Phylogenetic and Morphological Analyses Reveal Twelve New Species of the Genus Patellaria (Dothideomycetes, Ascomycota) from Mexico
Abstract
1. Introduction
2. Materials and Methods
2.1. Morphological Studies
2.2. Amplification and Sequencing
2.3. Phylogenetic Analysis
| Loci/Segment | Primer | Sequence 5′-3′ | T (°C) | Reference |
|---|---|---|---|---|
| ITS | ITS5 | GGAAGTAAAAGTCGTAACAAGG | 58 | [33] |
| ITS4 | TCCTCCGCTTATTGATATGC | 58 | [33] | |
| nLSU | LROR | ACCCGCTGAACTTAAGC | 48 | [33] |
| LR3 | GGTCCGTGTTTCAAGAC | 48 | [33] | |
| 0mtSSU | MS1 | CAGCAGTCAAGAATATTAGTCAATG | 63 | [33] |
| MS2 | GCGGATTATCGAATTAAATAAC | 53 | [33] |
3. Results
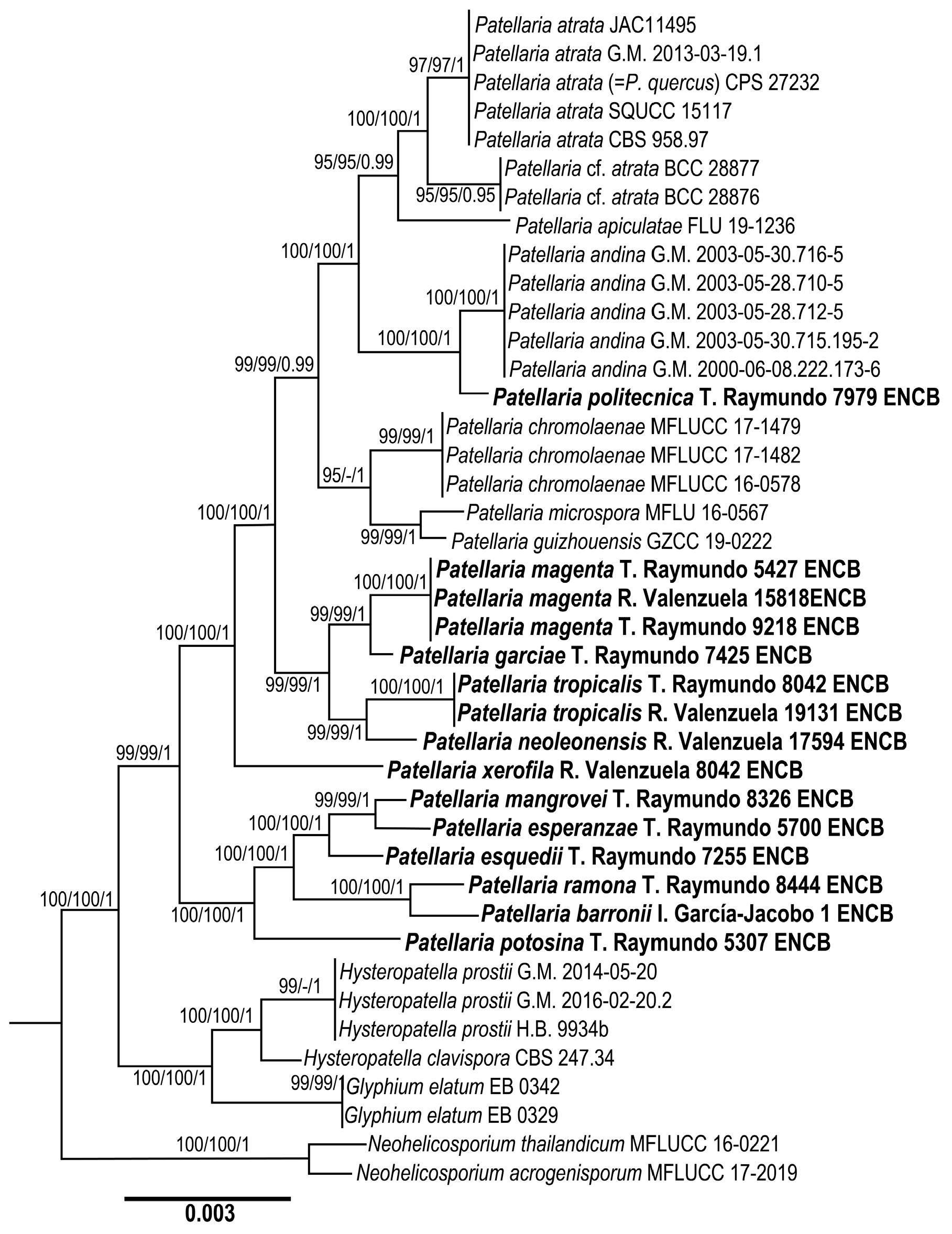
