Gapless near Telomer-to-Telomer Assembly of Neurospora intermedia, Aspergillus oryzae, and Trichoderma asperellum from Nanopore Simplex Reads
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction and Sequencing
2.2. Snakemake Workflow for High-Quality Assembly
2.3. Phylogenetic Determination with Universal Fungal Core Genes
2.4. Whole-Genome Alignment
2.5. Gene Prediction and Functional Annotation
2.6. Validation of Workflow
3. Results
3.1. Snakemake Workflow Output and Assembly Stats
3.2. Phylogenetic Determination
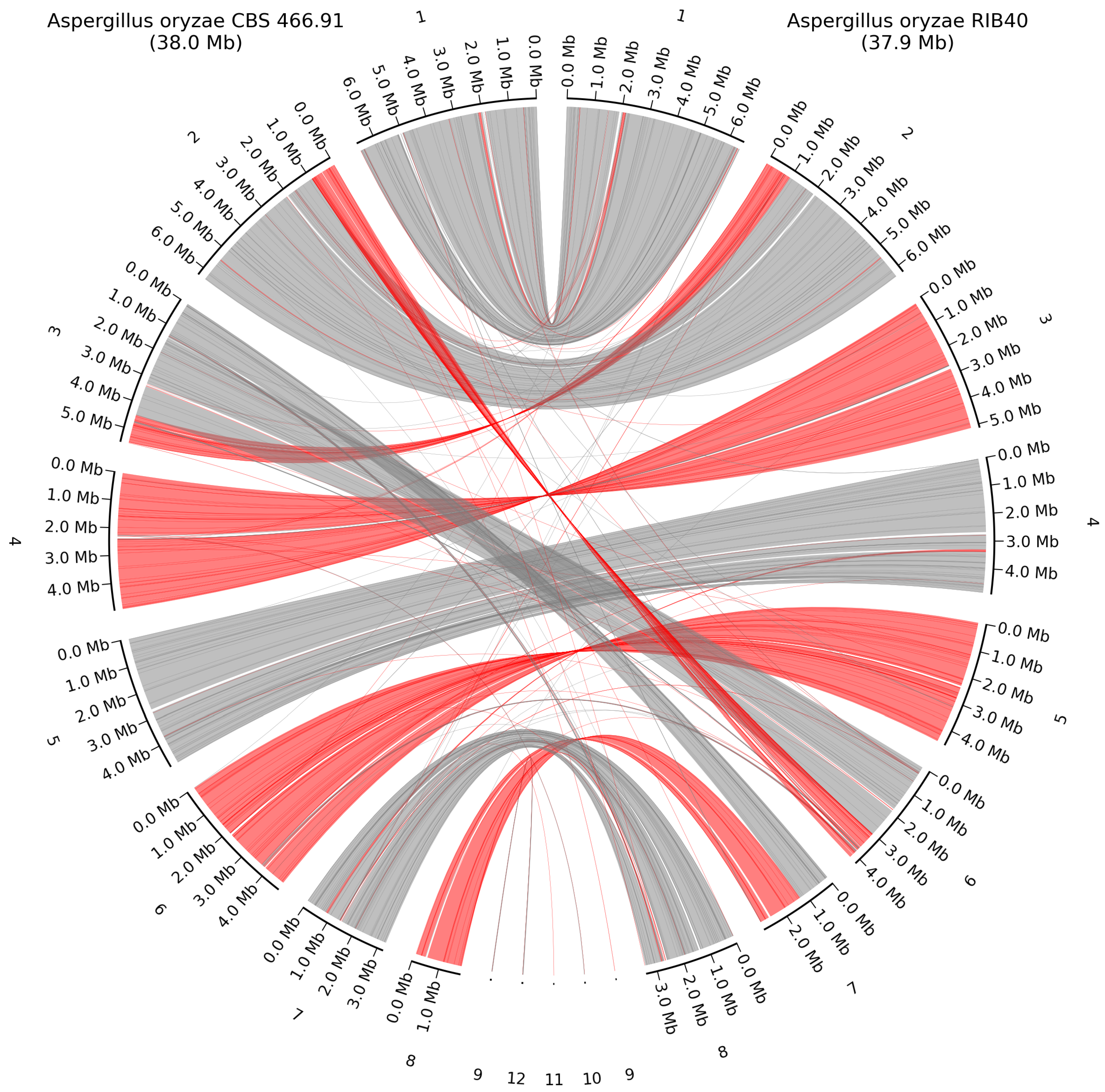
3.3. Whole-Genome Alignment with High Quality Closest Neighbor
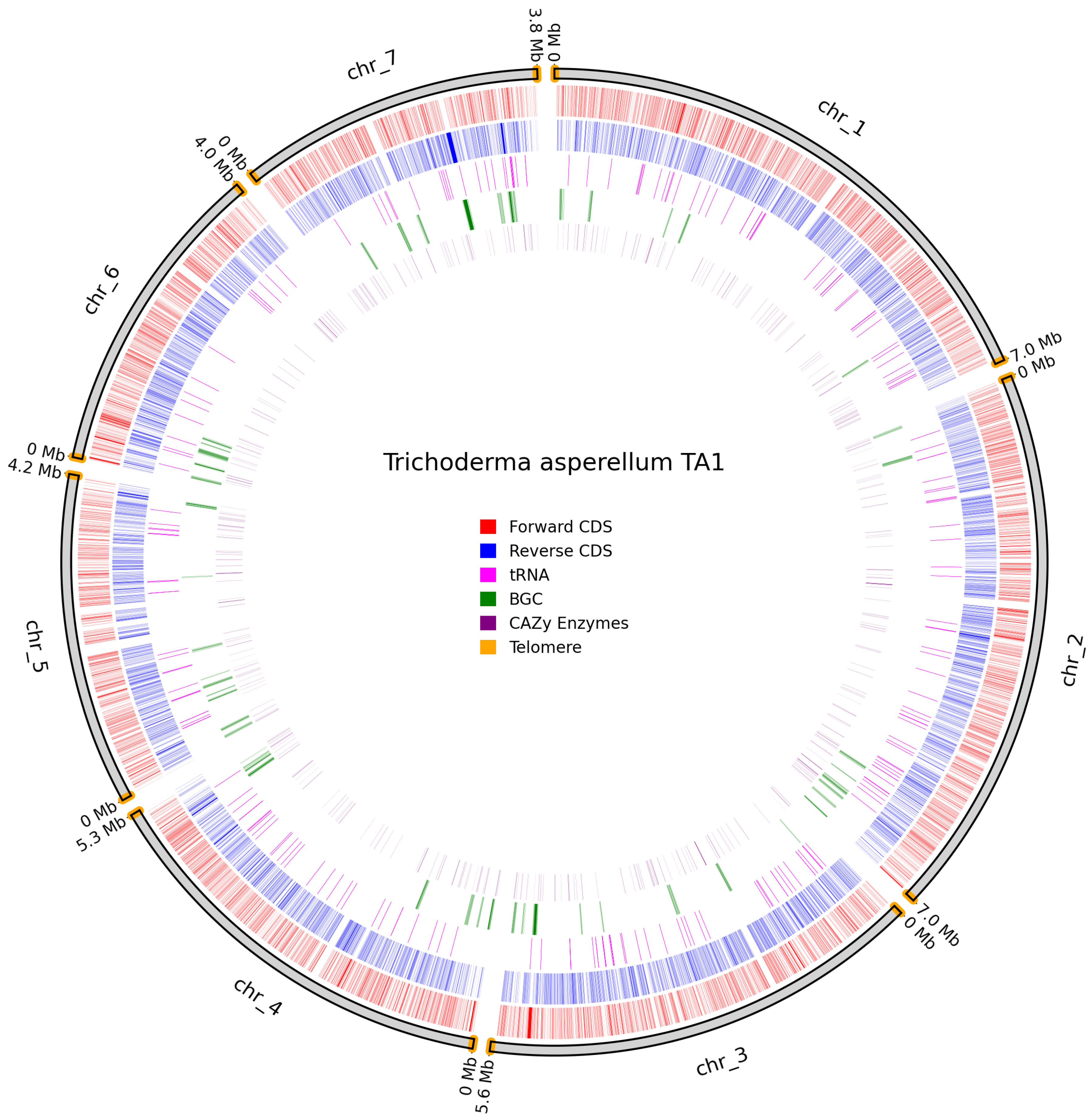
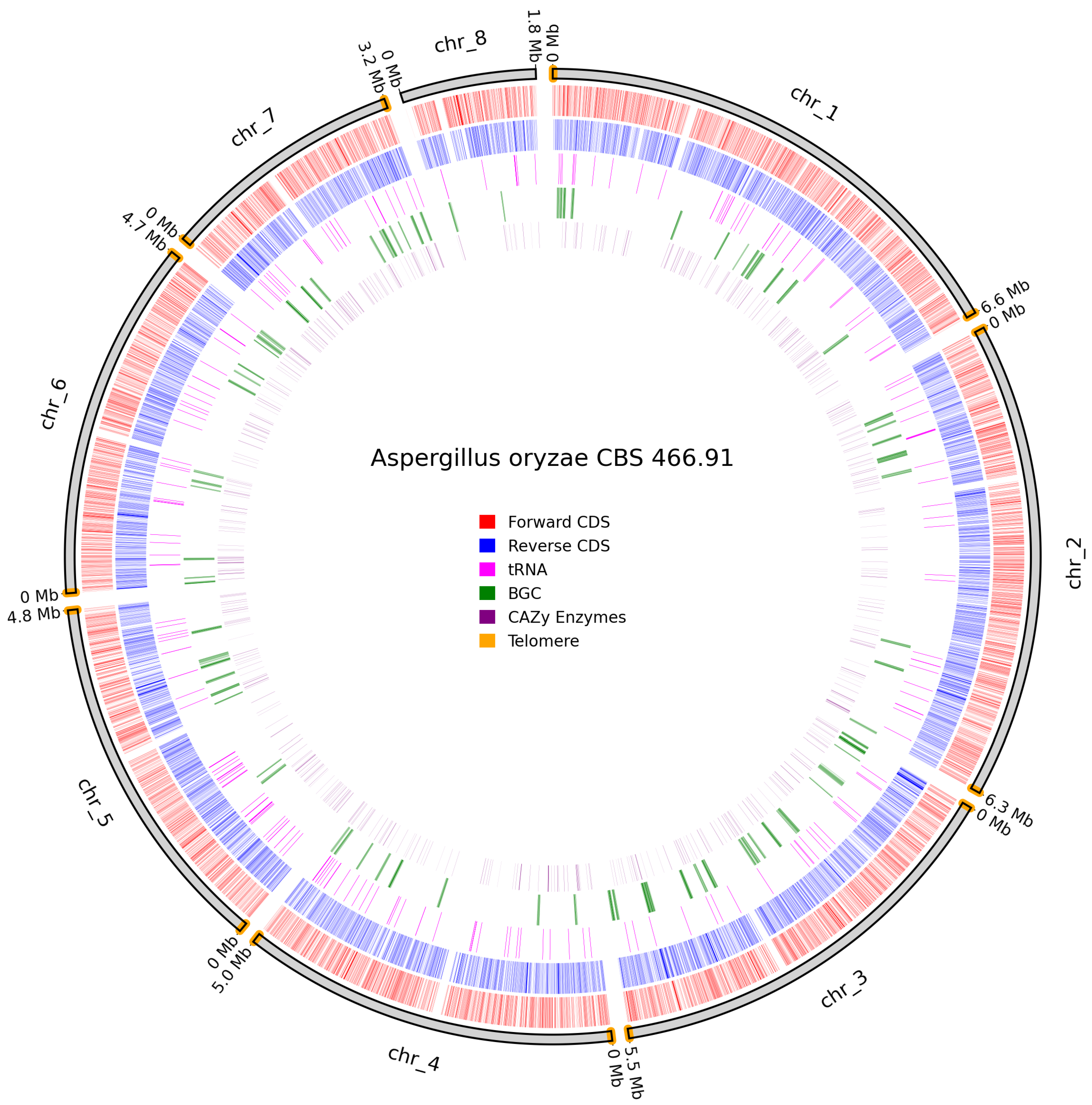
3.4. Predicted Genes and Telomeres
3.5. Workflow Validation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BGCs | Biosynthetic gene clusters |
| BUSCO | Benchmarking Universal Single-Copy Orthologs |
| CAZymes | Carbohydrate-Active Enzymes |
| CTAB | Cetyltrimethylammonium bromide |
| GSI | Genealogical Sorting Index |
| CDS | Coding sequence |
| HERRO | Haplotype-aware error correction |
| HMW | High molecular weight DNA |
| HPC | High-performance computing |
| HQ | High quality |
| NRPSs | Nonribosomal peptide synthetases |
| NUMTs | Nuclear mitochondrial DNA segments |
| PKSs | Polyketide synthases |
| RiPPs | Post-translationally modified peptides |
| T2T | Telomere to telomere |
| Tidk | Telomere identification toolkit |
| UFCG | Universal Fungal Core Genes |
| YES | Yeast extract sucrose |
| YPG | Yeast extract Peptone Glucose |
References
- Galagan, J.E.; Calvo, S.E.; Borkovich, K.A.; Selker, E.U.; Read, N.D.; Jaffe, D.; FitzHugh, W.; Ma, L.J.; Smirnov, S.; Purcell, S.; et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature 2003, 422, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Schwarze, K.; Buchanan, J.; Fermont, J.M.; Dreau, H.; Tilley, M.W.; Taylor, J.M.; Antoniou, P.; Knight, S.J.; Camps, C.; Pentony, M.M.; et al. The complete costs of genome sequencing: A microcosting study in cancer and rare diseases from a single center in the United Kingdom. Genet. Med. 2020, 22, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Heather, J.M.; Chain, B. The sequence of sequencers: The history of sequencing DNA. Genomics 2016, 107, 1–8. [Google Scholar] [CrossRef]
- Satam, H.; Joshi, K.; Mangrolia, U.; Waghoo, S.; Zaidi, G.; Rawool, S.; Thakare, R.P.; Banday, S.; Mishra, A.K.; Das, G.; et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology 2023, 12, 997. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Genome assembly in the telomere-to-telomere era. Nat. Rev. Genet. 2024, 25, 658–670. [Google Scholar] [CrossRef]
