Identification, Subcellular Localization, and Infection-Related Expression of a Novel Haloacid Dehalogenase Gene (VmHAD) from Valsa mali Vm1
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Preparation
2.2. Fungal Total RNA Extraction and First-Strand cDNA Synthesis
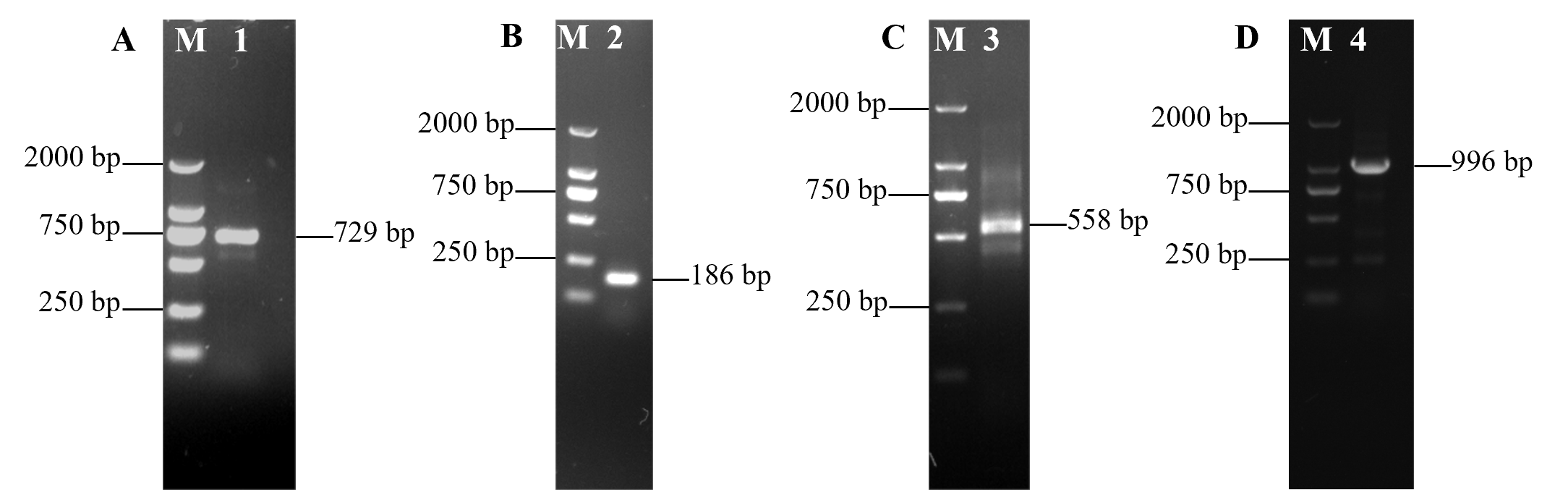
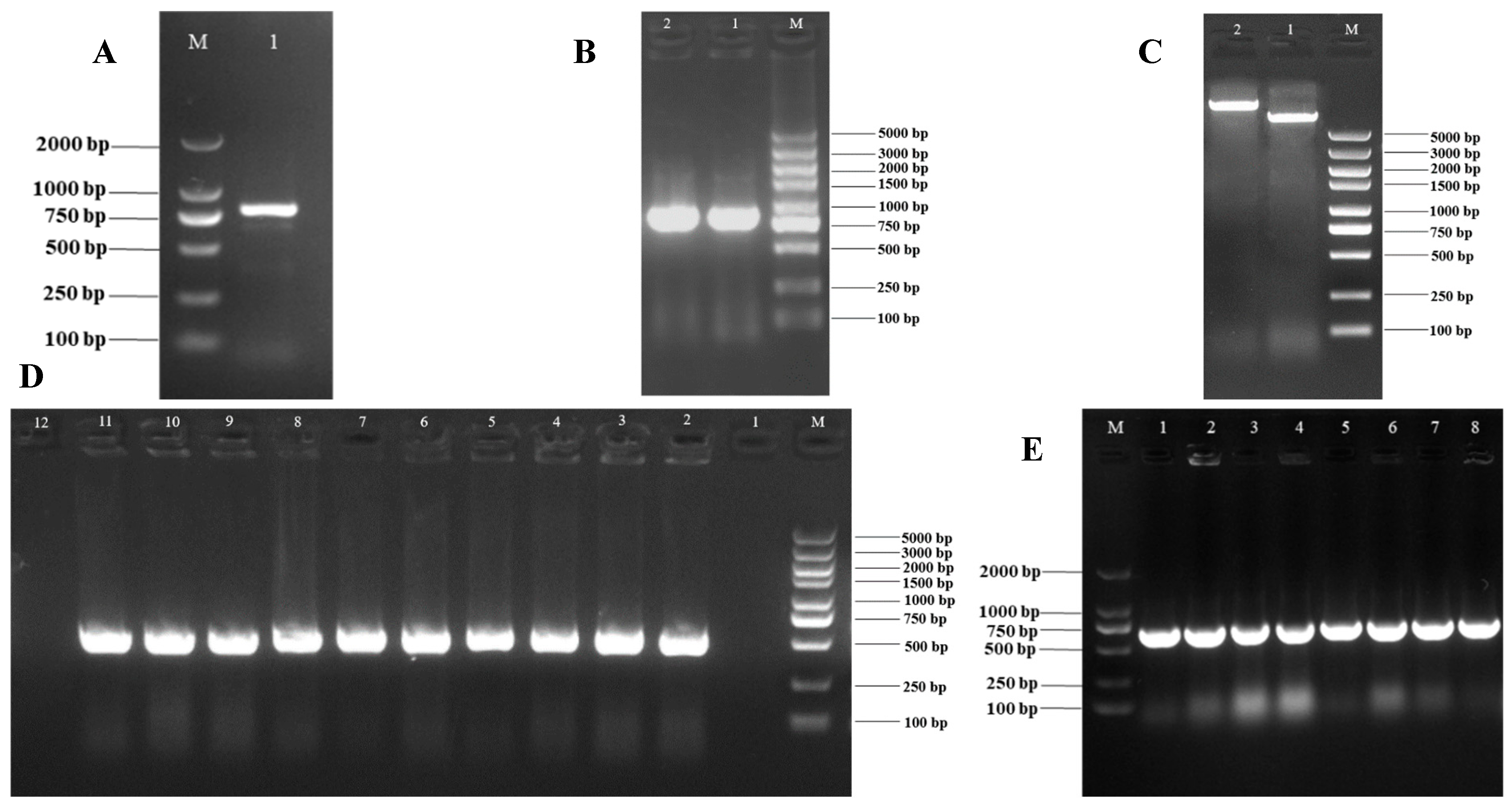
2.3. Haloacid Dehalogenase Gene (VmHAD) of Valsa mali Vm1 Cloning and Characterization
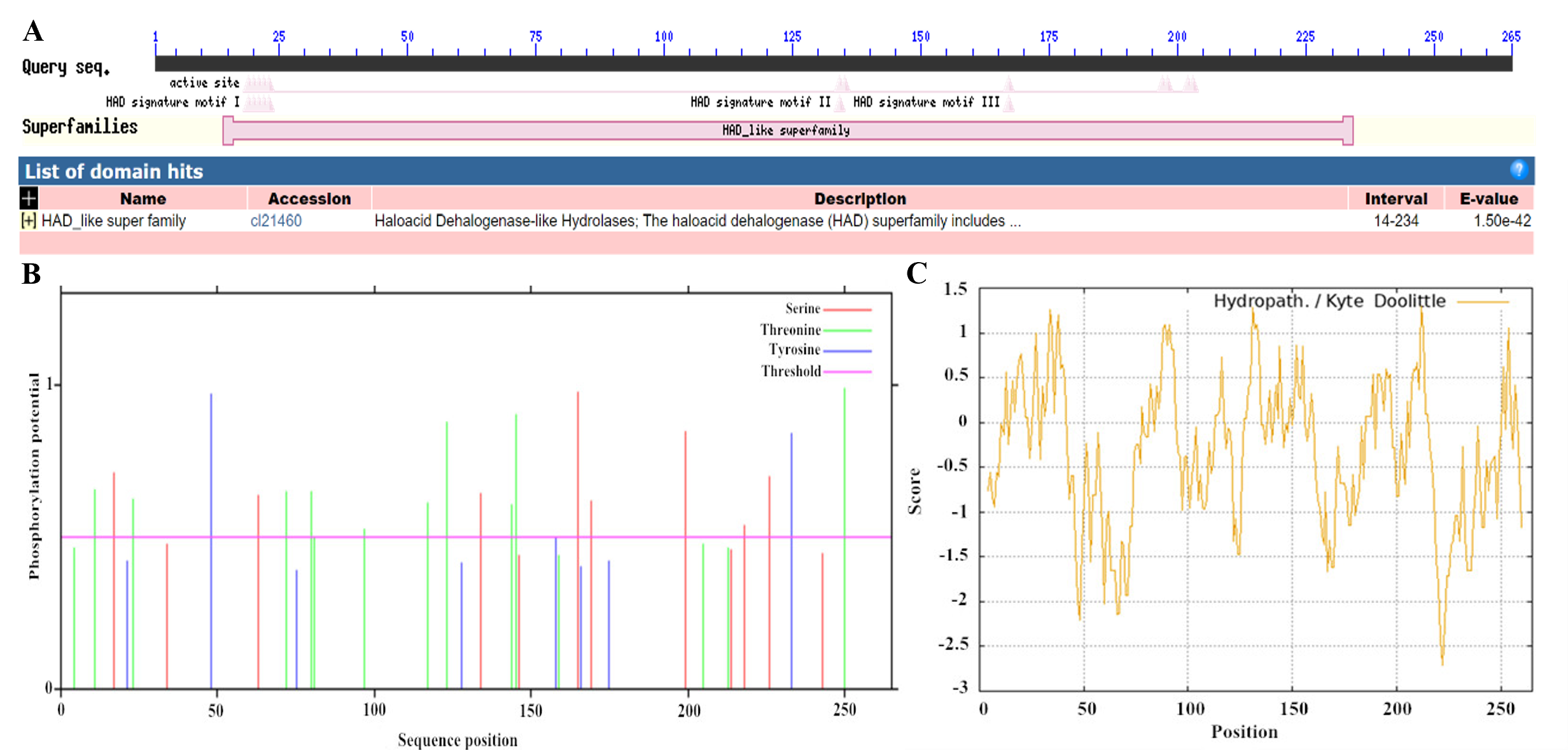
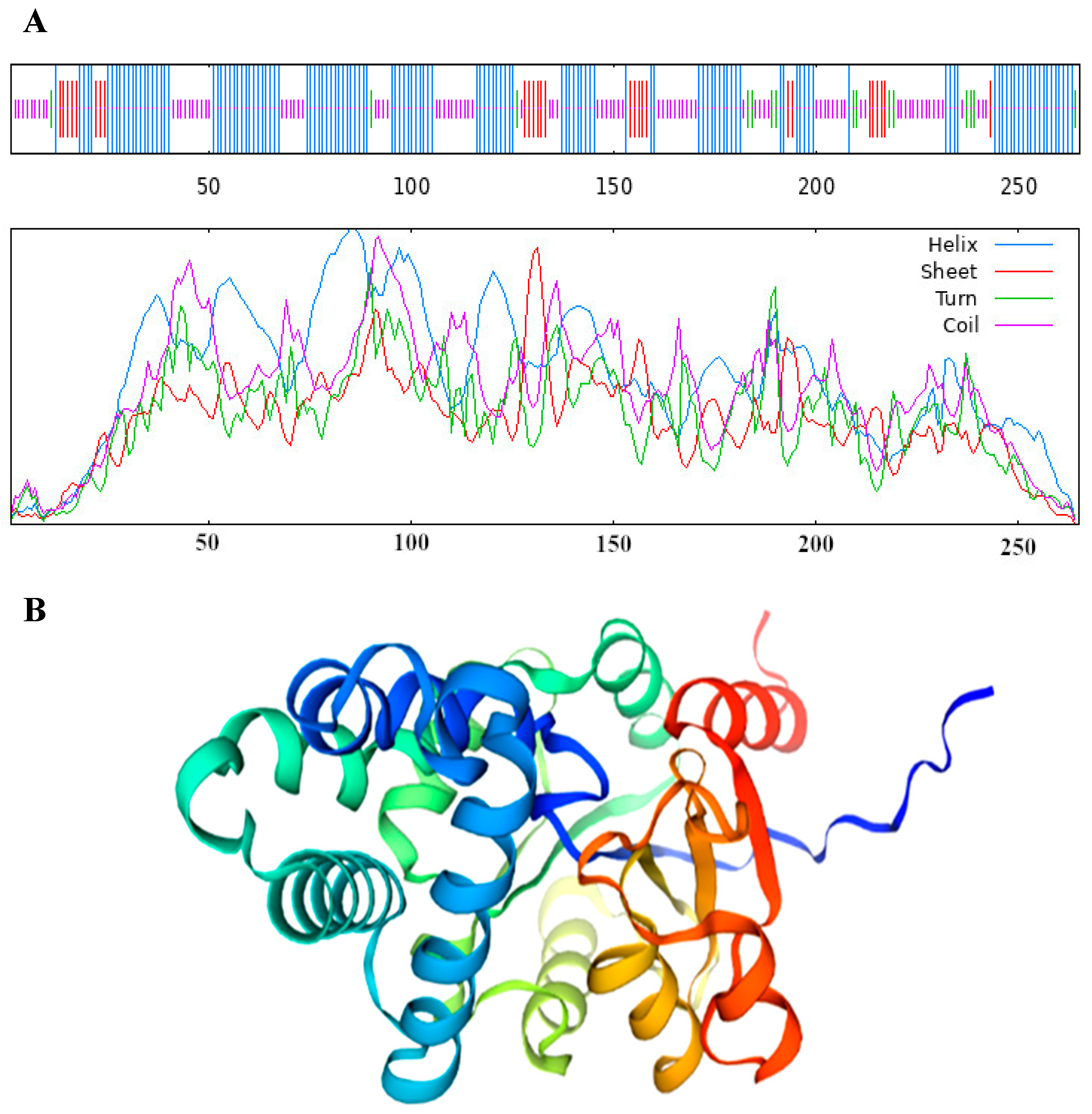
2.4. VmHAD Bioinformatic Analysis
2.5. VmHAD Gene Recombinant Vector Construction and Transformation
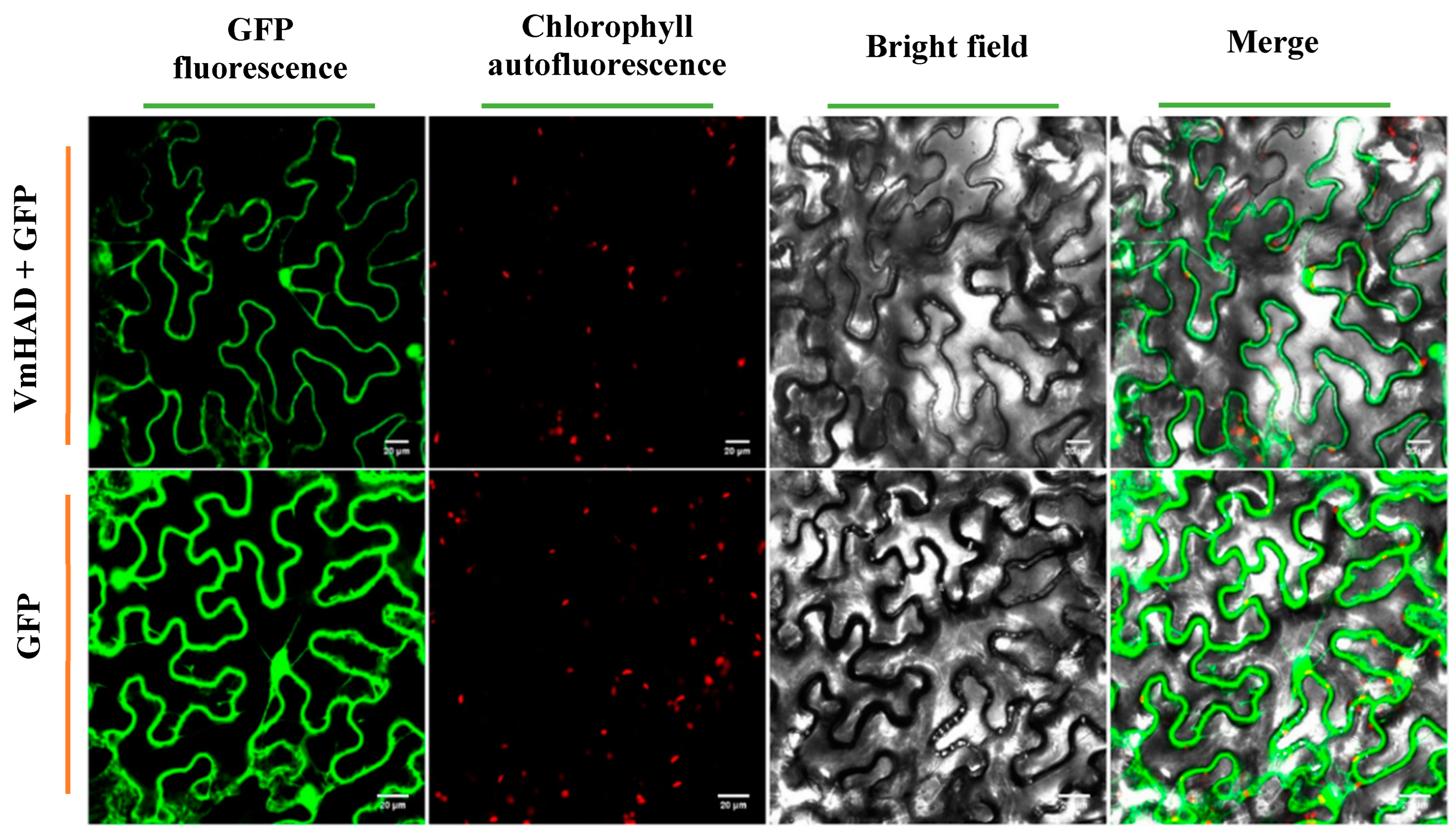
2.6. VmHAD Subcellular Location Analysis by Transient Expression
2.7. VmHAD Gene Expression Characteristics Analysis During the Valsa mali Vm1 Infection Process by RT-qPCR
2.8. Statistical Analysis
3. Results
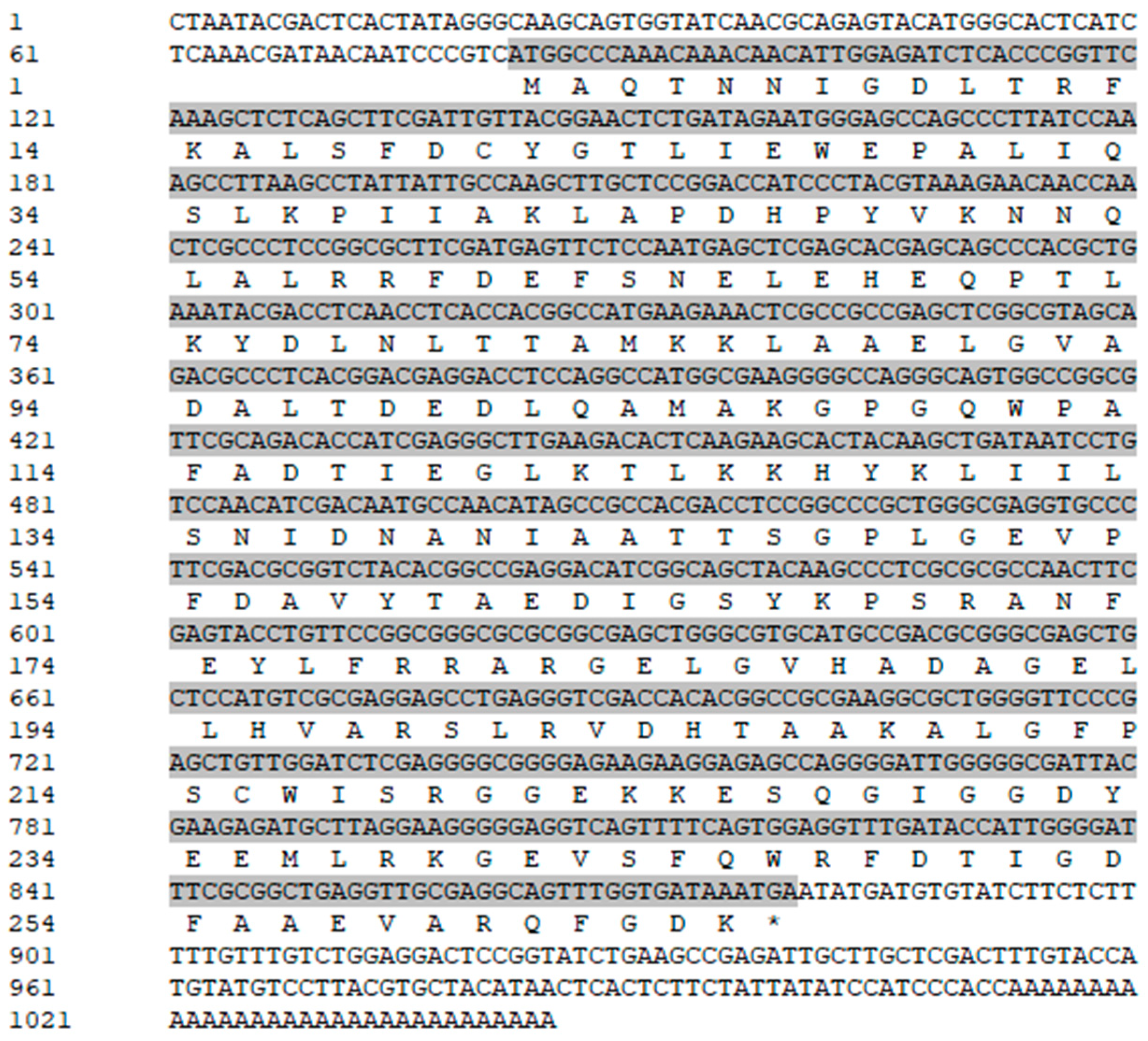
3.1. Full-Length cDNA Sequence of VmHAD Gene Cloning and Characterization
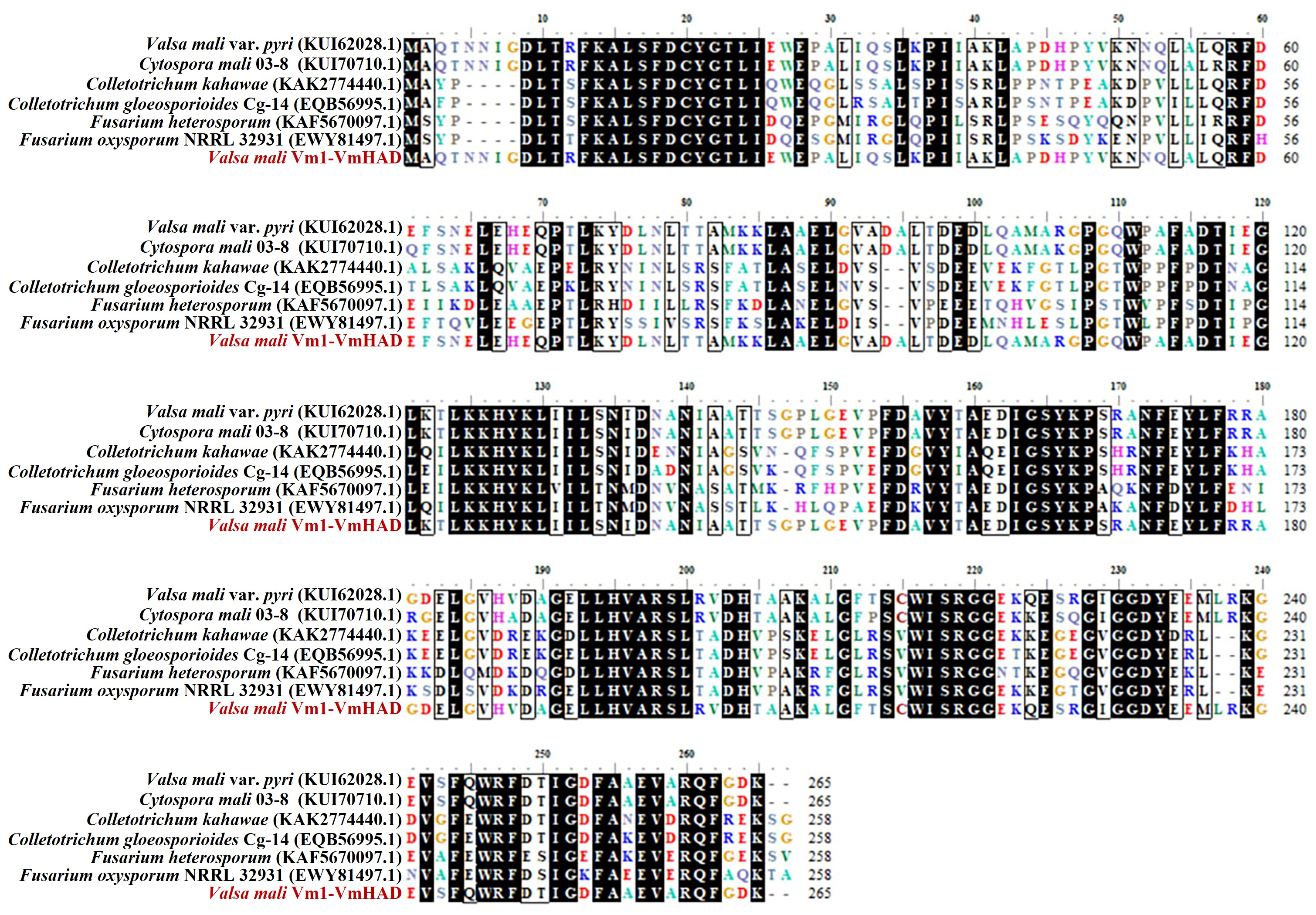
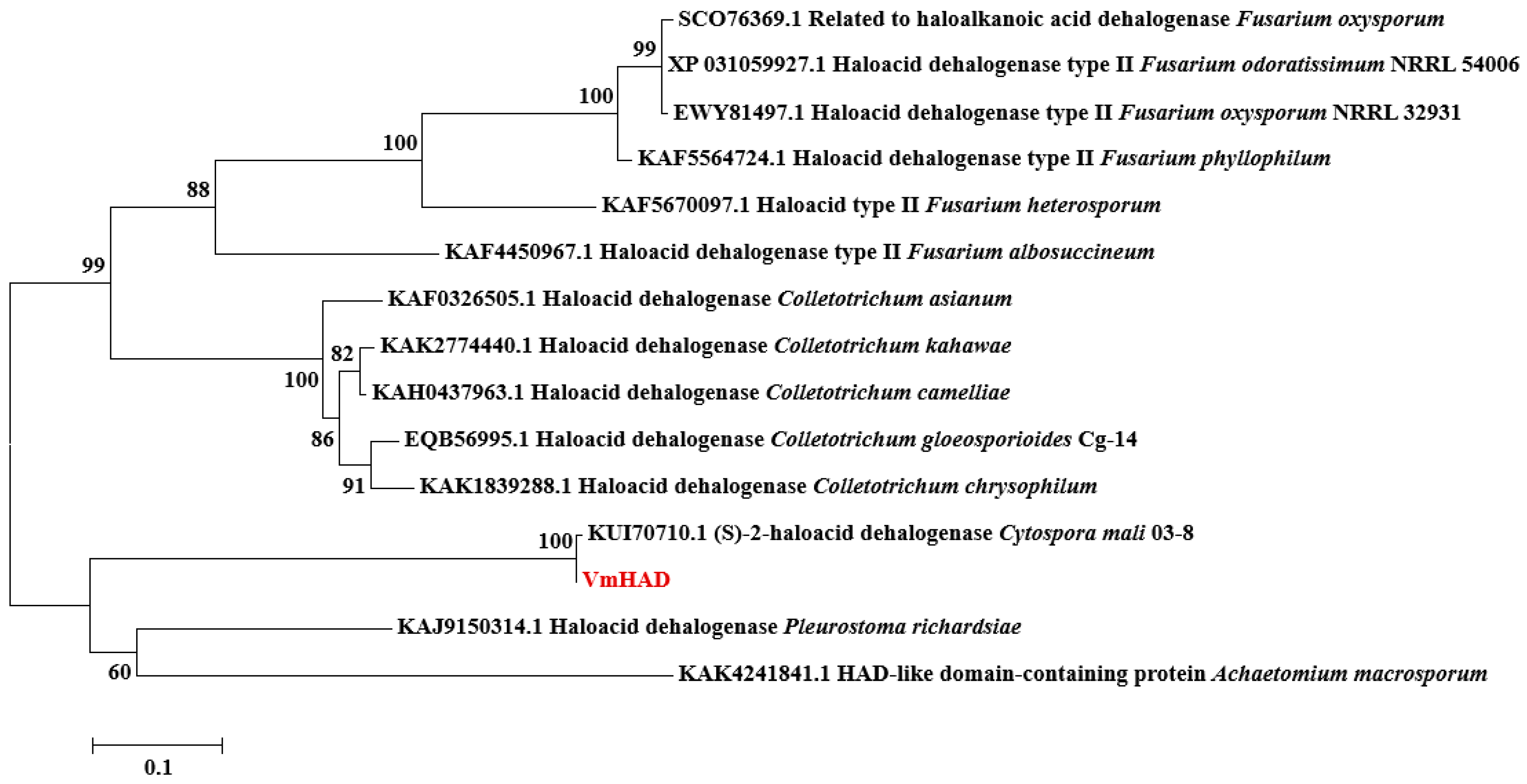
3.2. VmHAD Bioinformation Analysis
3.3. Subcellular Localization Vector of VmHAD Gene Construction and Transformation
3.4. VmHAD Subcellular Location Analysis
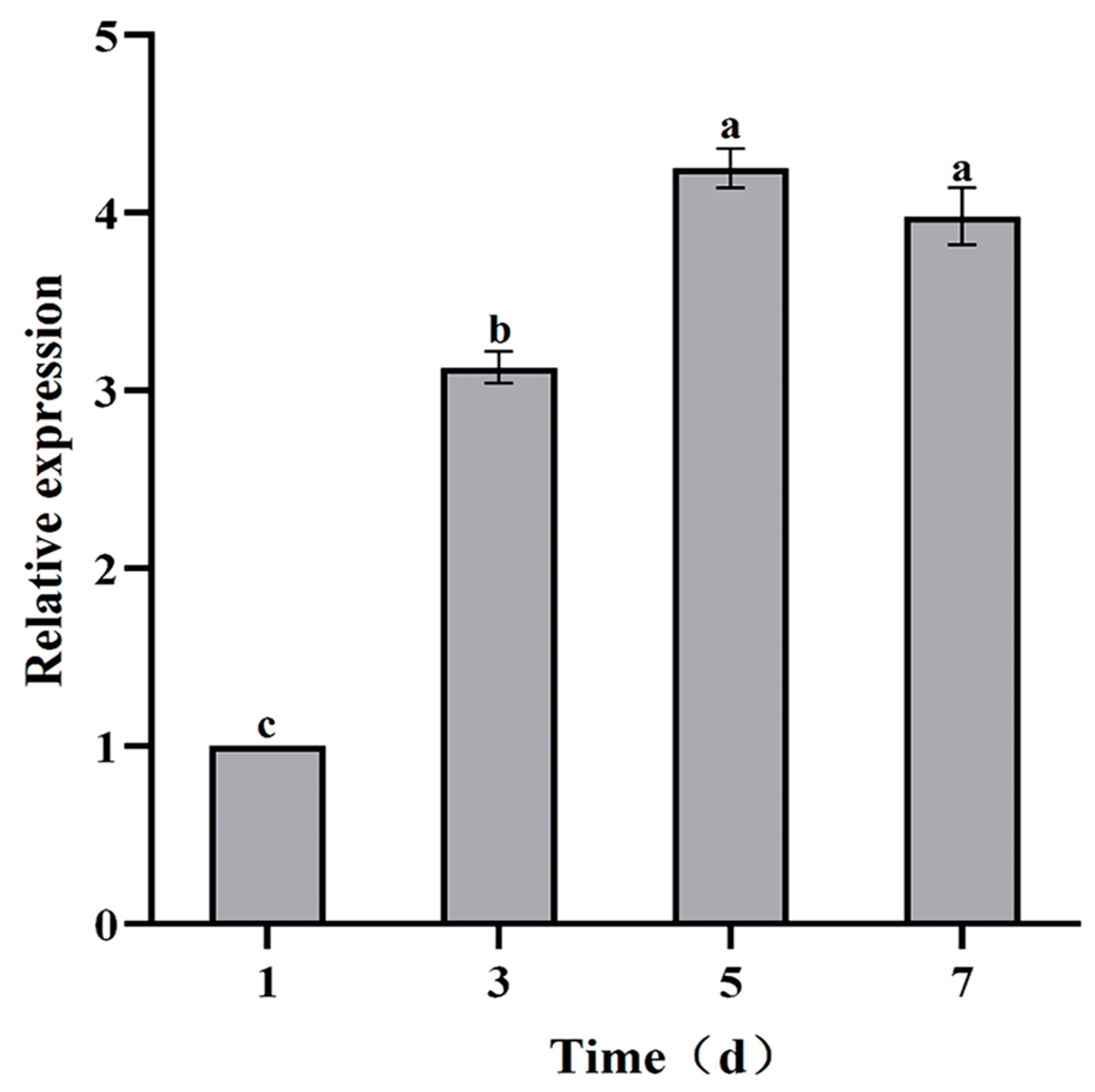
3.5. VmHAD Gene Expression Characteristics Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cornille, A.; Giraud, T.; Smulders, M.J.M.; Roldan-Ruiz, I.; Gladieux, P. The domestication and evolutionary ecology of apples. Trends Genet. 2014, 30, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Duan, N.B.; Bai, Y.; Sun, H.H.; Wang, N.; Ma, Y.M.; Li, M.J.; Wang, X.; Jiao, C.; Legall, N.; Mao, L.Y.; et al. Genome re-sequencing reveals the history of apple and supports a two-stage model for fruit enlargement. Nat. Commun. 2017, 8, 249. [Google Scholar] [CrossRef]
