascomycetes). N. Am. Flora 1990, 13, 1–129. [Google Scholar]
- Loilong, A.; Sakayaroj, J.; Rungjindamai, N.; Choeyklin, R.; Jones, E.B.G. Biodiversity of fungi on the palm Nypa fruticans. In Marine Fungi and Fungal-Like Organisms; Jones, E.B.G., Pang, K.L., Eds.; de Gruyter: Berlin, Germany, 2012; pp. 273–290. [Google Scholar]
- Reinking, O.A. Host Index of Diseases of Economic Plants in the Philippines. Philipp. Agric. 1919, 8, 38–54. [Google Scholar]
- Yue, J.Z.; Eriksson, O. Studies on Chinese Ascomycetes. 3. Astrosphaeriella lageniformis. Mycotaxon 1986, 27, 93–98. [Google Scholar] [CrossRef]
- Chen, C.Y.; Huang, J.W. Astrosphaeriella linguiformis, a new species on bamboo. Mycotaxon 2006, 98, 119–123. [Google Scholar]
- Rogers, J.D. Astrosphaeriella longispora, a new tropical species with large ascospores. Sydowia 2003, 55, 355–358. [Google Scholar]
- Chen, C.Y.; Hsieh, W.H. Astrosphaeriella from Taiwan, including two new species. Bot. Bull. Acad. Sin. 2004, 45, 171–178. [Google Scholar]
- Aptroot, A. Redisposition of some species excluded from Didymosphaeria (Ascomycotina). Nova Hedwig. 1995, 60, 325–379. [Google Scholar] [CrossRef]
- Aptroot, A. Additional lichen records from Australia 70. Species of Anisomeridium and Mycomicrothelia, with a note on Arthopyrenia. Australas. Lichenol. 2009, 64, 22–25. [Google Scholar]
- Scheinpflug, H. Untersuchungen über die Gattung Didymosphaeria Fuck. und einige verwandte Gattungen. Berichte Schweiz. Bot. Ges. 1958, 68, 325–385. [Google Scholar]
- Fairman, C.E. New or rare Pyrenomycetaceae from western New York. Proc. Rochester Acad. Sci. 1905, 4, 215–224. [Google Scholar]
- Hyde, K.D.; Goh, T.K. Fungi on submerged wood in the Riviere St Marie-Louis, The Seychelles. S. Afr. J. Bot. 1998, 64, 330–336. [Google Scholar] [CrossRef]
- Hsieh, W.H.; Chen, C.Y.; Wang, C.L. Taiwan Ascomycetes. Pyrenomycetes and Loculoascomycetes; China Graphics: Taichung, Taiwan, 2000; p. 244. [Google Scholar]
- Wong, M.K.M.; Hyde, K.D. Diversity of fungi on six species of Gramineae and one species of Cyperaceae in Hong Kong. Mycol. Res. 2001, 105, 1485–1491. [Google Scholar] [CrossRef]
- Wang, Y.Z.; Aptroot, A.; Hyde, K.D. Revision of the ascomycete genus Amphisphaeria. Fungal Divers. 2004, 168. [Google Scholar]
- Ren, J.; Zeng, C.; Jie, C.Y.; Jiang, Y.L.; Hyde, K.D.; Wang, Y. A new Thai species of Astrosphaeriella (Dothideomycetes, Ascomycota) from submerged wood in freshwater. Sydowia 2013, 65, 33–43. [Google Scholar]
- Niranjan, M.; Sarma, V.V. New species and new records of Astrosphaeriellaceae from Andaman Islands, India. Kavaka 2020, 54, 38–42. [Google Scholar] [CrossRef]
- San Martin, F.E.; Lavin, P.A. Four species and one new variety of the genus Astrosphaeriella (Dothideales, Melanommataceae) in Mexico. Acta Bot. Mex. 1999, 46, 19–27. [Google Scholar] [CrossRef]
- Shen, H.W.; Luo, Z.L.; Bao, D.F.; Luan, S.; Bhat, D.J.; Boonmee, S.; Wang, W.P.; Su, X.J.; Li, Y.X.; Al-Otibi, F.; et al. Morphology and phylogeny reveal new species of Pleosporales from plateau lakes in Yunnan Province, China. Mycosphere 2024, 15, 6439–6524. [Google Scholar] [CrossRef]
- Saccardo, P.A. 1. Fungi Singaporenses Bakeriani. 2. Fungi Abellinenses novi. Bull. dell’Orto Bot. Regia Univ. Napoli 1918, 6, 39–73. [Google Scholar]
- Abdel-Wahab, M.A.; Jones, E.B.G. Three new marine ascomycetes from driftwood in Australia sand dunes. Mycoscience 2000, 41, 379–388. [Google Scholar] [CrossRef]