3.1. Phylogenetic Analysis
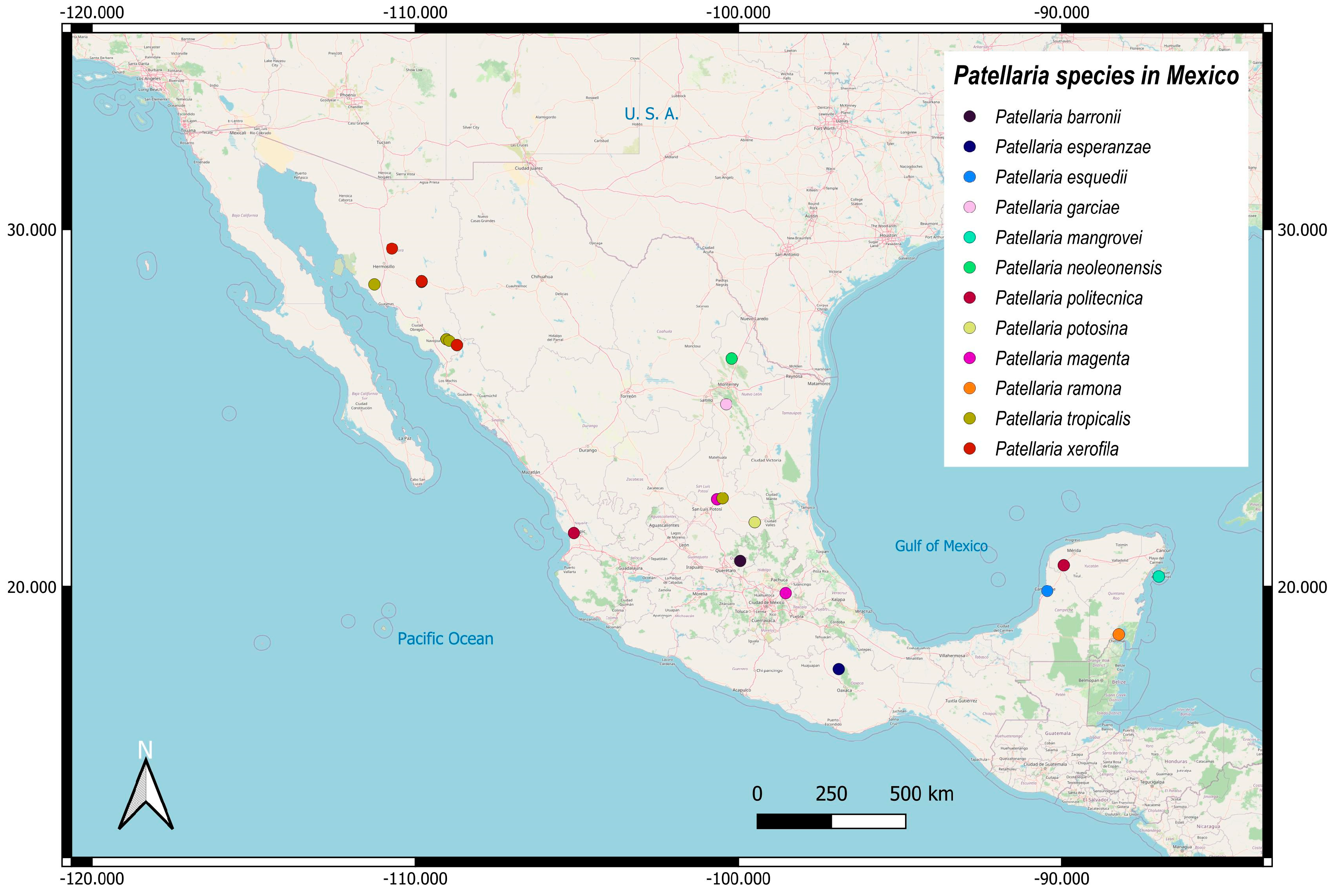
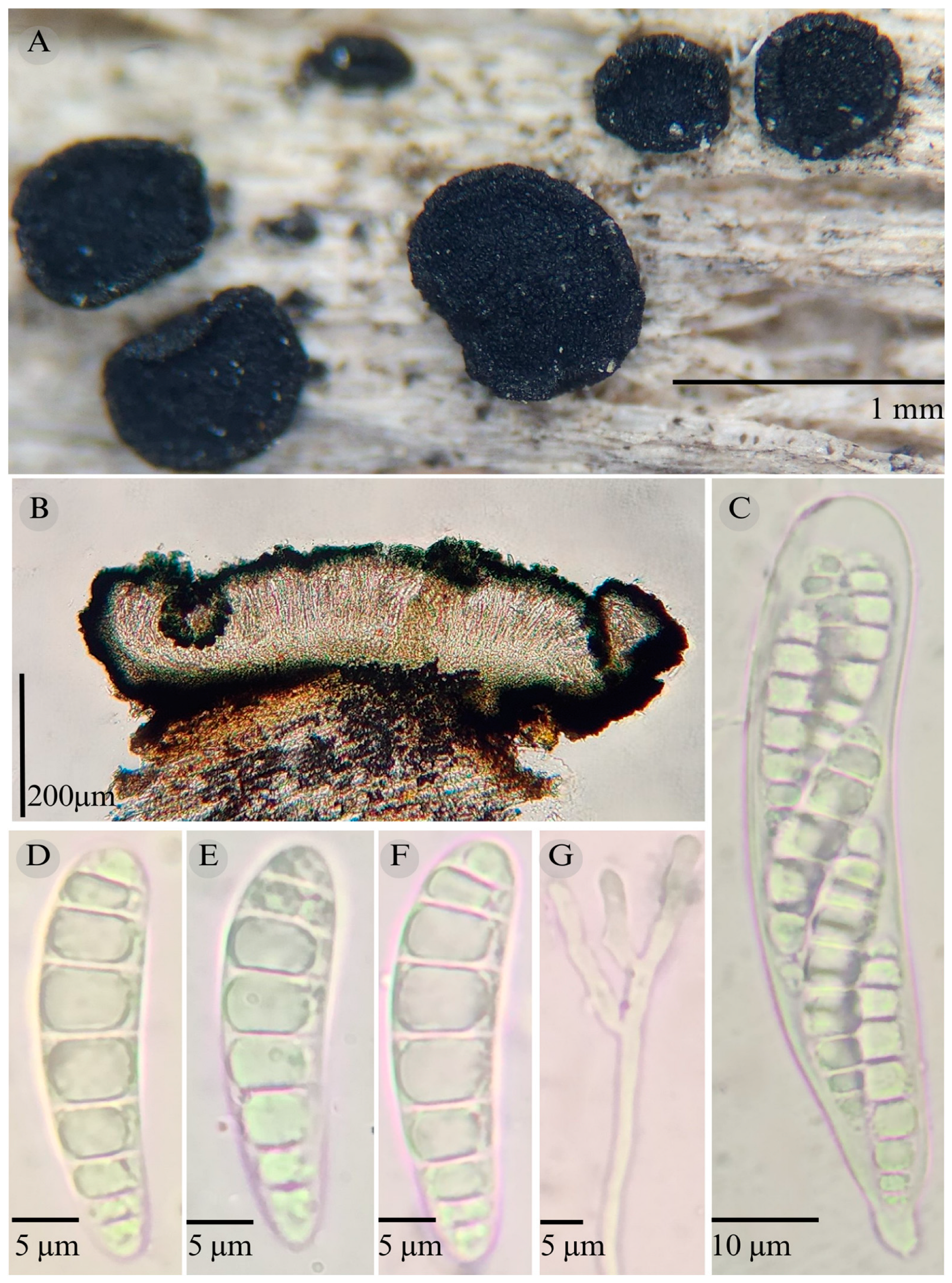
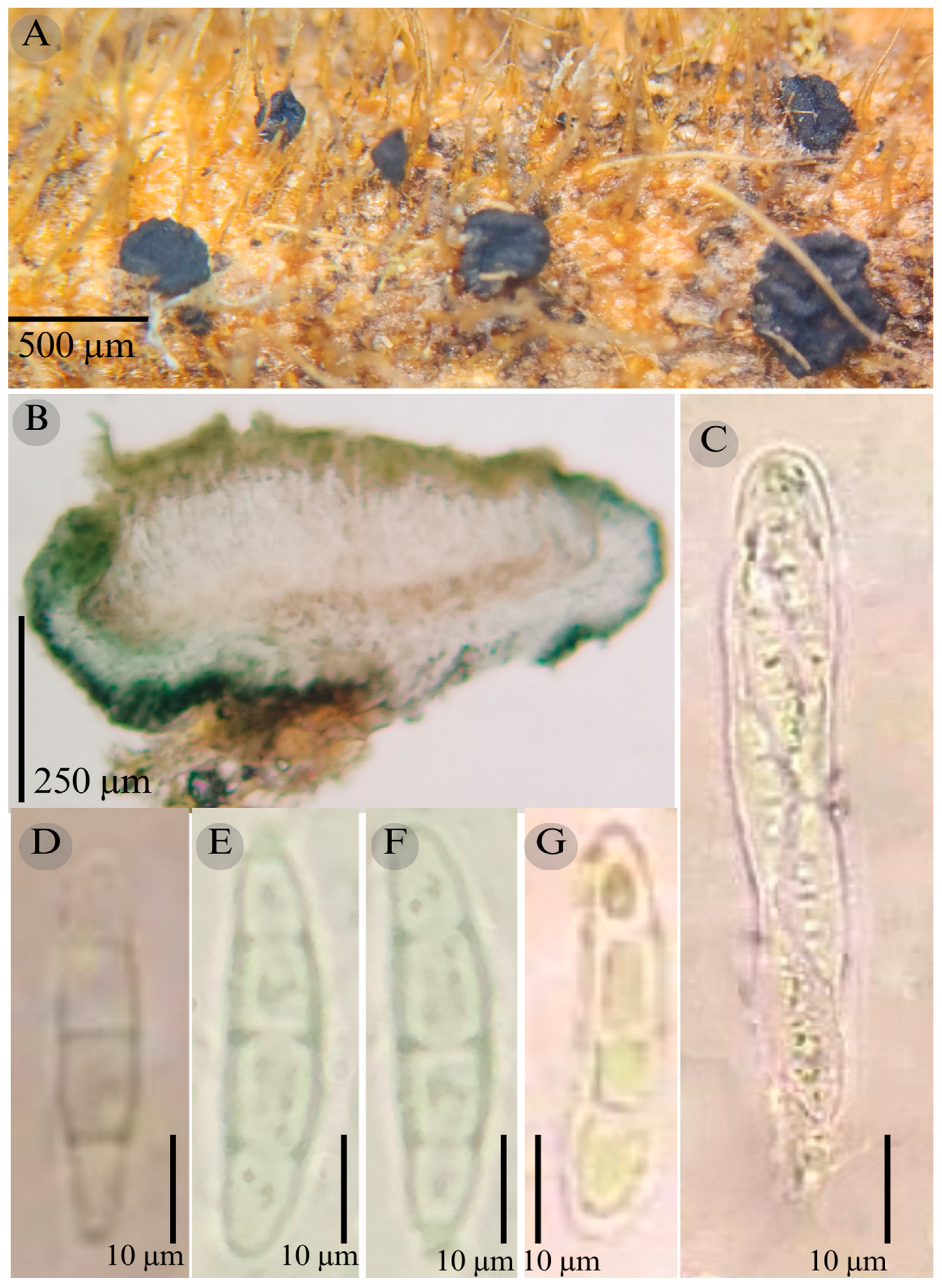