- Daccache, M.A.; Koubaa, M.; Maroun, R.G.; Salameh, D.; Louka, N.; Vorobiev, E. Impact of the physicochemical composition and microbial diversity in apple juice fermentation process: A review. Molecules 2020, 25, 3698. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.X.; Wei, X.Y.; Xiao, Y.X.; Sun, Y.; Biggs, A.R.; Gleason, M.L.; Shang, S.P.; Zhu, M.Q.; Guo, Y.Z.; Sun, G.Y. Management of Valsa canker on apple with adjustments to potassium nutrition. Plant Dis. 2016, 100, 884–889. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.T.; Hu, T.L.; Wang, Y.A.; Luo, Y.; Michailides, T.J.; Cao, K.Q. New understanding on infection processes of Valsa canker of apple in China. Eur. J. Plant Pathol. 2016, 146, 531–540. [Google Scholar] [CrossRef]
- Zhou, Y.B.; Wang, X.H.; Chen, K.M.; Han, C.Y.; Guan, H.P.; Wang, Y.; Zhao, Y.R. Feasibility and potential of terahertz spectral and imaging technology for apple Valsa canker detection: A preliminary investigation. Spectrochim. Acta 2025, 327, 125308. [Google Scholar] [CrossRef]
- Cao, K.Q.; Guo, L.Y.; Li, B.H.; Sun, G.Y.; Chen, H.J. Investigations on the occurrence and control of apple canker in China. Plant Protect. 2009, 35, 114–117. [Google Scholar]
- Vasilyeva, L.; Kim, W.G. Valsa mali Miyabe et Yamada, the causal fungus of apple tree canker in east Asia. Mycobiology 2000, 28, 153–157. [Google Scholar] [CrossRef]
- Wang, X.L.; Shi, C.M.; Gleason, M.L.; Huang, L.L. Fungal species associated with apple Valsa canker in east Asia. Phytopathol. Res. 2020, 2, 14. [Google Scholar] [CrossRef]
- Abe, K.; Kotoda, N.; Kato, H.; Soejima, J. Resistance sources to Valsa canker (Valsa ceratosperma) in a germplasm collection of diverse Malus species. Plant Breed. 2007, 126, 449–453. [Google Scholar] [CrossRef]
- Wang, X.L.; Zang, R.; Yin, Z.Y.; Kang, Z.S.; Huang, L.L. Delimiting cryptic pathogen species causing apple Valsa canker with multilocus data. Ecol. Evol. 2014, 4, 1369–1380. [Google Scholar] [CrossRef]
- Li, Z.P.; Yin, Z.Y.; Fan, Y.Y.; Xu, M.; Kang, Z.S.; Huang, L.L. Candidate effector proteins of the necrotrophic apple canker pathogen Valsa mali can suppress BAX-induced PCD. Front. Plant Sci. 2015, 6, 579. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.J.; Li, X.S.; Bozorov, T.A.; Ma, R.; Ma, J.B.; Zhang, Y.H.; Yang, H.L.; Li, L.; Zhang, D.Y. Characterization and pathogenicity of six Cytospora strains causing stem canker of wild apple in the Tianshan Forest, China. Forest Pathol. 2020, 50, e12587. [Google Scholar] [CrossRef]