- Wang, H.; Yao, G.; Chen, W.; Ayhan, D.H.; Wang, X.; Sun, J.; Yi, S.; Meng, T.; Chen, S.; Geng, X.; et al. A gap-free genome assembly of Fusarium oxysporum f. sp. conglutinans, a vascular wilt pathogen. Sci. Data 2024, 11, 925. [Google Scholar] [CrossRef]
- Li, Z.; Yang, J.; Ji, X.; Liu, J.; Yin, C.; Bhadauria, V.; Zhao, W.; Peng, Y.L. First telomere-to-telomere gapless assembly of the rice blast fungus Pyricularia oryzae. Sci. Data 2024, 11, 380. [Google Scholar] [CrossRef]
- Yao, G.; Chen, W.; Sun, J.; Wang, X.; Wang, H.; Meng, T.; Zhang, L.; Guo, L. Gapless genome assembly of Fusarium verticillioides, a filamentous fungus threatening plant and human health. Sci. Data 2023, 10, 229. [Google Scholar] [CrossRef]
- Amarasinghe, S.L.; Su, S.; Dong, X.; Zappia, L.; Ritchie, M.E.; Gouil, Q. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 2020, 21, 30. [Google Scholar] [CrossRef]
- Koren, S.; Bao, Z.; Guarracino, A.; Ou, S.; Goodwin, S.; Jenike, K.M.; Lucas, J.; McNulty, B.; Park, J.; Rautiainen, M.; et al. Gapless assembly of complete human and plant chromosomes using only nanopore sequencing. Genome Res. 2024, 34, 1919–1930. [Google Scholar] [CrossRef]
- Stanojevic, D.; Lin, D.; Nurk, S.; Florez De Sessions, P.; Sikic, M. Telomere-to-Telomere Phased Genome Assembly Using HERRO-Corrected Simplex Nanopore Reads. bioRxiv 2024. [Google Scholar] [CrossRef]
- Sigova, E.A.; Dvorianinova, E.M.; Arkhipov, A.A.; Rozhmina, T.A.; Kudryavtseva, L.P.; Kaplun, A.M.; Bodrov, Y.V.; Pavlova, V.A.; Borkhert, E.V.; Zhernova, D.A.; et al. Nanopore Data-Driven T2T Genome Assemblies of Colletotrichum lini Strains. J. Fungi 2024, 10, 874. [Google Scholar] [CrossRef]
- Cechova, M. Probably Correct: Rescuing Repeats with Short and Long Reads. Genes 2020, 12, 48. [Google Scholar] [CrossRef]
- Maiti, A.K.; Bouvagnet, P. Assembling and gap filling of unordered genome sequences through gene checking. Genome Biol. 2001, 2, preprint0008.1. [Google Scholar] [CrossRef]
- Khaldi, N.; Seifuddin, F.T.; Turner, G.; Haft, D.; Nierman, W.C.; Wolfe, K.H.; Fedorova, N.D. SMURF: Genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 2010, 47, 736–741. [Google Scholar] [CrossRef]
- Robey, M.T.; Caesar, L.K.; Drott, M.T.; Keller, N.P.; Kelleher, N.L. An interpreted atlas of biosynthetic gene clusters from 1,000 fungal genomes. Proc. Natl. Acad. Sci. USA 2021, 118, e2020230118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Leahy, I.; Collemare, J.; Seidl, M.F. Genomic Localization Bias of Secondary Metabolite Gene Clusters and Association with Histone Modifications in Aspergillus. Genome Biol. Evol. 2024, 16, evae228. [Google Scholar] [CrossRef] [PubMed]
