Biological Evaluation and Potential Applications of Secondary Metabolites from Fungi Belonging to the Cordycipitaceae Family with a Focus on Parengyodontium spp.
Abstract
1. Introduction
2. The Chemical Diversity of Cordycipitaceae Family: Comparison Between Entomopathogenic and Saprobic/Mycoparasitic Lineages
3. The Chemical Diversity in Parengyodontium Genus (Focus Section)
3.1. Identified Secondary Metabolites Classified by Chemical Family
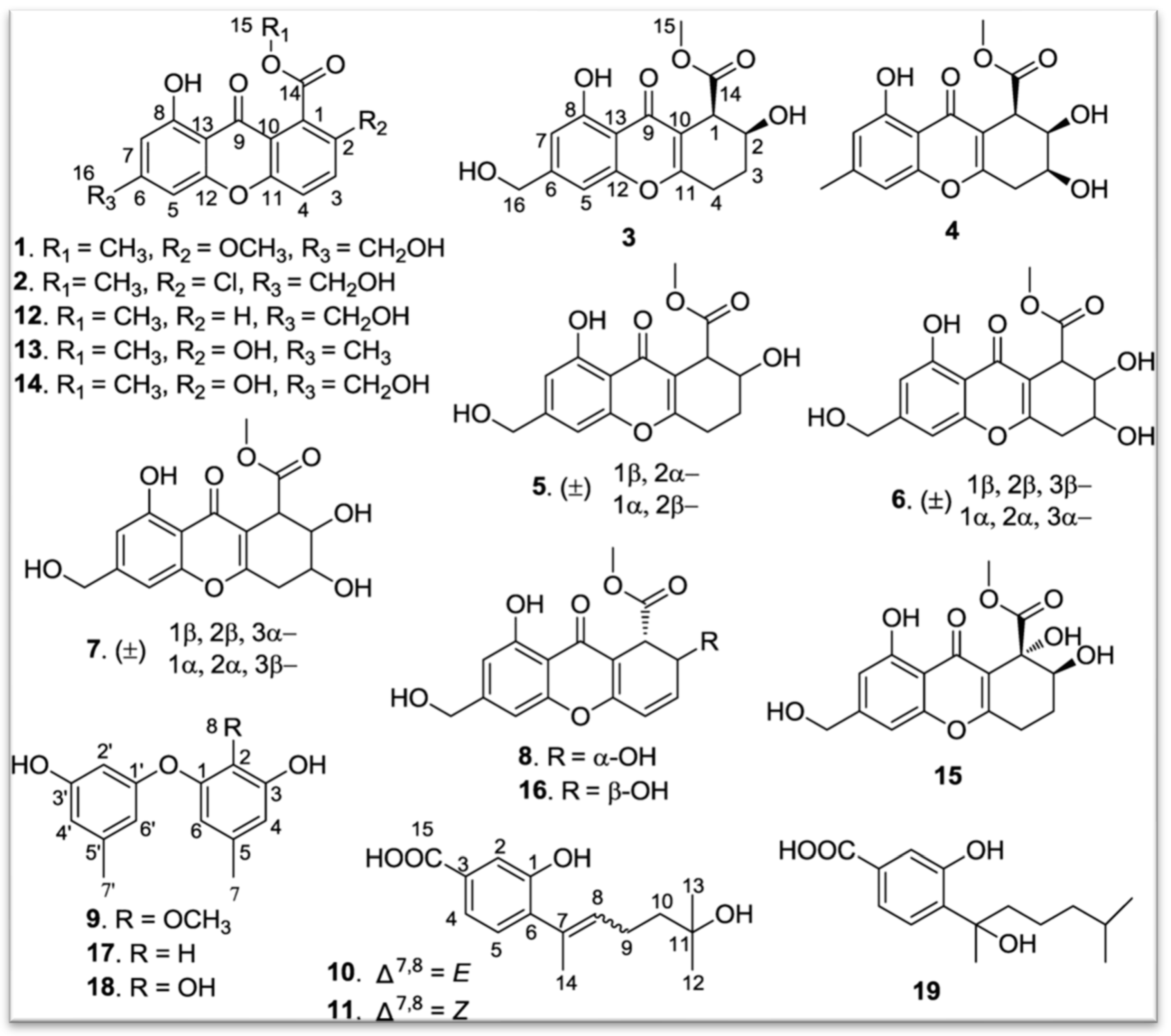
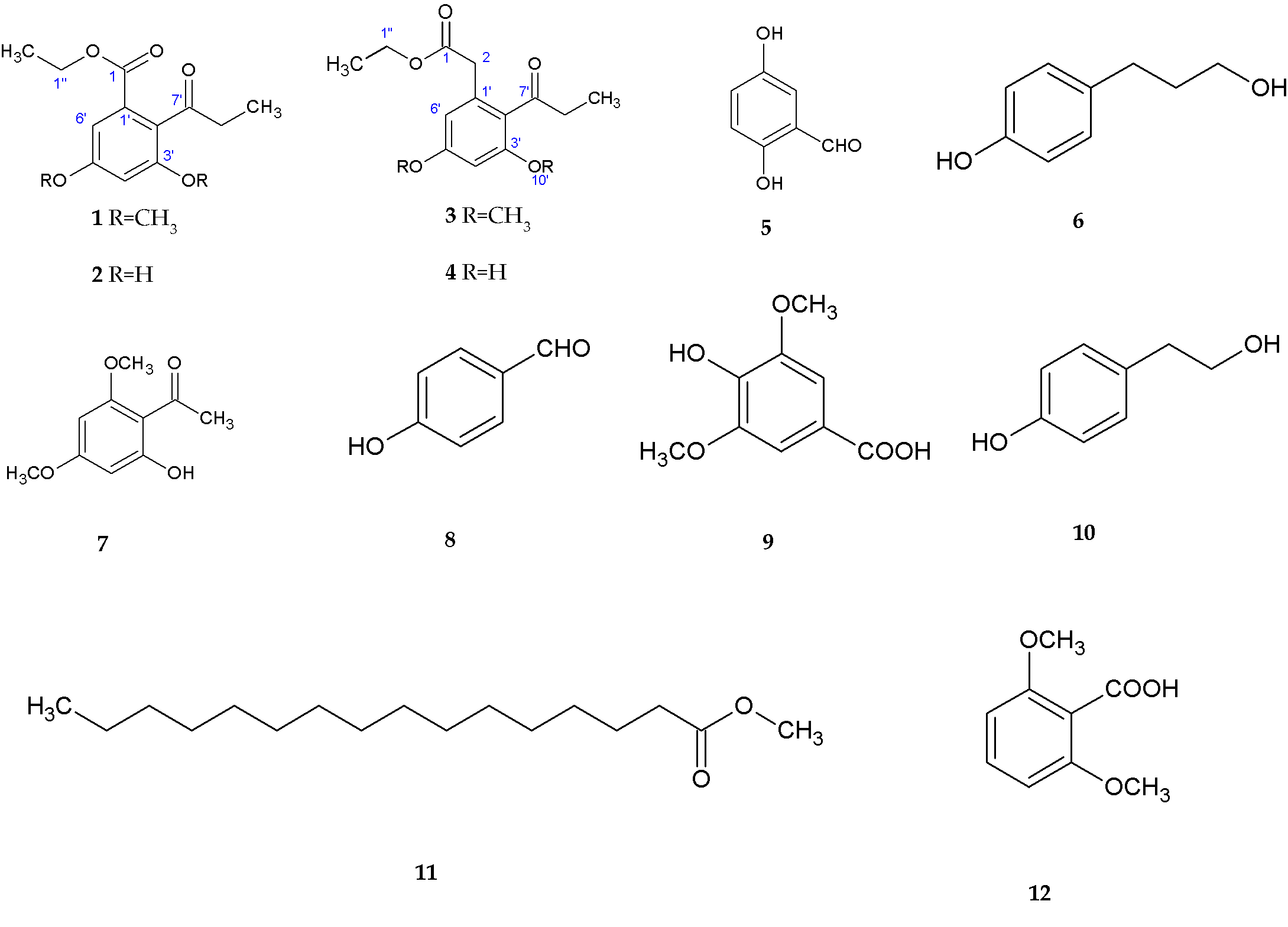
3.1.1. Polyketides
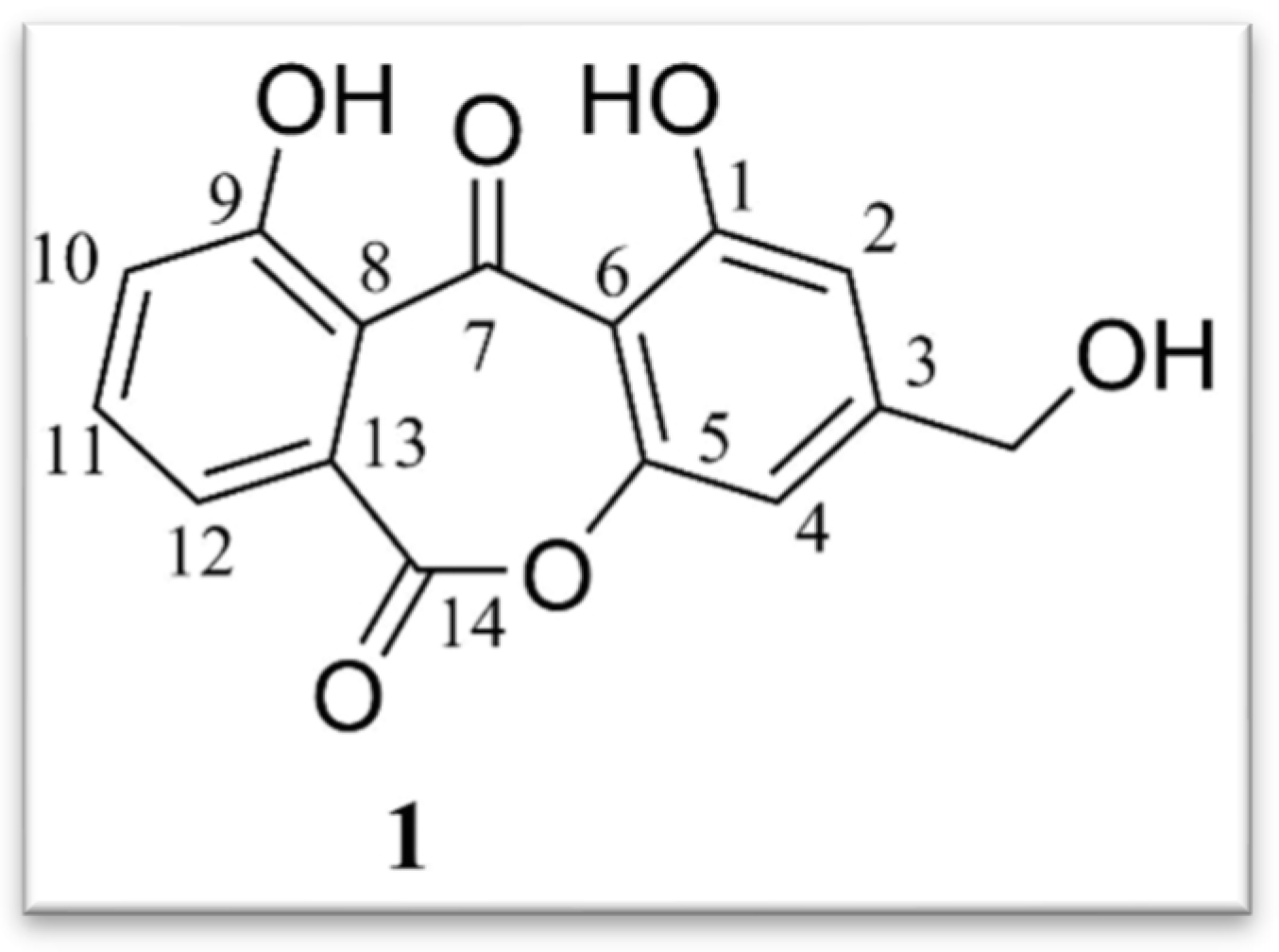
- -
- -
- -
3.1.2. Terpene Compounds
- -
- Cytochalasin: Genomic analysis of Parengyodontium torokii predicted the biosynthesis of cytochalasine K, a terpenoid compound [29]. This metabolite was identified by LC-MS in a fungal extract, confirming the in silico predictions [29]. Cytochalasin compounds are known for their structural diversity and biological activities, particularly anti-cancer activities [87].
- -