- Ke, X.W.; Yin, Z.Y.; Song, N.; Dai, Q.Q.; Voegele, R.T.; Liu, Y.Y.; Wang, H.Y.; Gao, X.N.; Kang, Z.S.; Huang, L.L. Transcriptome profiling to identify genes involved in pathogenicity of Valsa mali on apple tree. Fungal Genet. Biol. 2014, 68, 31–38. [Google Scholar] [CrossRef]
- Yin, Z.Y.; Liu, H.Q.; Li, Z.P.; Ke, X.W.; Dou, D.L.; Gao, X.N.; Song, N.; Dai, Q.Q.; Wu, Y.X.; Xu, J.R.; et al. Genome sequence of Valsa canker pathogens uncovers a potential adaptation of colonization of woody bark. New Phytol. 2015, 208, 1202–1216. [Google Scholar] [CrossRef]
- Sun, G.C.; Xie, S.C.; Tang, L.; Zhao, C.; Zhang, M.; Hang, L.L. Comparative genomics of five Valsa species gives insights on their pathogenicity evolution. G3 2023, 13, jkac312. [Google Scholar] [CrossRef]
- Feng, H.; Wang, C.L.; He, Y.T.; Tang, L.; Han, P.L.; Liang, J.H.; Huang, L.L. Apple Valsa canker: Insights into pathogenesis and disease control. Phytopathol. Res. 2023, 5, 45. [Google Scholar] [CrossRef]
- Wang, C.X.; Li, C.; Li, B.H.; Li, G.F.; Dong, X.L.; Wang, G.P.; Zhang, Q.M. Toxins produced by Valsa mali var. mali and their relationship with pathogenicity. Toxins 2014, 6, 1139–1154. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Gao, X.N.; Chen, J.L.; Yin, Z.Y.; Feng, H.; Huang, L.L. The feruloyl esterase genes are required for full pathogenicity of the apple tree canker pathogen Valsa mali. Mol. Plant Pathol. 2018, 19, 1353–1363. [Google Scholar] [CrossRef] [PubMed]
- McCombe, C.L.; Greenwood, J.R.; Solomon, P.S.; Williams, S.J. Molecular plant immunity against biotrophic, hemibiotrophic, and necrotrophic fungi. Essays Biochem. 2022, 66, 581–593. [Google Scholar] [CrossRef]
- Wang, F.; Ye, L. Biotrophic fungal pathogens: A critical overview. Appl. Biochem. Biotechnol. 2023, 195, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.L.; Li, T.; Shi, X.P.; Saleem, M.; Li, B.H.; Liang, W.X.; Wang, C.X. Deletion of endo-β-1, 4-xylanase VmXyl1 impacts the virulence of Valsa mali in apple tree. Front. Plant Sci. 2018, 9, 663. [Google Scholar] [CrossRef]