- Greco, C.; Keller, N.P.; Rokas, A. Unearthing fungal chemodiversity and prospects for drug discovery. Curr. Opin. Microbiol. 2019, 51, 22–29. [Google Scholar] [CrossRef]
- Zhu, S.; Xu, H.; Liu, Y.; Hong, Y.; Yang, H.; Zhou, C.; Tao, L. Computational advances in biosynthetic gene cluster discovery and prediction. Biotechnol. Adv. 2025, 79, 108532. [Google Scholar] [CrossRef]
- Lange, L.; Barrett, K.; Meyer, A.S. New Method for Identifying Fungal Kingdom Enzyme Hotspots from Genome Sequences. J. Fungi 2021, 7, 207. [Google Scholar] [CrossRef]
- Füting, P.; Barthel, L.; Cairns, T.C.; Briesen, H.; Schmideder, S. Filamentous fungal applications in biotechnology: A combined bibliometric and patentometric assessment. Fungal Biol. Biotechnol. 2021, 8, 23. [Google Scholar] [CrossRef]
- Wijaya, C.H.; Nuraida, L.; Nuramalia, D.R.; Hardanti, S.; Świąder, K. Oncom: A Nutritive Functional Fermented Food Made from Food Process Solid Residue. Appl. Sci. 2024, 14, 10702. [Google Scholar] [CrossRef]
- Qi, G.; Hao, L.; Xin, T.; Gan, Y.; Lou, Q.; Xu, W.; Song, J. Analysis of Whole-Genome facilitates rapid and precise identification of fungal species. Front. Microbiol. 2024, 15, 1336143. [Google Scholar] [CrossRef]
- Bartholomew, H.P.; Gottschalk, C.; Cooper, B.; Bukowski, M.R.; Yang, R.; Gaskins, V.L.; Luciano-Rosario, D.; Fonseca, J.M.; Jurick, W.M. Omics-Based Comparison of Fungal Virulence Genes, Biosynthetic Gene Clusters, and Small Molecules in Penicillium expansum and Penicillium chrysogenum. J. Fungi 2024, 11, 14. [Google Scholar] [CrossRef]
- Salazar-Cerezo, S.; de Vries, R.P.; Garrigues, S. Strategies for the Development of Industrial Fungal Producing Strains. J. Fungi 2023, 9, 834. [Google Scholar] [CrossRef]
- Petersen, C.; Sørensen, T.; Westphal, K.R.; Fechete, L.I.; Sondergaard, T.E.; Sørensen, J.L.; Nielsen, K.L. High molecular weight DNA extraction methods lead to high quality filamentous ascomycete fungal genome assemblies using Oxford Nanopore sequencing. Microb. Genom. 2022, 8, 000816. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Holt, K.E. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019, 20, 129. [Google Scholar] [CrossRef]
- Nanoporetech. GitHub-Dorado. 2022. Available online: https://github.com/nanoporetech/dorado (accessed on 14 August 2025).
- Köster, J.; Rahmann, S. Snakemake—A scalable bioinformatics workflow engine. Bioinformatics 2012, 28, 2520–2522. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Bonenfant, Q.; Noé, L.; Touzet, H. Porechop_ABI: Discovering unknown adapters in Oxford Nanopore Technology sequencing reads for downstream trimming. Bioinform. Adv. 2022, 3, vbac085. [Google Scholar] [CrossRef] [PubMed]