- Other terpenes: Gene clusters for the production of other terpenes or related compounds such as squalestatin S1 have been identified in the species P. torokii [29].
3.1.3. Alkaloids and Other Chemical Families
- -
- Indole alkaloids: A new indole alkaloid, 1-(4-hydroxybenzoyl)indole-3-carbaldehyde (Figure 7), was isolated from a strain of Engyodontium album derived from a marine sponge [52]. Alkaloids are a family of nitrogen-containing compounds known for their major pharmacological properties. They include, but are not limited to, morphine (analgesic), quinine (antimalarial), atropine (anticholinergic), etc.
- -
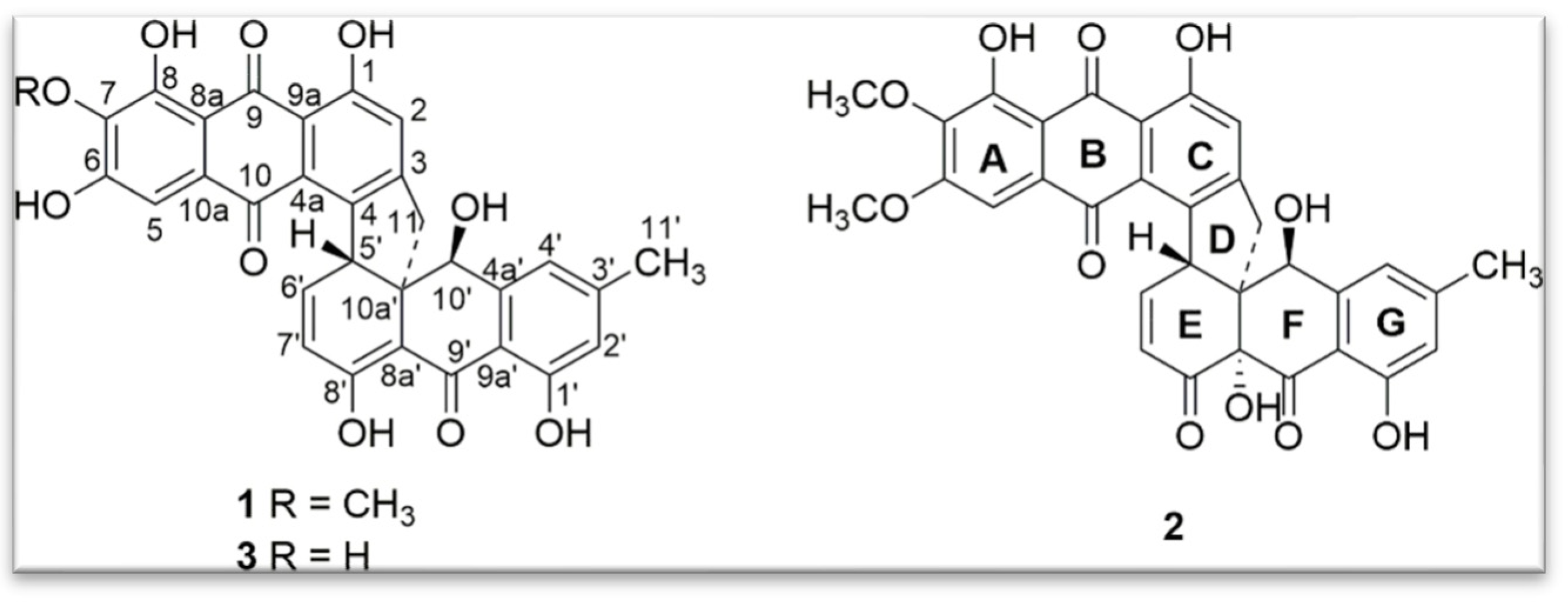
- Torrubiellin derivatives: A strain of P. album isolated from the leaves of Avicennia marina (in mangroves) produces new torrubiellin derivatives, named parengyomarin A (1) and B (2), in addition to the already known torrubiellin B (3) (Figure 8). Other compounds such as emodin and emodic acid have also been identified in extracts of this fungus [42].