- Huang, Y.; Yu, C.L.; Sun, C.C.; Saleem, M.; Li, P.L.; Li, B.H.; Wang, C.X. β-glucosidase VmGlu2 contributes to the virulence of Valsa mali in apple tree. Front. Microbiol. 2021, 12, 695112. [Google Scholar] [CrossRef]
- Cui, X.Y.; Li, X.K.; Li, S.; Huang, Y.; Liu, N.; Lian, S.; Li, B.H.; Wang, C.X. Xylanase VmXyl2 is involved in the pathogenicity of Valsa mali by regulating xylanase activity and inducing cell necrosis. Front. Plant Sci. 2024, 15, 1342714. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.L.; Sun, C.L.; Gao, L.Y.; Saleem, M.; Li, B.H.; Wang, C.C. Hydroxybenzoate hydroxylase genes underlying protocatechuic acid production in Valsa mali are required for full pathogenicity in apple trees. Mol. Plant Pathol. 2021, 22, 1370–1382. [Google Scholar] [CrossRef]
- Aravind, L.; Galperin, M.Y.; Koonin, E.V. The catalytic domain of the P-type ATPase has the haloacid dehalogenase fold. Trends Biochem. Sci. 1998, 23, 127–129. [Google Scholar] [CrossRef]
- Collet, J.F.; Stroobant, V.; Pirard, M.; Delpierre, G.; Van Schaftingen, E. A new class of phosphotransferases phosphorylated on an aspartate residue in an amino-terminal DXDX (T/V) motif. J. Biol. Chem. 1998, 273, 14107–14112. [Google Scholar] [CrossRef]
- Pandey, B.K.; Mehra, P.; Verma, L.; Bhadouria, J.; Giri, J. OsHAD1, a haloacid dehalogenase-like ATPase, enhances phosphate accumulation. Plant Physiol. 2017, 174, 2316–2332. [Google Scholar] [CrossRef]
- May, A.; Berger, S.; Hertel, T.; Kock, M. The Arabidopsis thaliana phosphate starvation responsive gene AtPPsPase1 encodes a novel type of inorganic pyrophosphatase. Biochim. Biophys. Acta 2011, 1810, 178–185. [Google Scholar] [CrossRef]
- Wang, W.; Kim, R.; Jancarik, J.; Yokota, H.; Kim, S.H. Crystal structure of phosphoserine phosphatase from Methanococcus jannaschii, a hyperthermophile, at 1.8 A resolution. Structure 2001, 9, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Yakunin, A.F.; Kuznetsova, E.; Xu, X.; Pennycooke, M.; Gu, J.; Cheung, F.; Proudfoot, M.; Arrowsmith, C.H.; Joachimiak, A.; et al. Structure and function-based characterization of a new phosphoglycolate phosphatase from Thermoplasma acidophilum. J. Biol. Chem. 2004, 279, 517–526. [Google Scholar] [CrossRef]