- Dennis, R.W.G. Fungus Flora of Venezuela and Adjacent Countries; Kew Bulletin Additional Series III; Stationery Office Books: Norwich, UK, 1970; pp. 1–531. [Google Scholar]
- Zhao, G.C.; Zhao, R.L. The Higher Microfungi from Forests of Yunnan Province; Yunnan Science and Technology Press: Kunming, China, 2012; pp. 1–572. [Google Scholar]
- Cash, E.K. A Record of the Fungi Named by J.B. Ellis (Part 1); United States Department of Agriculture: Beltsville, MD, USA, 1952; Volume 2, pp. 1–165. [Google Scholar]
- Patel, U.S.; Pandey, A.K.; Rajak, R.C. Additions to fungi of India. Kavaka 1997, 25, 61–65. [Google Scholar]
- Barr, M.E. On the Massariaceae in North America. Mycotaxon 1979, 9, 17–37. [Google Scholar] [CrossRef]
- Hawksworth, D.L. A new species of Caryospora from Eugenia in East Africa. Trans. Br. Mycol. 1982, 79, 69–74. [Google Scholar] [CrossRef]
- Boise, J.R. New and interesting fungi (Loculoascomycetes) from the Amazon. Acta Amaz. 1984, 14, 49–53. [Google Scholar] [CrossRef]
- McKenzie, E.H.C.; Buchanan, P.K.; Johnston, P.R. Checklist of fungi on Nothofagus species in New Zealand. New Zealand J. Bot. 2000, 38, 635–720. [Google Scholar] [CrossRef]
- Liu, L.L.; Long, Q.D.; Kang, J.C.; Zhang, X.; Hyde, K.D.; Shen, X.C.; Li, Q.R. Morphology, and phylogeny of Mycopepon. Mycosphere 2018, 9, 779–789. [Google Scholar] [CrossRef]
- Matsushima, T. Matsushima Mycological Memoirs No. 2; Matsushima Fungus Collection: Kobe, Japan, 1981; p. 68. [Google Scholar]
- Guo, Z.X.; Yu, Z.T. Notes on the genus Pithomyces (Hyphomycetes) from China. Mycotaxon 2007, 85, 241–245. [Google Scholar]
- Satya, H.N. A new species of Pithomyces with bulbils. Curr. Sci. 1975, 44, 522–523. [Google Scholar]
- Holubová-Jechová, V.; Mercado Sierra, A. Studies on Hyphomycetes from Cuba II. Hyphomycetes from the Isla de la Juventud. Česká Mykologie 1984, 38, 96–120. [Google Scholar] [CrossRef]
- Morgan-Jones, G. Notes on hyphomycetes. LIV. Concerning Pithomyces clavisporus, a new species, P. graminicola and P. pavgii. Mycotaxon 1987, 30, 29–37. [Google Scholar] [CrossRef]
- Last, F.T.; Deighton, F.C. The non-parasitic microflora on the surfaces of living leaves. Trans. Br. Mycol. Soc. 1965, 31, 143–150. [Google Scholar] [CrossRef]
- Gusmão, L.F.P.; Monteiro, J.S.; Castañeda Ruíz, R.F. Gonatophragmiopsis verrucosa gen. & sp. nov. and Pithomyces dimorphosporus sp. nov. from Brazil. Mycotaxon 2017, 132, 565–574. [Google Scholar] [CrossRef]
- Narayan, P.; Kamal. New foliicolous hyphomycetes from India. Can. J. Bot. 1986, 64, 201–207. [Google Scholar] [CrossRef]
- Turner, P.D. Microorganisms associated with oil palm (Elaeis guineensis Jacq.). Phytopathol. Pap. 1971, 14, 1–58. [Google Scholar]
- Pande, A.; Rao, V.G. A Compendium Fungi on Legumes from India; Scientific Publishers: Jodhpur, India, 1998; p. 188. [Google Scholar]
- Ellis, M.B. More Dematiaceous Hyphomycetes; Commonwealth Mycological Institute: Surrey, UK, 1976; p. 507. [Google Scholar]
- Mena Portales, J.; Mercado Sierra, A. Hyphomycetes of Topes-de-Collantes Cuba I. Holoblastic species. Acta Bot. Hung. 1986, 32, 189–206. [Google Scholar]
- Holubová-Jechová, V.; Mercado Sierra, A. Studies on Hyphomycetes from Cuba IV. Dematiaceous Hyphomycetes from the Province Pinar del Rio. Česká Mykol. 1986, 4, 142–164. [Google Scholar] [CrossRef]