- De Coster, W.; Rademakers, R. NanoPack2: Population-scale evaluation of long-read sequencing data. Bioinformatics 2023, 39, btad311. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Cheng, H.; Concepcion, G.T.; Feng, X.; Zhang, H.; Li, H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat. Methods 2021, 18, 170–175. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Hall, M. Rasusa: Randomly subsample sequencing reads to a specified coverage. J. Open Source Softw. 2022, 7, 3941. [Google Scholar] [CrossRef]
- lh3. GitHub-lh3/seqtk: Toolkit for Processing Sequences in FASTA/Q Formats—github.com. 2016. Available online: https://github.com/lh3/seqtk (accessed on 21 March 2025).
- Shen, W.; Sipos, B.; Zhao, L. SeqKit2: A Swiss army knife for sequence and alignment processing. iMeta 2024, 3, e191. [Google Scholar] [CrossRef] [PubMed]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simão, F.A.; Zdobnov, E.M. BUSCO Update: Novel and Streamlined Workflows along with Broader and Deeper Phylogenetic Coverage for Scoring of Eukaryotic, Prokaryotic, and Viral Genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Gilchrist, C.L.M.; Chun, J.; Steinegger, M. UFCG: Database of universal fungal core genes and pipeline for genome-wide phylogenetic analysis of fungi. Nucleic Acids Res. 2022, 51, D777–D784. [Google Scholar] [CrossRef] [PubMed]
- O’Leary, N.A.; Cox, E.; Holmes, J.B.; Anderson, W.R.; Falk, R.; Hem, V.; Tsuchiya, M.T.N.; Schuler, G.D.; Zhang, X.; Torcivia, J.; et al. Exploring and retrieving sequence and metadata for species across the tree of life with NCBI Datasets. Sci. Data 2024, 11, 732. [Google Scholar] [CrossRef]
- Svedberg, J.; Vogan, A.A.; Rhoades, N.A.; Sarmarajeewa, D.; Jacobson, D.J.; Lascoux, M.; Hammond, T.M.; Johannesson, H. An introgressed gene causes meiotic drive inNeurospora sitophila. Proc. Natl. Acad. Sci. USA 2021, 118, e2026605118. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef]
- Kurtz, S.; Phillippy, A.; Delcher, A.L.; Smoot, M.; Shumway, M.; Antonescu, C.; Salzberg, S.L. Versatile and open software for comparing large genomes. Genome Biol. 2004, 5. [Google Scholar] [CrossRef] [PubMed]
- Shimoyama, Y. pyCirclize: Circular Visualization in Python. 2022. Available online: https://github.com/moshi4/pyCirclize (accessed on 8 April 2025).
- Krzywinski, M.; Schein, J.; Birol, İ.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.M.; Stajich, J. Funannotate v1.8.1: Eukaryotic Genome Annotation. 2020. Available online: https://zenodo.org/records/4054262 (accessed on 24 March 2025).
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Zdobnov, E.M.; Apweiler, R. GitHub-ebi-pf-team/Interproscan: Genome-Scale Protein Function Classification—github.com. 2001. Available online: https://github.com/ebi-pf-team/interproscan (accessed on 8 April 2025).