- Morais, M.C.; Zhang, W.H.; Baker, A.S.; Zhang, G.F.; Dunaway-Mariano, D.; Allen, K.N. The crystal structure of Bacillus cereus phosphonoacetaldehyde hydrolase: insight into catalysis of phosphorus bond cleavage and catalytic diversification within the HAD enzyme superfamily. Biochemistry 2000, 39, 10385–10396. [Google Scholar] [CrossRef]
- Lahiri, S.D.; Zhang, G.F.; Dunaway-Mariano, D.; Allen, K.N. Caught in the act: The structure of phosphorylated β-phosphoglucomutase from Lactococcus lactis. Biochemistry 2002, 41, 8351–8359. [Google Scholar] [CrossRef]
- Huang, H.; Patskovsky, Y.; Toro, R.; Farelli, J.D.; Pandya, C.; Almo, S.C.; Allen, K.N.; Dunaway-Mariano, D. Divergence of structure and function in the haloacid dehalogenase enzyme superfamily: Bacteroides thetaiotaomicron BT2127 is an inorganic pyrophosphatase. Biochemistry 2011, 50, 8937–8949. [Google Scholar] [CrossRef]
- Roberts, A.; Lee, S.Y.; McCullagh, E.; Silversmith, R.E.; Wemmer, D.E. YbiV from Escherichia coli K12 is a HAD phosphatase. Proteins 2005, 58, 790–801. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, L.W.; Dunaway-Mariano, D.; Allen, K.N. Structure and activity analyses of Escherichia coli K-12 NagD provide insight into the evolution of biochemical function in the haloalkanoic acid dehalogenase superfamily. Biochemistry 2006, 45, 1183–1193. [Google Scholar] [CrossRef]
- Hisano, T.; Hata, Y.; Fujii, T.; Liu, J.Q.; Kurihara, T.; Esaki, N.; Soda, K. Crystal structure of L-2-haloacid dehalogenase from Pseudomonas sp. YL. An alpha/beta hydrolase structure that is different from the alpha/beta hydrolase fold. J. Biol. Chem. 1996, 271, 20322–20330. [Google Scholar] [CrossRef] [PubMed]
- Schmidberger, J.W.; Wilce, J.A.; Weightman, A.J.; Wilce, M.C.J. Purification, crystallization and preliminary crystallographic analysis of DehI, a group I α-haloacid dehalogenase from Pseudomonas putida strain PP3. Acta Crystallogr. Sect. F 2008, 64, 596–598. [Google Scholar] [CrossRef] [PubMed]