- -
- Other compounds: Metabolomic analysis of P.torokii identified several other molecules, including cyclic peptides such as cyclo(L-Leu-L-Pro) and (3β,22E)-cyclo(L-Pro-L-Leu), fatty acids (6,9-octadecadienoic acid), and compounds such as cephalochromin and betulinan [29]. In silico predictions have also suggested the presence of equistetin, cephalosporin C, EQ-4, curcupallide-B, pyranonigrin E and dimethylcoprogen [29].
4. Potential Applications of Secondary Metabolites Produced by Parengyodontium spp.
- -
- Anticancer activities: Polyketides, such as Engyodontiumones, have shown selective cytotoxicity against the human histiocytic lymphoma cell line U937, with IC50 values of 4.9 and 8.8 µM for compounds 8 and 16, respectively [85]. Cytochalasin K, identified in P. torokii, has been shown to influence the final stages of mitosis and have a marked synergistic effect on cancer cells [29]. Cytotoxic polyketides (Xanthoquinodin JBIR-99) have been isolated from Parengyodontium album [50]. Alternaphenol B2 from P. album showed selective inhibitory activity against mutant isocitrate dehydrogenase R132H (IDH1m), a relevant target for cancer treatment, with an IC50 of 41.9 µM [51].
- -
- Antibacterial activities: Several metabolites exhibited antibacterial properties. Compounds 8, 15, and 16 from P. album showed moderate antibacterial activity against Escherichia coli and Bacillus subtilis [85]. A phenylacetate derivative (compound 3) from P. album exhibited inhibitory activity against methicillin-resistant Staphylococcus aureus (MRSA) and Vibrio vulnificus, with MICs of 7.8 and 15.6 µg/mL, respectively [56]. Torrubielline derivatives have also demonstrated antibacterial activities [42]. Fungal mycelium extracts possess antimicrobial properties, with superior efficacy against Gram-positive bacteria [54].
- -
- Antilaryngeal activities: Compound 15, a polyketide from P. album DFFSCS021, showed potent antilaryngeal activity against the establishment of barnacle larvae (Balanus amphitrite) [85]. This property suggests potential for the development of biofoulants.
- -
- Enzymes and other applications: Genomic analyses of P. torokii have revealed the presence of enzyme families such as GH33 glycosyl hydrolases (sialidases) and GT20 and GT34 glycosyltransferases. These enzymes may have biotechnological applications, particularly for the modification of glycoconjugates or the biosynthesis of disaccharides and oligosaccharides [29]. In addition, the genus Parengyodontium is of interest in bioremediation, as evidenced by P. album, which is capable of biodegrading certain synthetic plastics such as polyethylene [44]. This result suggests the presence of enzymes such as laccases, oxidases, and peroxidases [44]. Laccases are multi-copper oxidases widely found in fungi, plants and bacteria. Fungal laccases are particularly valued because they oxidize a wide range of phenolic and non-phenolic substrates (often with redox mediators) while reducing O2 to H2O, enabling applications ranging from lignin modification to green synthesis and pollutant removal [88,89]. Peroxidases, including lignin peroxidase (LiP), manganese peroxidase (MnP), versatile peroxidase (VP), and dye-decolorizing peroxidases (DyPs), are heme enzymes that use H2O2 to attack lignin and recalcitrant aromatic compounds. Recent work highlights engineered VPs, MnP-mediated oxidation via Mn3+ chelates, and DyP diversity across fungi for lignin/dye transformation [90,91,92]. Oxidases generate H2O2 from O2. An example is glucose oxidase, which oxidizes β-D-glucose to D-glucono-δ-lactone and H2O2 and remains central in biosensors and food applications [93,94,95]. In white-rot systems, aryl-alcohol oxidase supplies H2O2 to ligninolytic peroxidases and can also act as a quinone reductase, enhancing the degradation of lignin by peroxidases [96,97].
5. Discussion: Perspectives, Limitations and Future Directions
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Atli, B.; Ozcakir, B.; Isik, B.; Mursaliyeva, V.; Mammadov, R. Secondary Metabolites in Fungi. Nat. Prod. Biotechnol. 2022, 2, 114–138. [Google Scholar] [CrossRef]
- Bills, G.F.; Gloer, J.B. Biologically Active Secondary Metabolites from the Fungi. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Avalos, J.; Limón, M.C. Fungal Secondary Metabolism. Encyclopedia 2021, 2, 1–13. [Google Scholar] [CrossRef]
- Demain, A.L.; Fang, A. The Natural Functions of Secondary Metabolites. In History of Modern Biotechnology I; Fiechter, A., Ed.; Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2000; Volume 69, pp. 1–39. ISBN 978-3-540-67793-2. [Google Scholar] [CrossRef]
- Keller, N.P. Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef]
- Bertrand, S.; Bohni, N.; Schnee, S.; Schumpp, O.; Gindro, K.; Wolfender, J.-L. Metabolite induction via microorganism co-culture: A potential way to enhance chemical diversity for drug discovery. Biotechnol. Adv. 2014, 32, 1180–1204. [Google Scholar] [CrossRef]
- Brakhage, A.A. Regulation of fungal secondary metabolism. Nat. Rev. Microbiol. 2013, 11, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, S.K.; Verekar, S.A.; Bhave, S.V. Endophytic fungi: A reservoir of antibacterials. Front. Microbiol. 2015, 5, 715. [Google Scholar] [CrossRef] [PubMed]
- Kempken, F. Marine fungi: A treasure trove of novel natural products and for biological discovery. PLoS Pathog. 2023, 19, e1011624. [Google Scholar] [CrossRef] [PubMed]
- Macheleidt, J.; Mattern, D.J.; Fischer, J.; Netzker, T.; Weber, J.; Schroeckh, V.; Valiante, V.; Brakhage, A.A. Regulation and Role of Fungal Secondary Metabolites. Annu. Rev. Genet. 2016, 50, 371–392. [Google Scholar] [CrossRef]