- Zdobnov, E.M.; Apweiler, R. InterProScan – an integration platform for the signature-recognition methods in InterPro. Bioinformatics 2001, 17, 847–848. [Google Scholar] [CrossRef]
- Brown, M.R.; Manuel Gonzalez de La Rosa, P.; Blaxter, M. tidk: A toolkit to rapidly identify telomeric repeats from genomic datasets. Bioinformatics 2025, 41, btaf049. [Google Scholar] [CrossRef]
- Lyčka, M.; Bubeník, M.; Závodník, M.; Peska, V.; Fajkus, P.; Demko, M.; Fajkus, J.; Fojtová, M. TeloBase: A community-curated database of telomere sequences across the tree of life. Nucleic Acids Res. 2023, 52, D311–D321. [Google Scholar] [CrossRef]
- Lang, B.F.; Beck, N.; Prince, S.; Sarrasin, M.; Rioux, P.; Burger, G. Mitochondrial genome annotation with MFannot: A critical analysis of gene identification and gene model prediction. Front. Plant Sci. 2023, 14. [Google Scholar] [CrossRef]
- Schalamun, M.; Schmoll, M. Trichoderma—Genomes and genomics as treasure troves for research towards biology, biotechnology and agriculture. Front. Fungal Biol. 2022, 3, 1002161. [Google Scholar] [CrossRef] [PubMed]
- Wieloch, W. Chromosome visualisation in filamentous fungi. J. Microbiol. Methods 2006, 67, 1–8. [Google Scholar] [CrossRef]
- Han, D.M.; Baek, J.H.; Choi, D.G.; Jeon, M.S.; Eyun, S.i.; Jeon, C.O. Comparative pangenome analysis of Aspergillus flavus and Aspergillus oryzae reveals their phylogenetic, genomic, and metabolic homogeneity. Food Microbiol. 2024, 119, 104435. [Google Scholar] [CrossRef]
- Monteiro, J.; Pratas, D.; Videira, A.; Pereira, F. Revisiting the Neurospora crassa mitochondrial genome. Lett. Appl. Microbiol. 2021, 73, 495–505. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Z.; Yang, F.; Lin, R.; Liu, T. Assembly, Annotation, and Comparative Analysis of Mitochondrial Genomes in Trichoderma. Int. J. Mol. Sci. 2024, 25, 12140. [Google Scholar] [CrossRef]