- Ridder, I.S.; Rozeboom, H.J.; Kalk, K.H.; Janssen, D.B.; Dijkstra, B.W. Three-dimensional structure of L-2-haloacid dehalogenase from Xanthobacter autotrophicus GJ10 complexed with the substrate-analogue formate. J. Biol. Chem. 1997, 272, 33015–33022. [Google Scholar] [CrossRef]
- Kaur, H.; Rode, S.; KP, S.; Mahto, J.K.; Alam, M.S.; Gupta, D.N.; Kar, B.; Singla, J.; Kumar, P.; Sharma, A.K. Characterization of haloacid dehalogenase superfamily acid phosphatase from Staphylococcus lugdunensis. Arch. Biochem. Biophys. 2024, 753, 109888. [Google Scholar] [CrossRef]
- García-Martínez, J.; Castrillo, M.; Avalos, J. The gene cutA of Fusarium fujikuroi, encoding a protein of the haloacid dehalogenase family, is involved in osmotic stress and glycerol metabolism. Microbiology 2014, 160, 26–36. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Wen, C.Y.; Li, F.X.; Wei, C.X.; Du, H.Y.; Zhong, R.R.; Zhao, Y. Function analysis of haloacid dehalogenase type II gene FoHAD-type II in Fusarium oxysporum f. sp. momordicae. Acta Phytopathol. Sin. 2023, 53, 195–206. [Google Scholar]
- Fang, X.L.; Barbetti, M.J. Differential protein accumulations in isolates of the strawberry wilt pathogen Fusarium oxysporum f. sp. fragariae differing in virulence. J. Proteom. 2014, 108, 223–237. [Google Scholar] [CrossRef]
- Ren, W.C.; Zhang, Y.H.; Zhu, M.Q.; Liu, Z.Q.; Lian, S.; Wang, C.X.; Li, B.H.; Liu, N. The phosphatase cascade Nem1/Spo7-Pah1 regulates fungal development, lipid homeostasis, and virulence in Botryosphaeria dothidea. Microbiol. Spectr. 2023, 11, e0388122. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Tatusov, R.L. Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity. Application of an iterative approach to database search. J. Mol. Biol. 1994, 244, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Burroughs, A.M.; Allen, K.N.; Dunaway-Mariano, D.; Aravind, L. Evolutionary genomics of the HAD superfamily: Understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J. Mol. Biol. 2006, 361, 1003–1034. [Google Scholar] [CrossRef]