- Rateb, M.E.; Ebel, R. Secondary metabolites of fungi from marine habitats. Nat. Prod. Rep. 2011, 28, 290. [Google Scholar] [CrossRef]
- Kepler, R.M.; Luangsa-ard, J.J.; Hywel-Jones, N.L.; Quandt, C.A.; Sung, G.-H.; Rehner, S.A.; Aime, M.C.; Henkel, T.W.; Sanjuan, T.; Zare, R.; et al. A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 2017, 8, 335–353. [Google Scholar] [CrossRef]
- Wang, Y.-B.; Wang, Y.; Fan, Q.; Duan, D.-E.; Zhang, G.-D.; Dai, R.-Q.; Dai, Y.-D.; Zeng, W.-B.; Chen, Z.-H.; Li, D.-D.; et al. Multigene phylogeny of the family Cordycipitaceae (Hypocreales): New taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Divers. 2020, 103, 1–46. [Google Scholar] [CrossRef]
- Khonsanit, A.; Thanakitpipattana, D.; Mongkolsamrit, S.; Kobmoo, N.; Phosrithong, N.; Samson, R.A.; Crous, P.W.; Luangsa-ard, J.J. A phylogenetic assessment of Akanthomyces sensu lato in Cordycipitaceae (Hypocreales, Sordariomycetes): Introduction of new genera, and the resurrection of Lecanicillium. Fungal Syst. Evol. 2024, 14, 271–306. [Google Scholar] [CrossRef]
- Bu, J.; Wei, D.-P.; Liu, Z.-H.; Yang, Y.; Liu, Z.-L.; Kang, J.-C.; Peng, X.-C.; Xie, S.-W.; Zhang, H.-G.; He, Z.-J.; et al. Molecular phylogeny and morphology reveal four novel species in Cordycipitaceae in China. MycoKeys 2025, 116, 91–124. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.-H.; Li, D.; Shu, H.-L.; Liang, J.-D.; Zhao, J.-H.; Tian, W.-Y.; Han, Y.-F. Four new araneogenous species and a new genus in Hypocreales (Clavicipitaceae, Cordycipitaceae) from the karst region of China. MycoKeys 2025, 112, 335–359. [Google Scholar] [CrossRef]
- Kobmoo, N.; Tasanathai, K.; Araújo, J.P.M.; Noisripoom, W.; Thanakitpipattana, D.; Mongkolsamrit, S.; Himaman, W.; Houbraken, J.; Luangsa-Ard, J.J. New mycoparasitic species in the genera Niveomyces and Pseudoniveomyces gen. nov. (Hypocreales: Cordycipitaceae), with sporothrix-like asexual morphs, from Thailand. Fungal Syst. Evol. 2023, 12, 91–110. [Google Scholar] [CrossRef]
- Mongkolsamrit, S.; Noisripoom, W.; Tasanathai, K.; Kobmoo, N.; Thanakitpipattana, D.; Khonsanit, A.; Petcharad, B.; Sakolrak, B.; Himaman, W. Comprehensive treatise of Hevansia and three new genera Jenniferia, Parahevansia and Polystromomyces on spiders in Cordycipitaceae from Thailand. MycoKeys 2022, 91, 113–149. [Google Scholar] [CrossRef]
- Dong, Q.-Y.; Wang, Y.; Wang, Z.-Q.; Tang, D.-X.; Zhao, Z.-Y.; Wu, H.-J.; Yu, H. Morphology and Phylogeny Reveal Five Novel Species in the Genus Cordyceps (Cordycipitaceae, Hypocreales) From Yunnan, China. Front. Microbiol. 2022, 13, 846909. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, Y.; Dong, Q.; Fan, Q.; Dao, V.-M.; Yu, H. Morphological and Phylogenetic Characterization Reveals Five New Species of Samsoniella (Cordycipitaceae, Hypocreales). JoF 2022, 8, 747. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, X.-C.; Wu, L.-X.; Wang, Y.; Xu, A.; Lin, W.-F. Blackwellomyces kaihuaensis and Metarhizium putuoense (Hypocreales), Two New Entomogenous Fungi from Subtropical Forests in Zhejiang Province, Eastern China. Forests 2023, 14, 2333. [Google Scholar] [CrossRef]
- Sung, G.-H.; Hywel-Jones, N.L.; Sung, J.-M.; Luangsa-ard, J.J.; Shrestha, B.; Spatafora, J.W. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef]
- Aini, A.N.; Mongkolsamrit, S.; Noisripoom, W.; Himaman, W.; Houbraken, J.; Luangsa-ard, J.J. Diversity of Akanthomyces on moths (Lepidoptera) in Thailand. MycoKeys 2020, 72, 1–24. [Google Scholar] [CrossRef]
- Chen, M.-J.; Wang, T.; Lin, Y.; Huang, B. Morphological and molecular analyses reveal two new species of Gibellula from China. MycoKeys 2022, 90, 53–69. [Google Scholar] [CrossRef] [PubMed]
- Mendes-Pereira, T.; De Araújo, J.P.M.; Kloss, T.G.; Costa-Rezende, D.H.; De Carvalho, D.S.; Góes-Neto, A. Disentangling the Taxonomy, Systematics, and Life History of the Spider-Parasitic Fungus Gibellula (Cordycipitaceae, Hypocreales). JoF 2023, 9, 457. [Google Scholar] [CrossRef] [PubMed]
- Humber, R.A. Identification of entomopathogenic fungi. In Manual of Techniques in Invertebrate Pathology; Lacey, L.A., Ed.; Academic Press: Amsterdam, The Netherlands, 2012; pp. 151–187. [Google Scholar]
- Vega, F.E.; Goettel, M.S.; Blackwell, M.; Chandler, D.; Jackson, M.A.; Keller, S.; Pell, J.K. Fungal entomopathogens: New insights on their ecology. Fungal Ecol. 2009, 2, 149–159. [Google Scholar] [CrossRef]
- Wang, Y.; Fan, Q.; Wang, D.; Zou, W.-Q.; Tang, D.-X.; Hongthong, P.; Yu, H. Species Diversity and Virulence Potential of the Beauveria bassiana Complex and Beauveria scarabaeidicola Complex. Front. Microbiol. 2022, 13, 841604. [Google Scholar] [CrossRef]
- Parker, C.W.; Teixeira, M.d.M.; Singh, N.K.; Raja, H.A.; Cank, K.B.; Spigolon, G.; Oberlies, N.H.; Barker, B.M.; Stajich, J.E.; Mason, C.E.; et al. Genomic Characterization of Parengyodontium torokii sp. nov., a Biofilm-Forming Fungus Isolated from Mars 2020 Assembly Facility. J Fungi 2022, 8, 66. [Google Scholar] [CrossRef] [PubMed]
- Sung, G.-H.; Spatafora, J.W.; Zare, R.; Hodge, K.T.; Gams, W. A revision of Verticillium sect. Prostrata. II. Phylogenetic analyses of SSU and LSU nuclear rDNA sequences from anamorphs and teleomorphs of the Clavicipitaceae. Nova Hedwig. 2001, 72, 311–328. [Google Scholar] [CrossRef]
- Wei, D.; Wanasinghe, D.N.; Hyde, K.D.; Mortimer, P.E.; Xu, J.-C.; Xiao, Y.-P.; Bhunjun, C.S.; To-anun, C. The genus Simplicillium. MycoKeys 2019, 60, 69–92. [Google Scholar] [CrossRef]