- Asaf, S.; Khan, A.L.; Hamayun, M.; Khan, M.A.; Bilal, S.; Kang, S.M.; Lee, I.J. Complete mitochondrial genome sequence of Aspergillus oryzae RIB 127 and its comparative analysis with related species. Mitochondrial DNA Part B 2017, 2, 632–633. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Henikoff, S. What makes a centromere? Exp. Cell Res. 2020, 389, 111895. [Google Scholar] [CrossRef] [PubMed]
- Zaccaron, A.Z.; Stergiopoulos, I. The dynamics of fungal genome organization and its impact on host adaptation and antifungal resistance. J. Genet. Genom. 2025, 52, 628–640. [Google Scholar] [CrossRef]
- Hazkani-Covo, E.; Zeller, R.M.; Martin, W. Molecular Poltergeists: Mitochondrial DNA Copies (numts) in Sequenced Nuclear Genomes. PLoS Genet. 2010, 6, e1000834. [Google Scholar] [CrossRef]
- van den Boogaart, P.; Samallo, J.; Agsteribbe, E. Similar genes for a mitochondrial ATPase subunit in the nuclear and mitochondrial genomes of Neurospora crassa. Nature 1982, 298, 187–189. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Buckler, E.S.; Stitzer, M.C. New whole-genome alignment tools are needed for tapping into plant diversity. Trends Plant Sci. 2024, 29, 355–369. [Google Scholar] [CrossRef]
- Li, W.C.; Lin, T.C.; Chen, C.L.; Liu, H.C.; Lin, H.N.; Chao, J.L.; Hsieh, C.H.; Ni, H.F.; Chen, R.S.; Wang, T.F. Complete Genome Sequences and Genome-Wide Characterization of Trichoderma Biocontrol Agents Provide New Insights into their Evolution and Variation in Genome Organization, Sexual Development, and Fungal-Plant Interactions. Microbiol. Spectr. 2021, 9, e00663-21. [Google Scholar] [CrossRef] [PubMed]
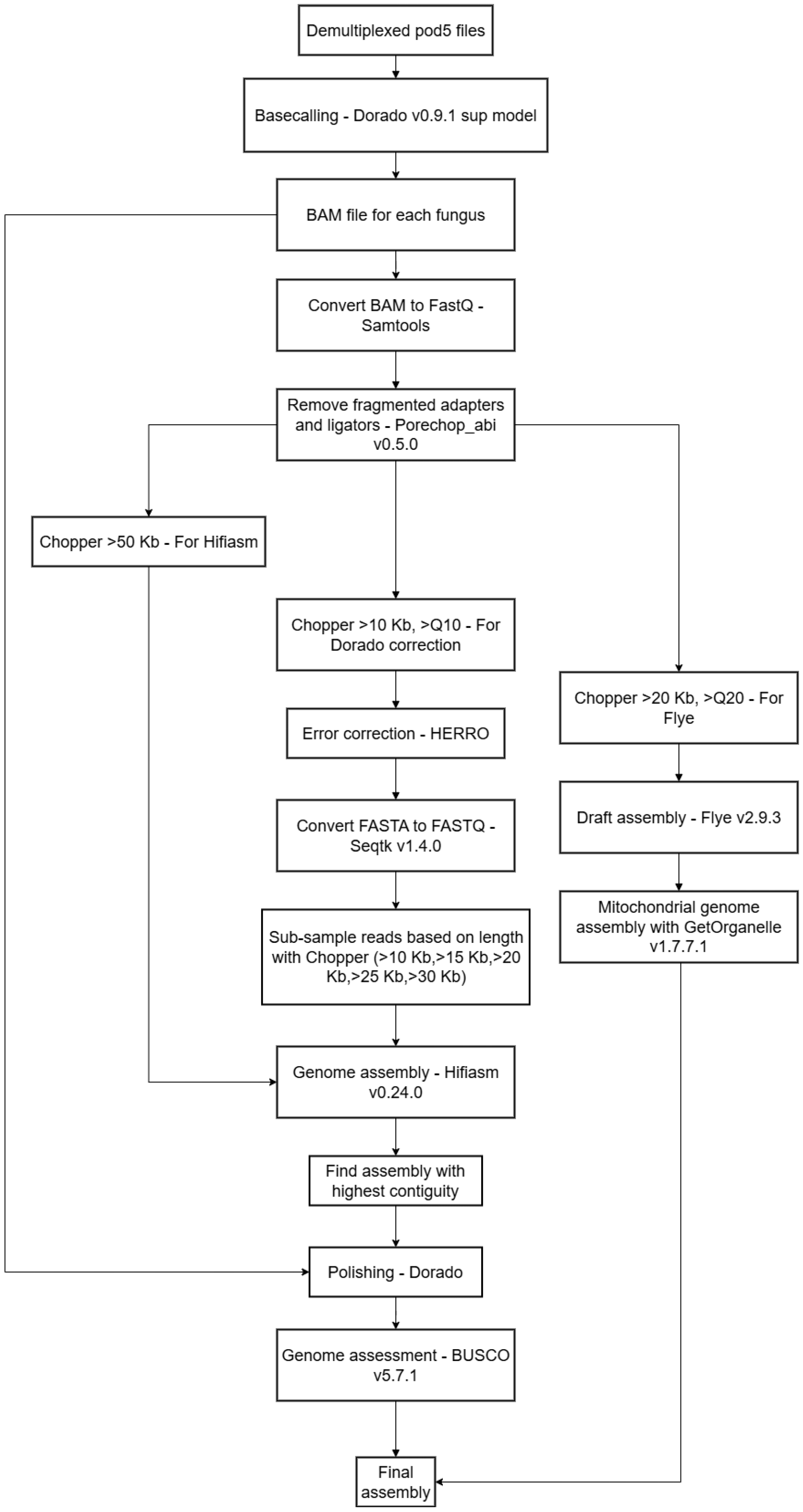

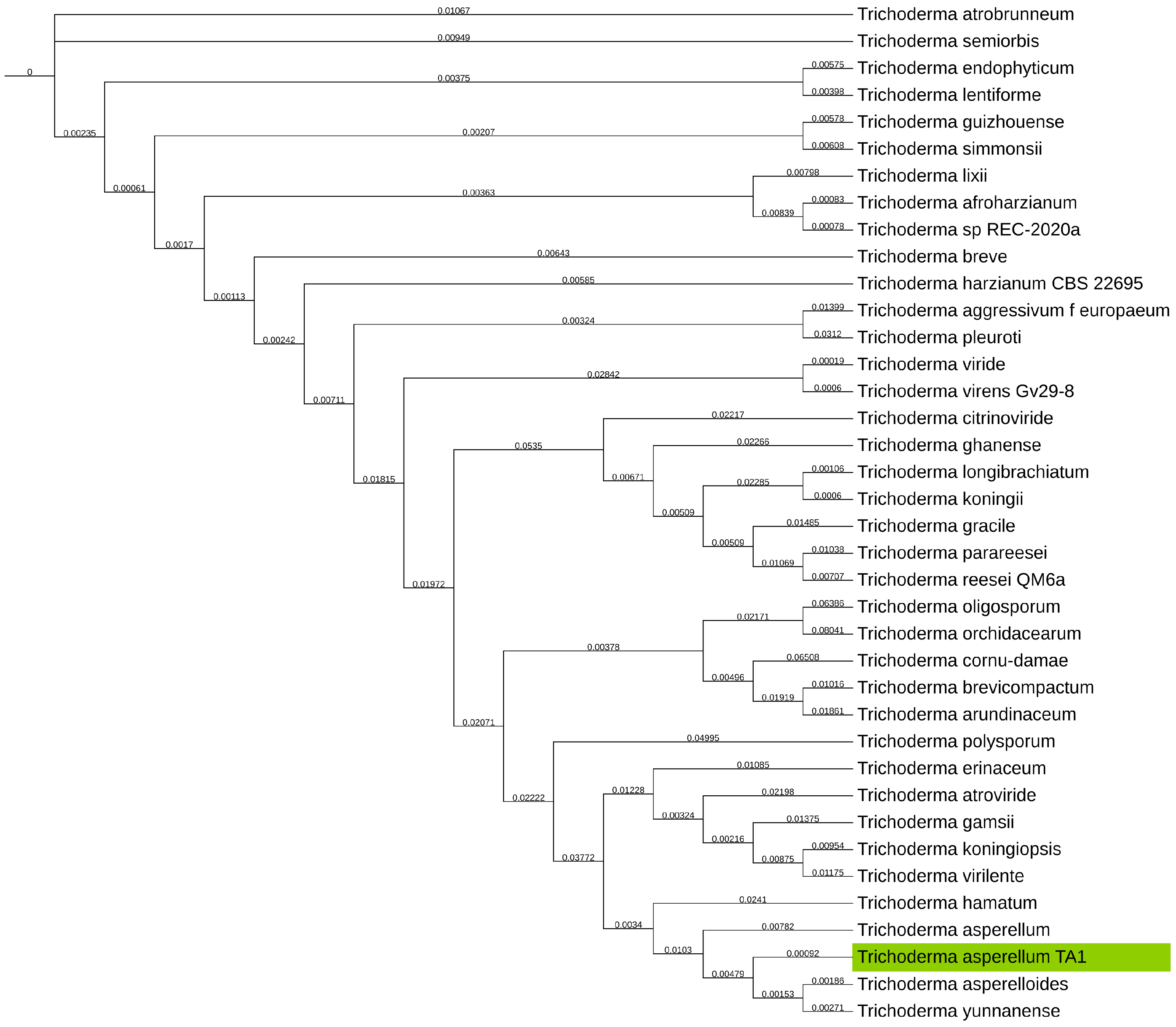