- Kuznetsova, E.; Nocek, B.; Brown, G.; Makarova, K.S.; Flick, R.; Wolf, Y.I.; Khusnutdinova, A.; Evdokimova, E.; Jin, K.; Tan, K.; et al. Functional diversity of haloacid dehalogenase superfamily phosphatases from Saccharomyces cerevisiae: Biochemical, structural, and evolutionary insights. J. Biol. Chem. 2015, 290, 18678–18698. [Google Scholar] [CrossRef] [PubMed]
- Pathira Kankanamge, L.S.; Ruffner, L.A.; Touch, M.M.; Pina, M.; Beuning, P.J.; Ondrechen, M.J. Functional annotation of haloacid dehalogenase superfamily structural genomics proteins. Biochem. J. 2023, 480, 1553–1569. [Google Scholar] [CrossRef]
- Zakary, S.; Oyewusi, H.A.; Huyop, F. Genomic analysis of Mesorhizobium loti strain tono reveals dehalogenases for bioremediation. J. Trop. Life Sci. 2021, 11, 67–77. [Google Scholar] [CrossRef]
- Rye, C.A.; Isupov, M.N.; Lebedev, A.A.; Littlechild, J.A. Biochemical and structural studies of a L-haloacid dehalogenase from the thermophilic archaeon Sulfolobus tokodaii. Extremophiles 2009, 13, 179–190. [Google Scholar] [CrossRef]
- Ohkouchi, Y.; Koshikawa, H.; Terashima, Y. Cloning and expression of DL-2-haloacid dehalogenase gene from Burkholderia cepacia. Water Sci. Technol. 2000, 42, 261–268. [Google Scholar] [CrossRef]
- Kumar, A.; Pillay, B.; Olaniran, A.O. L-2-haloacid dehalogenase from Ancylobacter aquaticus UV5: Sequence determination and structure prediction. Int. J. Biol. Macromol. 2016, 83, 216–225. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiao, T.L.; Liu, X.H.; Lin, F.C.; Wu, W.R. Functional characterization of a NEM1-like gene in Magnaporthe oryzae. Agric. Sci. China 2011, 10, 1385–1390. [Google Scholar] [CrossRef]
- Lee, S.; Choi, E.; Kim, T.; Hwang, J.; Lee, J. AtHAD1, a haloacid dehalogenase-like phosphatase, is involved in repressing the ABA response. Biochem. Bioph. Res. Commun. 2022, 587, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Winkelströter, L.K.; Dolan, S.K.; Fernanda Dos Reis, T.; Bom, V.L.P.; Alves de Castro, P.; Hagiwara, D.; Alowni, R.; Jones, G.W.; Doyle, S.; Brown, N.A.; et al. Systematic global analysis of genes encoding protein phosphatases in Aspergillus fumigatus. G3 2015, 5, 1525–1539. [Google Scholar] [CrossRef]