- Thanakitpipattana, D.; Mongkolsamrit, S.; Khonsanit, A.; Himaman, W.; Luangsa-ard, J.J.; Pornputtapong, N. Is Hyperdermium Congeneric with Ascopolyporus? Phylogenetic Relationships of Ascopolyporus spp. (Cordycipitaceae, Hypocreales) and a New Genus Neohyperdermium on Scale Insects in Thailand. JoF 2022, 8, 516. [Google Scholar] [CrossRef]
- Tsang, C.-C.; Chan, J.F.W.; Pong, W.-M.; Chen, J.H.K.; Ngan, A.H.Y.; Cheung, M.; Lai, C.K.C.; Tsang, D.N.C.; Lau, S.K.P.; Woo, P.C.Y. Cutaneous hyalohyphomycosis due to Parengyodontium album gen. et comb. nov. Med. Mycol. 2016, 54, 699–713. [Google Scholar] [CrossRef]
- Leplat, J.; François, A.; Bousta, F. Diversity of Parengyodontium spp. strains isolated from the cultural heritage environment: Phylogenetic diversity, phenotypical diversity, and occurrence. Mycologia 2022, 114, 825–840. [Google Scholar] [CrossRef]
- Belfiori, B.; Rubini, A.; Riccioni, C. Diversity of Endophytic and Pathogenic Fungi of Saffron (Crocus sativus) Plants from Cultivation Sites in Italy. Diversity 2021, 13, 535. [Google Scholar] [CrossRef]
- Lucero, M.E.; Barrow, J.R.; Osuna, P.; Reyes, I. Plant–fungal interactions in arid and semi-arid ecosystems: Large-scale impacts from microscale processes. J. Arid. Environ. 2006, 65, 276–284. [Google Scholar] [CrossRef]
- Wu, H.; Yang, H.-Y.; You, X.-L.; Li, Y.-H. Diversity of endophytic fungi from roots of Panax ginseng and their saponin yield capacities. SpringerPlus 2013, 2, 107. [Google Scholar] [CrossRef] [PubMed]
- Kachuei, R.; Emami, M.; Naeimi, B.; Diba, K. Isolation of keratinophilic fungi from soil in Isfahan province, Iran. J. Mycol. Méd. 2012, 22, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.; Zhuang, X.; Wu, J.; Cui, M.; Lv, D.; Liu, C.; Zhuang, G. Ascomycota Members Dominate Fungal Communities during Straw Residue Decomposition in Arable Soil. PLoS ONE 2013, 8, e66146. [Google Scholar] [CrossRef] [PubMed]
- Banchi, E.; Manna, V.; Muggia, L.; Celussi, M. Marine Fungal Diversity and Dynamics in the Gulf of Trieste (Northern Adriatic Sea). Microb. Ecol. 2024, 87, 78. [Google Scholar] [CrossRef]
- Lefkowitz, R.J. Identification of adenylate cyclase-coupled beta-adrenergic receptors with radiolabeled beta-adrenergic antagonists. Biochem. Pharmacol. 1975, 24, 1651–1658. [Google Scholar] [CrossRef]
- Liu, S.; Mao, Y.; Lu, H.; Zhao, Y.; Bilal, M.; Proksch, P.; Hu, P. Two new torrubiellin derivatives from the mangrove endophytic fungus Parengyodontium album. Phytochem. Lett. 2021, 46, 149–152. [Google Scholar] [CrossRef]
- Pindi, P.K. Diversity of fungi at various depths of marine water. Res. Biotechnol. 2012, 3. [Google Scholar]
- Vaksmaa, A.; Vielfaure, H.; Polerecky, L.; Kienhuis, M.V.M.; Van Der Meer, M.T.J.; Pflüger, T.; Egger, M.; Niemann, H. Biodegradation of polyethylene by the marine fungus Parengyodontium album. Sci. Total Environ. 2024, 934, 172819. [Google Scholar] [CrossRef]
- Teixeira, M.D.M.; Muszewska, A.; Travis, J.; Moreno, L.F.; Ahmed, S.; Roe, C.; Mead, H.; Steczkiewicz, K.; Lemmer, D.; De Hoog, S.; et al. Genomic characterization of Parengyodontium americanum sp. nov. Fungal Genet. Biol. 2020, 138, 103351. [Google Scholar] [CrossRef]
- Ciferri, O. Microbial Degradation of Paintings. Appl. Environ. Microbiol. 1999, 65, 879–885. [Google Scholar] [CrossRef]
- Dán, K.; Kocsubé, S.; Tóth, L.; Farkas, A.; Rákhely, G.; Galgóczy, L. Isolation and identification of fungal biodeteriogens from the wall of a cultural heritage church and potential applicability of antifungal proteins in protection. J. Cult. Herit. 2024, 67, 194–202. [Google Scholar] [CrossRef]
- Leplat, J.; François, A.; Bousta, F. Parengyodontium album, a frequently reported fungal species in the cultural heritage environment. Fungal Biol. Rev. 2020, 34, 126–135. [Google Scholar] [CrossRef]
- Gadd, G.M.; Fomina, M.; Pinzari, F. Fungal biodeterioration and preservation of cultural heritage, artwork, and historical artifacts: Extremophily and adaptation. Microbiol. Mol. Biol. Rev. 2024, 88, e00200-22. [Google Scholar] [CrossRef]
- Anaya-Eugenio, G.D.; Rebollar-Ramos, D.; González, M.D.C.; Raja, H.; Mata, R.; Carcache De Blanco, E.J. Apoptotic activity of xanthoquinodin JBIR-99, from Parengyodontium album MEXU 30054, in PC-3 human prostate cancer cells. Chem.-Biol. Interact. 2019, 311, 108798. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shen, W.; Li, G.; Song, Y.; Lu, X.; Wong, N.-K.; Yan, Y. Alternaphenol B2, a new IDH1 inhibitor from the coral-derived fungus Parengyodontium album SCSIO SX7W11. Nat. Prod. Res. 2023, 38, 3917–3923. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.-H.; Chen, H.-Q.; Form, I.; Konuklugil, B.; Proksch, P.; Wang, B.-G. New Chromone, Isocoumarin, and Indole Alkaloid Derivatives from three Sponge-derived Fungal Strains. Nat. Prod. Commun. 2016, 11, 1293–1296. [Google Scholar] [CrossRef]
- Wang, L.; Huang, Y.; Zhang, L.; Liu, Z.; Liu, W.; Xu, H.; Zhang, Q.; Zhang, H.; Yan, Y.; Liu, Z.; et al. Structures and absolute configurations of phomalones from the coral-associated fungus Parengyodontium album sp. SCSIO 40430. Org. Biomol. Chem. 2021, 19, 6030–6037. [Google Scholar] [CrossRef]
- Wong Chin, J.M.; Puchooa, D.; Bahorun, T.; Jeewon, R. Antimicrobial properties of marine fungi from sponges and brown algae of Mauritius. Mycology 2021, 12, 231–244. [Google Scholar] [CrossRef]
- Wu, B.; Wiese, J.; Wenzel-Storjohann, A.; Malien, S.; Schmaljohann, R.; Imhoff, J.F. Engyodontochones, Antibiotic Polyketides from the Marine Fungus Engyodontium album Strain LF069. Chem. Eur. J. 2016, 22, 7452–7462. [Google Scholar] [CrossRef]