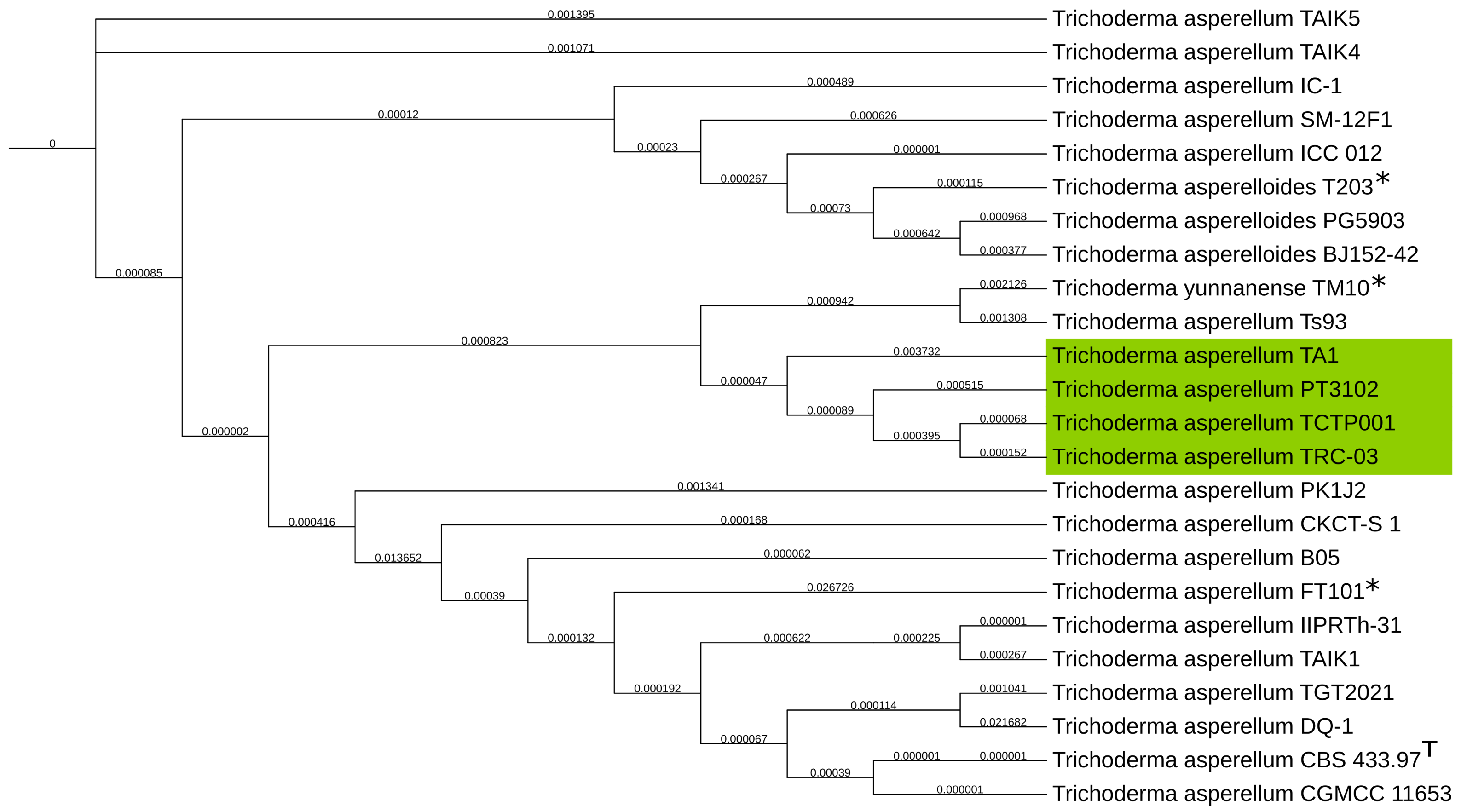


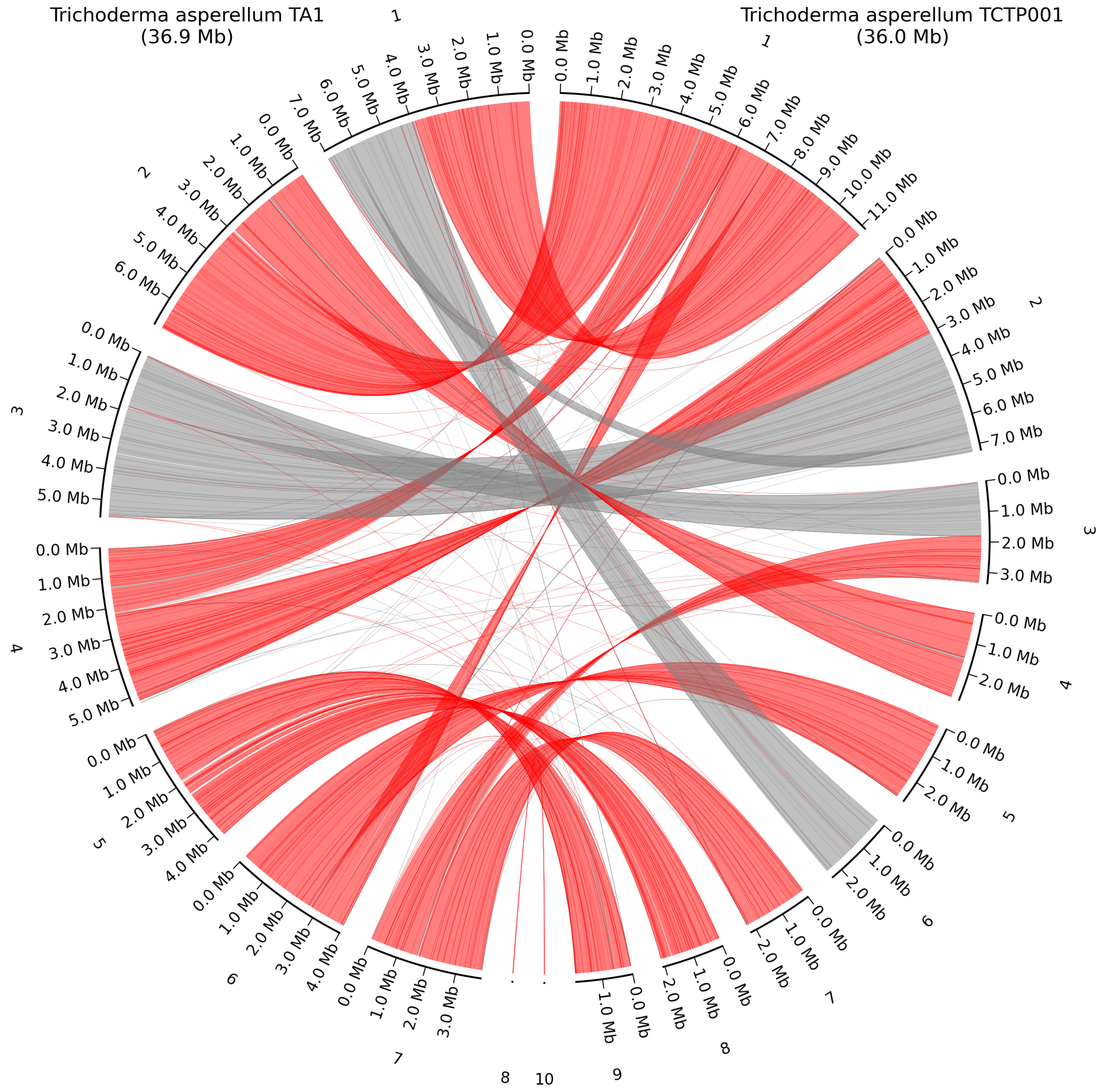

| Genus | Method | Coverage [X] | N50 [Kb] | Average Quality |
|---|---|---|---|---|
| Neurospora | Raw-Basecalled | 194.56 | 11.375 | 20.31 |
| Neurospora | Pre-Correction | 104.67 | 21.338 | 20.58 |
| Neurospora | Corrected | 94.05 | 20.317 | n/a |
| Neurospora | Contiguity | 33.68 | 34.920 | n/a |
| Neurospora | Ultra-long | 7.53 | 59.306 | 20.14 |
| Neurospora | Flye | 42.53 | 30.951 | 23.34 |
| Trichoderma | Raw-Basecalled | 66.34 | 22.114 | 20.35 |
| Trichoderma | Pre-Correction | 49.06 | 29.752 | 20.56 |
| Trichoderma | Corrected | 44.78 | 27.294 | n/a |
| Trichoderma | Contiguity | 43.60 | 27.993 | n/a |
| Trichoderma | Ultra-long | 9.37 | 61.816 | 20.27 |
| Trichoderma | Flye | 26.45 | 37.005 | 23.45 |
| Aspergillus | Raw-Basecalled | 238.93 | 11.980 | 20.94 |
| Aspergillus | Pre-Correction | 136.76 | 19.340 | 21.13 |
| Aspergillus | Corrected | 118.29 | 18.200 | n/a |
| Aspergillus | Contiguity | 114.65 | 18.633 | n/a |
| Aspergillus | Ultra-long | 7.60 | 59.144 | 20.40 |
| Aspergillus | Flye | 51.27 | 29.417 | 23.74 |
| Genus | Contigs | Size [Mb] | C [%] | S [%] | D [%] | F [%] | M [%] | n | DB |
|---|---|---|---|---|---|---|---|---|---|
| Neurospora | 7(+1) 1 | 40 | 99.4 | 99.1 | 0.3 | 0.3 | 0.3 | 758 | fungi_odb10 |
| Trichoderma | 7 | 37 | 98.8 | 98.5 | 0.3 | 0.3 | 0.9 | 758 | fungi_odb10 |
| Aspergillus | 8 | 38 | 98.7 | 98.3 | 0.4 | 0.5 | 0.8 | 758 | fungi_odb10 |
| Strain | Genes | CAZymes | PKSs | NRPSs/NRPSs-like | Terpene Synthases | RiPPs | BGCs |
|---|---|---|---|---|---|---|---|
| N. intermedia NRRL 2884 | 8790 | 345 | 8 | 5 | 3 | 4 | 21 |
| T. asperellum TA1 | 9805 | 401 | 17 | 18 | 10 | 3 | 51 |
| A. oryzae CBS 466.91 | 13,042 | 579 | 28 | 32 | 12 | 6 | 78 |