- Ahmad, S.; Iqbal, S.H.; Khalid, A.N. Fungi of Pakistan; Mycological Society of Pakistan. Department of Botany, University of the Punjab: Lahore, Pakistan, 1997; p. 248. [Google Scholar]
- Atkinson, G.F. Some fungi from Alabama. Bull. Cornell Univ. Sci. 1897, 3, 1–50. [Google Scholar]
- Rao, N.K.; Manoharachary, C. A new Pithomyces species on leaf litter from A. P., India. Trans. Br. Mycol. Soc. 1988, 91, 349–352. [Google Scholar]
- Matsushima, T. Matsushima Mycological Memoirs No. 5; Matsushima Fungus Collection: Kobe, Japan, 1987; p. 100. [Google Scholar]
- Kirk, P.M. New or interesting microfungi IX. Dematiaceous Hyphomycetes from Esher Common. Trans. Brit. Mycol. Soc. 1983, 80, 449–467. [Google Scholar] [CrossRef]
- Lee, S.; Melnik, V.; Taylor, J.E.; Crous, P.W. Diversity of saprobic hyphomycetes on Proteaceae and Restionaceae from South Africa. Fung. Divers. 2004, 17, 91–114. [Google Scholar]
- Filer, J.T.H. A new species of Pteridiospora. Mycologia 1969, 61, 167–169. [Google Scholar] [CrossRef]
- Luo, Z.L.; Bao, D.F.; Li, L.L.; Luan, S.; Su, H.Y. Xenoastrosphaeriella aquatica sp. nov. from freshwater habitat in Yunnan Province, China. Phytotaxa 2022, 544, 193–200. [Google Scholar] [CrossRef]
- Berkeley, M.J.; Curtis, M.A. Exotic fungi from the Schweinitzian Herbarium, principally from Surinam. J. Acad. Nat. Sci. Phila. 1853, 2, 277–294. [Google Scholar]
- Hyde, K.D. Fungi from decaying inter-tidal fronds of Nypa fruticans, including three new genera and four new species. Bot. J. Linn. Soc. 1992, 110, 95–110. [Google Scholar] [CrossRef]
- Liew, E.C.Y.; Aptroot, A.; Hyde, K.D. Phylogenetic significance of the pseudoparaphyses in Loculoascomycete taxonomy. Mol. Phylogenet. Evol. 2000, 16, 392–402. [Google Scholar] [CrossRef] [PubMed]
- Vohník, M.; Borovec, O.; Kolaříková, Z.; Sudová, R.; Réblová, M. Extensive sampling and high-throughput sequencing reveal Posidoniomyces atricolor gen. et sp. nov. (Aigialaceae, Pleosporales) as the dominant root mycobiont of the dominant Mediterranean seagrass Posidonia oceanica. MycoKeys 2019, 55, 59. [Google Scholar] [CrossRef]
- Hyde, K.D.; Norphanphoun, C.; Abreu, V.P.; Bazzicalupo, A.; Chethana, K.W.T.; Clericuzio, M.; Dayarathne, M.C.; Dissanayake, A.J.; Ekanayaka, A.H.; He, M.Q.; et al. Fungal diversity notes 603–708: Taxonomic and phylogenetic notes on genera and species. Fungal Divers. 2017, 87, 1–235. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tennakoon, D.S.; de Silva, N.I.; Xie, N.; Hongsanan, S. Global Diversity, Host Associations, and New Insights into Aigialaceae, Astrosphaeriellaceae, and Pseudoastrosphaeriellaceae. J. Fungi 2025, 11, 834. https://doi.org/10.3390/jof11120834
Tennakoon DS, de Silva NI, Xie N, Hongsanan S. Global Diversity, Host Associations, and New Insights into Aigialaceae, Astrosphaeriellaceae, and Pseudoastrosphaeriellaceae. Journal of Fungi. 2025; 11(12):834. https://doi.org/10.3390/jof11120834
Chicago/Turabian StyleTennakoon, Danushka S., Nimali I. de Silva, Ning Xie, and Sinang Hongsanan. 2025. "Global Diversity, Host Associations, and New Insights into Aigialaceae, Astrosphaeriellaceae, and Pseudoastrosphaeriellaceae" Journal of Fungi 11, no. 12: 834. https://doi.org/10.3390/jof11120834
APA StyleTennakoon, D. S., de Silva, N. I., Xie, N., & Hongsanan, S. (2025). Global Diversity, Host Associations, and New Insights into Aigialaceae, Astrosphaeriellaceae, and Pseudoastrosphaeriellaceae. Journal of Fungi, 11(12), 834. https://doi.org/10.3390/jof11120834