- Wang, W.; Chen, R.; Luo, Z.; Wang, W.; Chen, J. Two new benzoate derivatives and one new phenylacetate derivative from a marine-derived fungus Engyodontium album. Nat. Prod. Res. 2017, 31, 758–764. [Google Scholar] [CrossRef]
- Mongkolsamrit, S.; Noisripoom, W.; Tasanathai, K.; Khonsanit, A.; Thanakitpipattana, D.; Himaman, W.; Kobmoo, N.; Luangsa-ard, J.J. Molecular phylogeny and morphology reveal cryptic species in Blackwellomyces and Cordyceps (Cordycipitaceae) from Thailand. Mycol. Prog. 2020, 19, 957–983. [Google Scholar] [CrossRef]
- Mongkolsamrit, S.; Noisripoom, W.; Thanakitpipattana, D.; Wutikhun, T.; Spatafora, J.W.; Luangsa-ard, J.J. Disentangling cryptic species with Isaria-like morphs in Cordycipitaceae. Mycologia 2018, 110, 230–257. [Google Scholar] [CrossRef] [PubMed]
- Berestetskiy, A.; Hu, Q. The Chemical Ecology Approach to Reveal Fungal Metabolites for Arthropod Pest Management. Microorganisms 2021, 9, 1379. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Feng, M.-G.; Keller, N.P. Genomics-driven discovery of natural products in filamentous fungi. Nat. Prod. Rep. 2020, 37, 1164–1180. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Sun, W.; Feng, M.-G. Secondary metabolites from hypocrealean entomopathogenic fungi. Nat. Prod. Rep. 2020, 37, 1181–1206. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Orozco, R.; Wijeratne, E.M.K.; Espinosa-Artiles, P.; Gunatilaka, A.A.L.; Stock, S.P.; Molnár, I. Biosynthesis of the cyclooligomer depsipeptide bassianolide, an insecticidal virulence factor of Beauveria bassiana. Fungal Genet. Biol. 2009, 46, 353–364. [Google Scholar] [CrossRef]
- Fan, Y.; Liu, X.; Keyhani, N.O.; Tang, G.; Pei, Y.; Zhang, W.; Pei, Y.; Xiao, Y.; Fang, W.; Wang, C. Regulatory cascade and biological activity of Beauveria bassiana oosporein that limits bacterial growth after host death. Proc. Natl. Acad. Sci. USA 2017, 114, E1578–E1586. [Google Scholar] [CrossRef]
- Feng, P.; Shang, Y.; Cen, K.; Wang, C. Fungal biosynthesis of the bibenzoquinone oosporein to evade insect immunity. Proc. Natl. Acad. Sci. USA 2015, 112, 11365–11370. [Google Scholar] [CrossRef]
- Raghunandan, B.L.; Dave, A.; Baria, P.R.; Manjari. Applications of Bioactive Compounds from Fungal Entomopathogens. In Entomopathogenic Fungi; Deshmukh, S.K., Sridhar, K.R., Eds.; Springer Nature: Singapore, 2024; pp. 453–478. ISBN 978-981-97-5990-3. [Google Scholar]
- Safavi, S.A. In Vitro and In Vivo Induction, and Characterization of Beauvericin Isolated from Beauveria bassiana and Its Bioassay on Galleria mellonella Larvae. J. Agric. Sci. Technol. 2013, 15, 1–10. [Google Scholar]
- Wang, H.; Peng, H.; Li, W.; Cheng, P.; Gong, M. The Toxins of Beauveria bassiana and the Strategies to Improve Their Virulence to Insects. Front. Microbiol. 2021, 12, 705343. [Google Scholar] [CrossRef]
- Xu, Y.; Orozco, R.; Wijeratne, E.M.K.; Gunatilaka, A.A.L.; Stock, S.P.; Molnár, I. Biosynthesis of the Cyclooligomer Depsipeptide Beauvericin, a Virulence Factor of the Entomopathogenic Fungus Beauveria bassiana. Chem. Biol. 2008, 15, 898–907. [Google Scholar] [CrossRef]
- Sánchez-Gómez, T.; Harte, S.J.; Zamora, P.; Bareyre, M.; Díez, J.J.; Herrero, B.; Niño-Sánchez, J.; Martín-García, J. Nematicidal effect of Beauveria species and the mycotoxin beauvericin against pinewood nematode Bursaphelenchus xylophilus. Front. For. Glob. Change 2023, 6, 1229456. [Google Scholar] [CrossRef]
- Li, X.; Jiang, R.; Wang, S.; Li, C.; Xu, Y.; Li, S.; Li, Q.; Wang, L. Prospects for cordycepin biosynthesis in microbial cell factories. Front. Chem. Eng. 2024, 6, 1446454. [Google Scholar] [CrossRef]
- Peng, T.; Guo, J.; Tong, X. Advances in biosynthesis and metabolic engineering strategies of cordycepin. Front. Microbiol. 2024, 15, 1386855. [Google Scholar] [CrossRef] [PubMed]
- Qu, S.-L.; Li, S.-S.; Li, D.; Zhao, P.-J. Metabolites and Their Bioactivities from the Genus Cordyceps. Microorganisms 2022, 10, 1489. [Google Scholar] [CrossRef] [PubMed]
- Suparmin, A.; Kato, T.; Dohra, H.; Park, E.Y. Insight into cordycepin biosynthesis of Cordyceps militaris: Comparison between a liquid surface culture and a submerged culture through transcriptomic analysis. PLoS ONE 2017, 12, e0187052. [Google Scholar] [CrossRef]
- Chou, Y.-C.; Sung, T.-H.; Hou, S.-J.; Khumsupan, D.; Santoso, S.P.; Cheng, K.-C.; Lin, S.-P. Current Progress Regarding Cordyceps militaris, Its Metabolite Function, and Its Production. Appl. Sci. 2024, 14, 4610. [Google Scholar] [CrossRef]
- Xia, Y.; Luo, F.; Shang, Y.; Chen, P.; Lu, Y.; Wang, C. Fungal Cordycepin Biosynthesis Is Coupled with the Production of the Safeguard Molecule Pentostatin. Cell Chem. Biol. 2017, 24, 1479–1489. [Google Scholar] [CrossRef] [PubMed]
- Zheng, P.; Xia, Y.; Xiao, G.; Xiong, C.; Hu, X.; Zhang, S.; Zheng, H.; Huang, Y.; Zhou, Y.; Wang, C. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol. 2011, 12, R116. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Z.-Q.; Luo, R.; Souvanhnachit, S.; Thanarut, C.; Dao, V.M.; Yu, H. Species diversity and major host/substrate associations of the genus Akanthomyces (Hypocreales, Cordycipitaceae). MycoKeys 2024, 101, 113–141. [Google Scholar] [CrossRef]
- Zhou, Y.-M.; Zhi, J.-R.; Qu, J.-J.; Zou, X. Estimated divergence times of Lecanicillium in the family Cordycipitaceae provide insights into the attribution of Lecanicillium. Front. Microbiol. 2022, 13, 859886. [Google Scholar] [CrossRef] [PubMed]