- Liu, N.; Yun, Y.Z.; Yin, Y.N.; Hahn, M.; Ma, Z.H.; Chen, Y. Lipid droplet biogenesis regulated by the FgNem1/Spo7-FgPah1 phosphatase cascade plays critical roles in fungal development and virulence in Fusarium graminearum. New Phytol. 2019, 223, 412–429. [Google Scholar] [CrossRef]
- Yun, Y.Z.; Liu, Z.Y.; Yin, Y.N.; Jiang, J.H.; Chen, Y.; Xu, J.R.; Ma, Z.H. Functional analysis of the Fusarium graminearum phosphatome. New Phytol. 2015, 207, 119–134. [Google Scholar] [CrossRef]
| Physicochemical Properties | VmHAD Protein |
|---|---|
| Number of amino acids | 265 |
| Relative molecular mass | 29.4 kDa |
| Molecular formula | C1317H2049N357O395S6 |
| Theoretical isoelectric point (pI) | 5.47 |
| Total number of positively charged residues (Arg + Lys) | 31 |
| Total number of negatively charged residues (Asp + Glu) | 38 |
| Grand average of hydropathicity (GRAVY) | −0.358 |
| Aliphatic index (AI) | 83.36 |
| Instability index (II) (<40 stable, >40 unstable) | 38.20 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Chen, X.; Duan, C.; Liu, J.; Tao, F.; Xu, B. Identification, Subcellular Localization, and Infection-Related Expression of a Novel Haloacid Dehalogenase Gene (VmHAD) from Valsa mali Vm1. J. Fungi 2025, 11, 827. https://doi.org/10.3390/jof11120827
Zhang S, Chen X, Duan C, Liu J, Tao F, Xu B. Identification, Subcellular Localization, and Infection-Related Expression of a Novel Haloacid Dehalogenase Gene (VmHAD) from Valsa mali Vm1. Journal of Fungi. 2025; 11(12):827. https://doi.org/10.3390/jof11120827
Chicago/Turabian StyleZhang, Shuwu, Xingxu Chen, Cizhong Duan, Jia Liu, Fei Tao, and Bingliang Xu. 2025. "Identification, Subcellular Localization, and Infection-Related Expression of a Novel Haloacid Dehalogenase Gene (VmHAD) from Valsa mali Vm1" Journal of Fungi 11, no. 12: 827. https://doi.org/10.3390/jof11120827
APA StyleZhang, S., Chen, X., Duan, C., Liu, J., Tao, F., & Xu, B. (2025). Identification, Subcellular Localization, and Infection-Related Expression of a Novel Haloacid Dehalogenase Gene (VmHAD) from Valsa mali Vm1. Journal of Fungi, 11(12), 827. https://doi.org/10.3390/jof11120827