| Strain | Size [Kb] | GC Content [%] | Protein CDS | tRNA | rRNA |
|---|---|---|---|---|---|
| N. intermedia NRRL 2884 | 56.3 | 36 | 31 | 27 | 1 |
| T. asperellum TA1 | 29.7 | 28 | 18 | 27 | 1 |
| A. oryzae CBS 466.91 | 29.1 | 26 | 21 | 28 | 1 |
| Sample | Contigs | Size [Mb] | C [%] | S [%] | D [%] | F [%] | M [%] | DB |
|---|---|---|---|---|---|---|---|---|
| C. lini 394-2 1 | 13 | 53.56 | 96.8 | 96.6 | 0.2 | 0.6 | 2.6 | glomerellales_odb10 |
| C. lini 394-2 | 13 | 53.69 | 96.8 | 96.6 | 0.2 | 0.6 | 2.6 | glomerellales_odb10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Terp, M.; Nyitrai, M.; Rusbjerg-Weberskov, C.E.; Sondergaard, T.E.; Lübeck, M. Gapless near Telomer-to-Telomer Assembly of Neurospora intermedia, Aspergillus oryzae, and Trichoderma asperellum from Nanopore Simplex Reads. J. Fungi 2025, 11, 701. https://doi.org/10.3390/jof11100701
Terp M, Nyitrai M, Rusbjerg-Weberskov CE, Sondergaard TE, Lübeck M. Gapless near Telomer-to-Telomer Assembly of Neurospora intermedia, Aspergillus oryzae, and Trichoderma asperellum from Nanopore Simplex Reads. Journal of Fungi. 2025; 11(10):701. https://doi.org/10.3390/jof11100701
Chicago/Turabian StyleTerp, Mikael, Mark Nyitrai, Christian Enrico Rusbjerg-Weberskov, Teis E. Sondergaard, and Mette Lübeck. 2025. "Gapless near Telomer-to-Telomer Assembly of Neurospora intermedia, Aspergillus oryzae, and Trichoderma asperellum from Nanopore Simplex Reads" Journal of Fungi 11, no. 10: 701. https://doi.org/10.3390/jof11100701
APA StyleTerp, M., Nyitrai, M., Rusbjerg-Weberskov, C. E., Sondergaard, T. E., & Lübeck, M. (2025). Gapless near Telomer-to-Telomer Assembly of Neurospora intermedia, Aspergillus oryzae, and Trichoderma asperellum from Nanopore Simplex Reads. Journal of Fungi, 11(10), 701. https://doi.org/10.3390/jof11100701