- Helaly, S.E.; Kuephadungphan, W.; Phainuphong, P.; Ibrahim, M.A.A.; Tasanathai, K.; Mongkolsamrit, S.; Luangsa-ard, J.J.; Phongpaichit, S.; Rukachaisirikul, V.; Stadler, M. Pigmentosins from Gibellula sp. as antibiofilm agents and a new glycosylated asperfuran from Cordyceps javanica. Beilstein J. Org. Chem. 2019, 15, 2968–2981. [Google Scholar] [CrossRef]
- Phutthacharoen, K.; Llanos-López, N.A.; Toshe, R.; Noisripoom, W.; Khonsanit, A.; Luangsa-ard, J.J.; Hyde, K.D.; Ebada, S.S.; Stadler, M. Bioactive Bioxanthracene and Cyclodepsipeptides from the Entomopathogenic Fungus Blackwellomyces roseostromatus BCC56290. Antibiotics 2024, 13, 585. [Google Scholar] [CrossRef]
- Yoneyama, T.; Iguchi, M.; Yoshii, K.; Elshamy, A.I.; Ban, S.; Noji, M.; Umeyama, A. Xanthone glucoside from an insect pathogenic fungus Conoideocrella luteorostrata NBRC106950. Nat. Prod. Res. 2021, 36, 6349–6353. [Google Scholar] [CrossRef]
- Chen, W.-H.; Chen, L.; Ren, X.-X.; Zhao, J.-H.; Han, F.-Y.; Liang, Z.-Q. Taxonomic and phylogenetic characterizations reveal four new species of Simplicillium (Cordycipitaceae, Hypocreales) from China. Sci. Rep. 2021, 11, 14893. [Google Scholar] [CrossRef]
- Zhu, M.; Duan, X.; Cai, P.; Li, Y.-F.; Qiu, Z. Deciphering the genome of Simplicillium aogashimaense to understand its mechanisms against the wheat powdery mildew fungus Blumeria graminis f. sp. tritici. Phytopathol. Res. 2022, 4, 16. [Google Scholar] [CrossRef]
- Baró Robaina, Y.; González Marrero, I.; Lorenzo Nicao, M.E.; Ruiz, R.F.C.; Li, D.W.; de la Cal, A.P.; Gharsa, H.B.; Manfrino, R.G.; Schuster, C.; Leclerque, A. First Description of Simplicillium lanosoniveum, a Potential Antagonist of the Coffee Leaf Rust from Cuba. Appl. Microbiol. 2024, 4, 275–283. [Google Scholar] [CrossRef]
- Yao, Q.; Wang, J.; Zhang, X.; Nong, X.; Xu, X.; Qi, S. Cytotoxic Polyketides from the Deep-Sea-Derived Fungus Engyodontium album DFFSCS021. Mar. Drugs 2014, 12, 5902–5915. [Google Scholar] [CrossRef]
- Baerson, S.R.; Rimando, A.M. A Plethora of Polyketides: Structures, Biological Activities, and Enzymes. In Polyketides; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 2007; Volume 955, pp. 2–14. ISBN 978-0-8412-3978-4. [Google Scholar] [CrossRef]
- Formánek, B.; Dupommier, D.; Volfová, T.; Rimpelová, S.; Škarková, A.; Herciková, J.; Rösel, D.; Brábek, J.; Perlíková, P. Synthesis and migrastatic activity of cytochalasin analogues lacking a macrocyclic moiety. RSC Med. Chem. 2024, 15, 322–343. [Google Scholar] [CrossRef]
- Aza, P.; Camarero, S. Fungal Laccases: Fundamentals, Engineering and Classification Update. Biomolecules 2023, 13, 1716. [Google Scholar] [CrossRef]
- Brugnari, T.; Braga, D.M.; Dos Santos, C.S.A.; Torres, B.H.C.; Modkovski, T.A.; Haminiuk, C.W.I.; Maciel, G.M. Laccases as green and versatile biocatalysts: From lab to enzyme market—An overview. Bioresour. Bioprocess. 2021, 8, 131. [Google Scholar] [CrossRef]
- Adamo, M.; Comtet-Marre, S.; Büttner, E.; Kellner, H.; Luis, P.; Vallon, L.; Prego, R.; Hofrichter, M.; Girlanda, M.; Peyret, P.; et al. Fungal dye-decolorizing peroxidase diversity: Roles in either intra- or extracellular processes. Appl. Microbiol. Biotechnol. 2022, 106, 2993–3007. [Google Scholar] [CrossRef] [PubMed]
- Barber-Zucker, S.; Mindel, V.; Garcia-Ruiz, E.; Weinstein, J.J.; Alcalde, M.; Fleishman, S.J. Stable and Functionally Diverse Versatile Peroxidases Designed Directly from Sequences. J. Am. Chem. Soc. 2022, 144, 3564–3571. [Google Scholar] [CrossRef]
- Kumar, A.; Arora, P.K. Biotechnological Applications of Manganese Peroxidases for Sustainable Management. Front. Environ. Sci. 2022, 10, 875157. [Google Scholar] [CrossRef]
- Bauer, J.A.; Zámocká, M.; Majtán, J.; Bauerová-Hlinková, V. Glucose Oxidase, an Enzyme “Ferrari”: Its Structure, Function, Production and Properties in the Light of Various Industrial and Biotechnological Applications. Biomolecules 2022, 12, 472. [Google Scholar] [CrossRef] [PubMed]
- Guoqiang, G.; Liang, Q.; Yani, Z.; Pengyun, W.; Fanzhuo, K.; Yuyang, Z.; Zhiyuan, L.; Xing, N.; Xue, Z.; Qiongya, L.; et al. Recent advances in glucose monitoring utilizing oxidase electrochemical biosensors integrating carbon-based nanomaterials and smart enzyme design. Front. Chem. 2025, 13, 1591302. [Google Scholar] [CrossRef]
- Wang, H.; Sun, Z.; Qi, Y.; Hu, Y.; Ni, Z.; Li, C. Update application of enzyme in food processing, preservation, and detection. Food Bioeng. 2024, 3, 380–394. [Google Scholar] [CrossRef]
- Ferreira, P.; Carro, J.; Balcells, B.; Martínez, A.T.; Serrano, A. Expanding the Physiological Role of Aryl-Alcohol Flavooxidases as Quinone Reductases. Appl. Environ. Microbiol. 2023, 89, e01844-22. [Google Scholar] [CrossRef] [PubMed]
- Urlacher, V.B.; Koschorreck, K. Pecularities and applications of aryl-alcohol oxidases from fungi. Appl. Microbiol. Biotechnol. 2021, 105, 4111–4126. [Google Scholar] [CrossRef] [PubMed]
- Chellappan, S.; Jasmin, C.; Basheer, S.M.; Elyas, K.K.; Bhat, S.G.; Chandrasekaran, M. Production, purification and partial characterization of a novel protease from marine Engyodontium album BTMFS10 under solid state fermentation. Process Biochem. 2006, 41, 956–961. [Google Scholar] [CrossRef]
- Chellappan, S.; Jasmin, C.; Basheer, S.M.; Kishore, A.; Elyas, K.K.; Bhat, S.G.; Chandrasekaran, M. Characterization of an extracellular alkaline serine protease from marine Engyodontium album BTMFS10. J. Ind. Microbiol. Biotechnol. 2011, 38, 743–752. [Google Scholar] [CrossRef]
- Jasmin, C.; Chellappan, S.; Sukumaran, R.K.; Elyas, K.K.; Bhat, S.G.; Chandrasekaran, M. Molecular cloning and homology modelling of a subtilisin-like serine protease from the marine fungus, Engyodontium album BTMFS10. World J. Microbiol. Biotechnol. 2010, 26, 1269–1279. [Google Scholar] [CrossRef]
- Sarkar, S.; Pramanik, A.; Mitra, A.; Mukherjee, J. Bioprocessing Data for the Production of Marine Enzymes. Mar. Drugs 2010, 8, 1323–1372. [Google Scholar] [CrossRef]

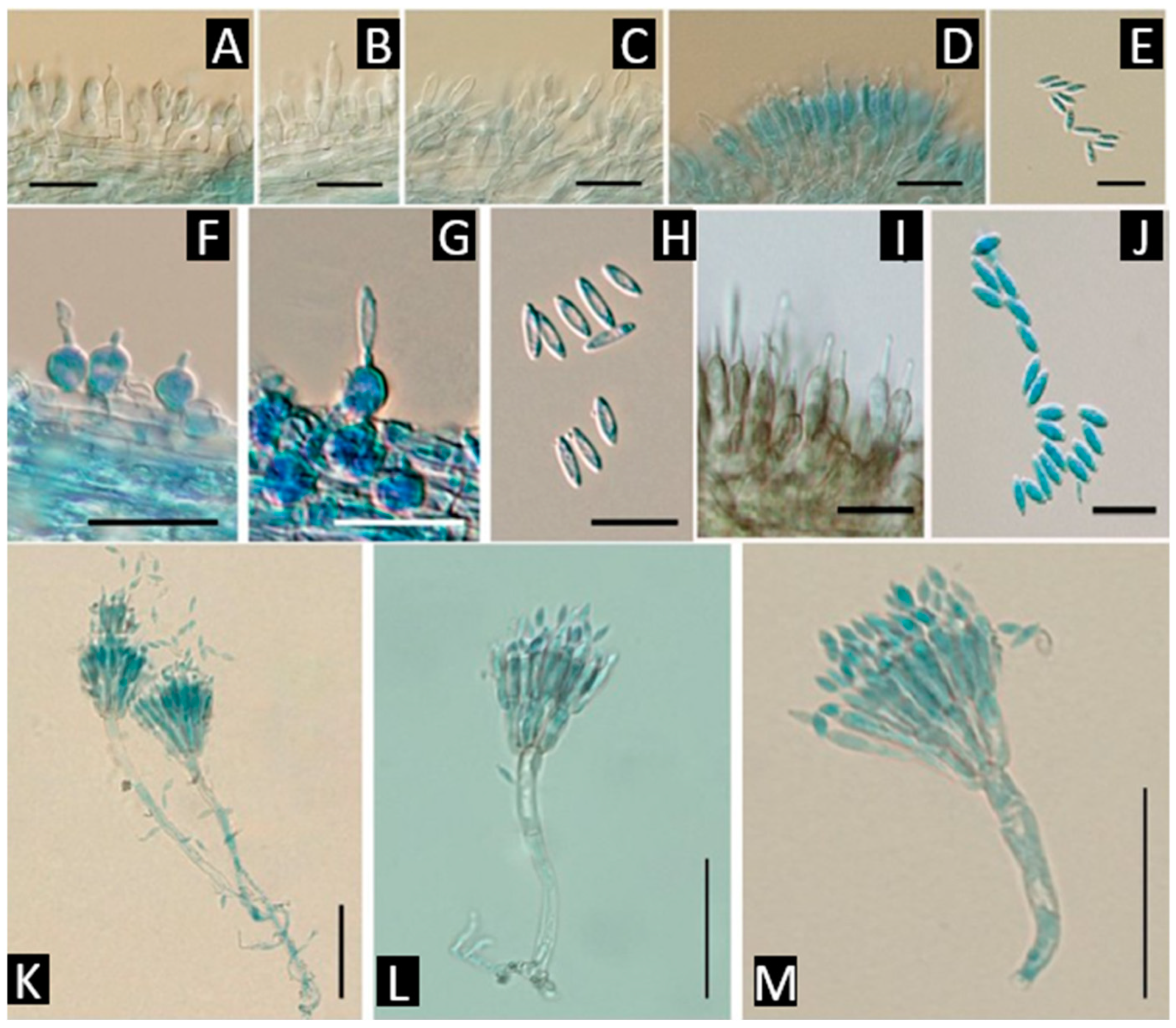
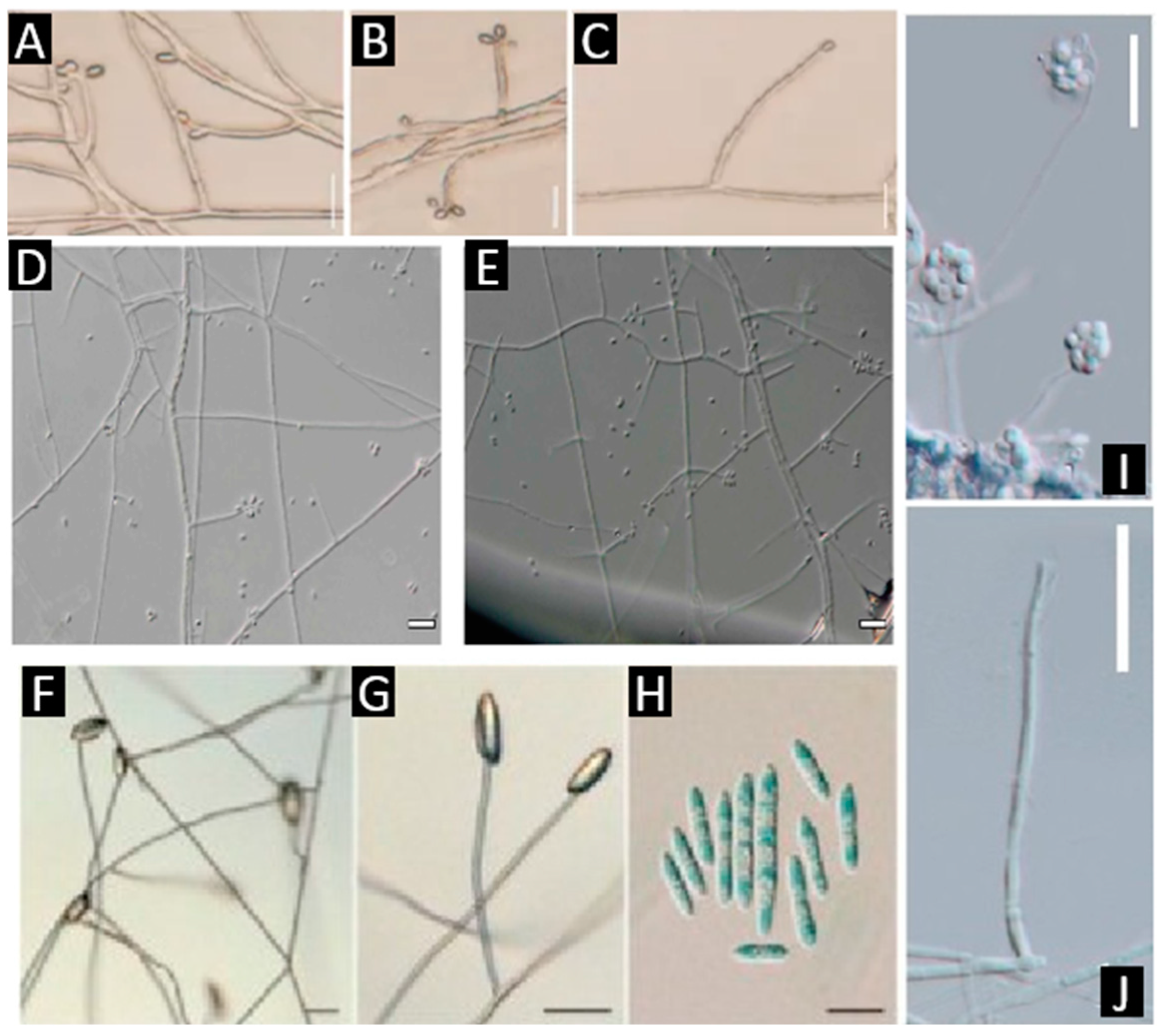

| Genus (Representative Species) | Lifestyle | Dominant Metabolite Classes | Flagship Examples | Ecological/Functional Roles | References |
|---|---|---|---|---|---|
| Beauveria (e.g., B. bassiana) | Entomopathogen (insects) | Depsipeptides (bassianolide, beauvericin); Quinones (oosporein) | Bassianolide; Beauvericin; Oosporein | Virulence (membrane-active), immune modulation, cadaver defense; nematicidal/biocontrol effects | [62,63,64,67,69] |
| Cordyceps (e.g., C. militaris) | Entomopathogen (insects) | Nucleosides (cordycepin); Xanthones; Polysaccharides; Peptides | Cordycepin; Militarinones (rep.); Xanthones | Host manipulation, signaling/interference; broader bioactivities; production/engineering model | [70,71,72,73,74] |
| Akanthomyces | Entomopathogen (insects/arachnids) | Polyketides; Peptides (putative); Phenopicolinic-type derivatives (reported historically) | Representative polyketides/peptides (var.) | Pathogenesis and competitive interactions on arthropod hosts; genus-level idiosyncrasies | [77] |
| Lecanicillium | Entomopathogen (insects) | Polyketides; Peptides; (chemistry less mapped than Beauveria/Cordyceps) | Representative polyketides/peptides (var.) | Insect infection; potential overlaps with Akanthomyces toolkits | [78] |
| Gibellula | Entomopathogen (spiders) | Anthraquinones; Antibiofilm compounds | Pigmentosins | Antibiofilm/antimicrobial activity during host colonization and microbiome control | [79] |
| Blackwellomyces | Entomopathogen (insects/arachnids) | Bioxanthracenes; Cyclodepsipeptides | Bioxanthracene derivatives; Cyclodepsipeptides | Antimicrobial/cytotoxic activities likely aiding infection and post-host defense | [80] |
| Simplicillium | Mycoparasite (on fungi) | Polyketides; NRPS/PKS-derived antimicrobials (putative) | Genomic BGC inventory (NRPS/PKS); species-level yet emerging | Antagonism of fungal pathogens (e.g., powdery mildew, coffee rust); niche competition | [31,82,83,84] |
| Parengyodontium (e.g., P. album) | Saprobe/Opportunistic | Aromatic polyketides (anthraquinones; xanthoquinodines) | Engyodontochones A–F; JBIR-99 | Antibacterial/antibiofilm, cytotoxic activities; oxidative polyethylene degradation | [44,50,55] |
| Genus (Representative Species and Strain) | Genome Size/GC%/Number of Genes |
|---|---|
| Beauveria bassiana (ARSEF 2860) | 33.7 Mb/51.5/10364 |
| Cordyceps militaris (CM01) | 32.2 Mb/51.5/9651 |
| Akanthomyces muscarius (Ve6) | 36.2 Mb/53/12347 |
| Lecanicillium saksenae (MUC18310) | 33.5 Mb/52.2/nd |
| Simplicillium aogashimaense (ZM-2020) | 30.3 Mb/49/nd |
| Parengyodontium torokii (FJII-L10-SW-P1) | 30.4 Mb/50.5/nd |
| Cordyceps militaris (ATCC 34164) | 33.6 Mb/51/9362 |
| Beauveria bassiana (HN6) | 37.1 Mb/49/nd |
| Cordyceps militaris (CH1) | 32.4 Mb/51.5/nd |
| Cordyceps javanica (Apopka 97) | 35.1 Mb/53/10519 |
| Cordyceps gunnii (Cg-01) | 30.3 Mb/54/nd |
| Cordyceps javanica (Bd01) | 34 Mb/53/nd |
| Species (Strain) | Total Regions | PKS (Any) | NRPS (Any) | Terpenes | Hybrids (≥2) |
|---|---|---|---|---|---|
| Beauveria bassiana (ARSEF 2860) | 51 | 15 | 25 | 15 | 7 |
| Cordyceps militaris (CM01) | 33 | 11 | 18 | 8 | 6 |
| Akanthomyces muscarius (Ve6) | 50 | 13 | 28 | 14 | 6 |
| Cordyceps javanica (Apopka97) | 48 | 18 | 25 | 8 | 7 |
| Cordyceps militaris (ATCC 34164) | 37 | 13 | 19 | 8 | 6 |
| Anticancer | Antibacterial | Antilaryngeal | Enzymes & Bioremediation |
|---|---|---|---|
| Polyketides (Engyodontiumones): selective cytotoxicity (U937) [85] Cytochalasin K in P. torokii: impacts late mitosis; synergy on cancer cells [29] Cytotoxic polyketides: Xanthoquinodin JBIR-99[50] Alternaphenol B2 (P. album): IDH1 R132H inhibitor [51] | P. album compounds 8, 15, 16: moderate activity vs. E. coli & B. subtilis [85] Phenylacetate derivative: active versus MRSA & V. vulnificus [56] Torrubielline derivatives [42] Fungal mycelial extracts [54] | Compound 15 (P. album DFFSCS021): anti-settlement of Balanus amphitrite (Potential for biofouling control) [85] | P. torokii genomics: GH33 sialidases; GT20/GT34 glycosyltransferases [29] P. album biodegrades UV-pretreated polyethylene [44] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marin, D.; Petit, P.; Pruneau, L. Biological Evaluation and Potential Applications of Secondary Metabolites from Fungi Belonging to the Cordycipitaceae Family with a Focus on Parengyodontium spp. J. Fungi 2025, 11, 764. https://doi.org/10.3390/jof11110764
Marin D, Petit P, Pruneau L. Biological Evaluation and Potential Applications of Secondary Metabolites from Fungi Belonging to the Cordycipitaceae Family with a Focus on Parengyodontium spp. Journal of Fungi. 2025; 11(11):764. https://doi.org/10.3390/jof11110764
Chicago/Turabian StyleMarin, Dylan, Philippe Petit, and Ludovic Pruneau. 2025. "Biological Evaluation and Potential Applications of Secondary Metabolites from Fungi Belonging to the Cordycipitaceae Family with a Focus on Parengyodontium spp." Journal of Fungi 11, no. 11: 764. https://doi.org/10.3390/jof11110764
APA StyleMarin, D., Petit, P., & Pruneau, L. (2025). Biological Evaluation and Potential Applications of Secondary Metabolites from Fungi Belonging to the Cordycipitaceae Family with a Focus on Parengyodontium spp. Journal of Fungi, 11(11), 764. https://doi.org/10.3390/jof11110764