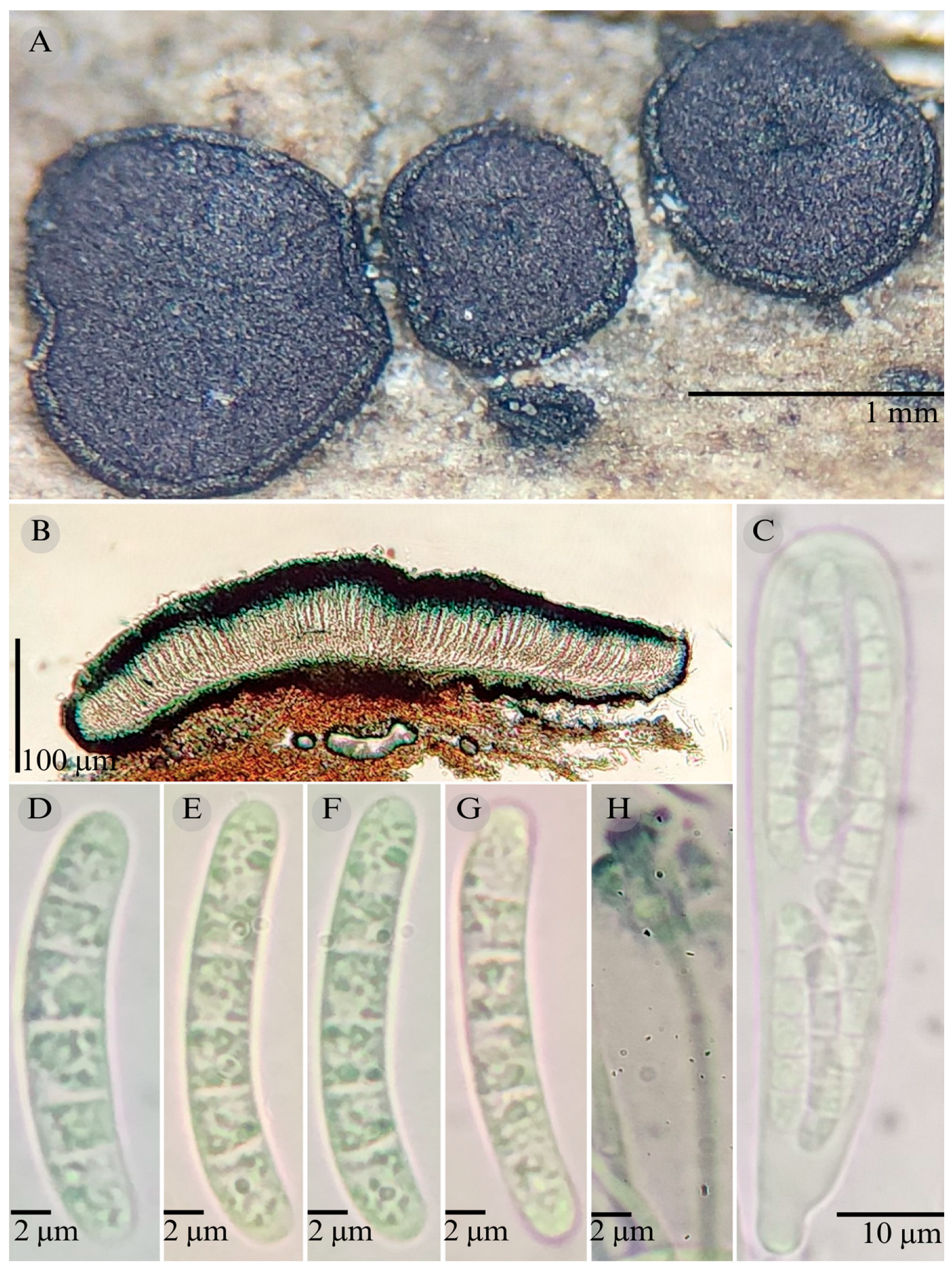
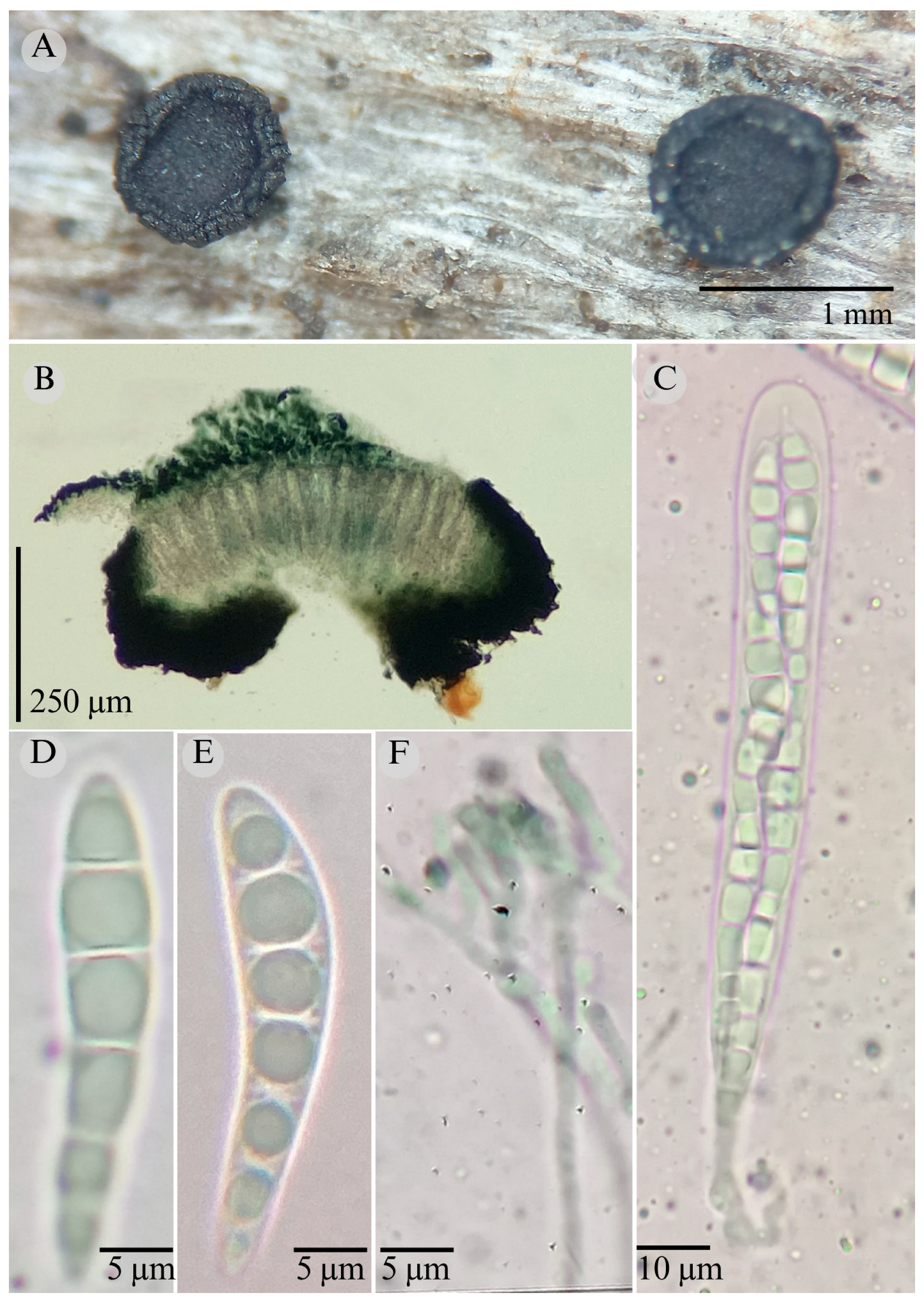
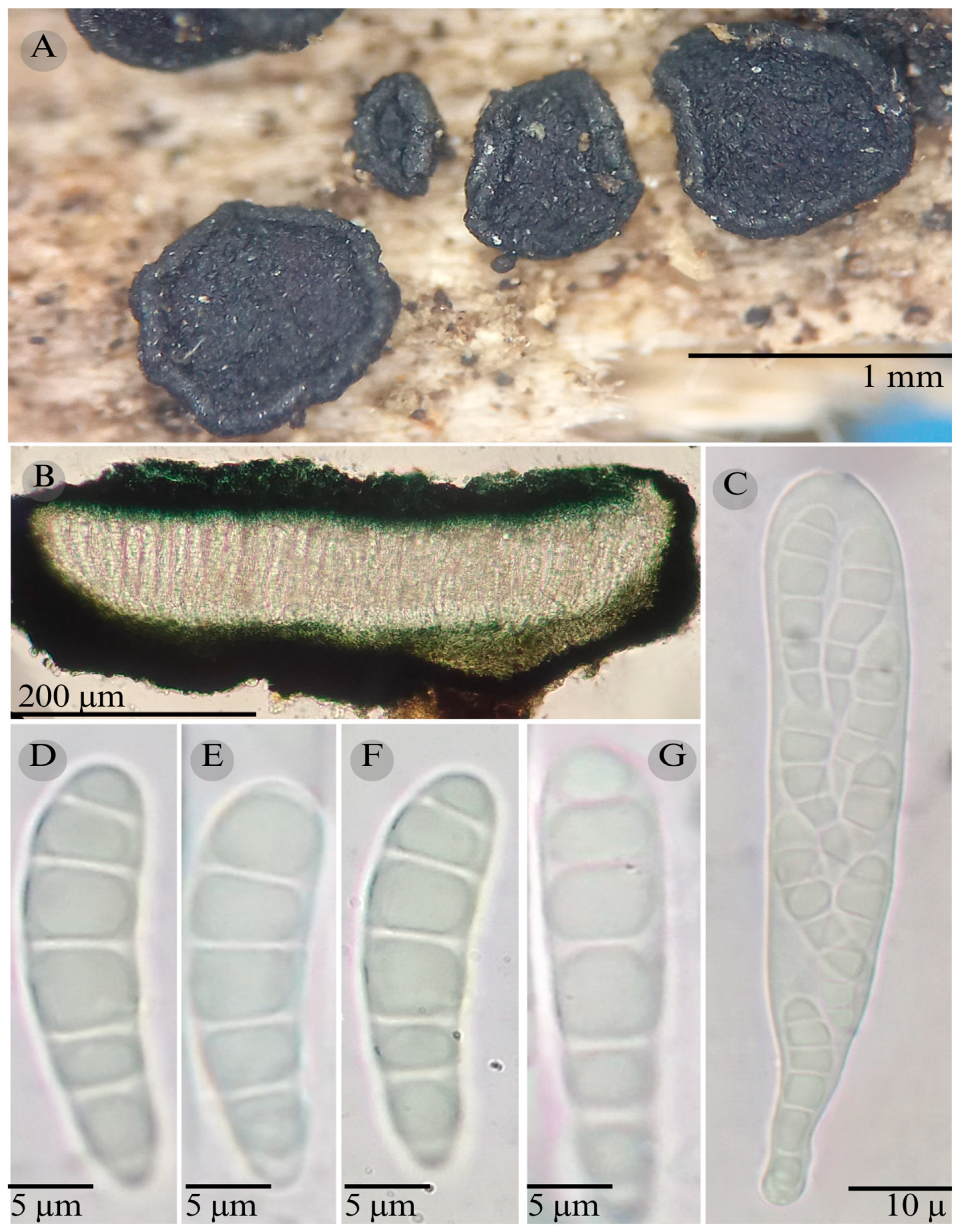
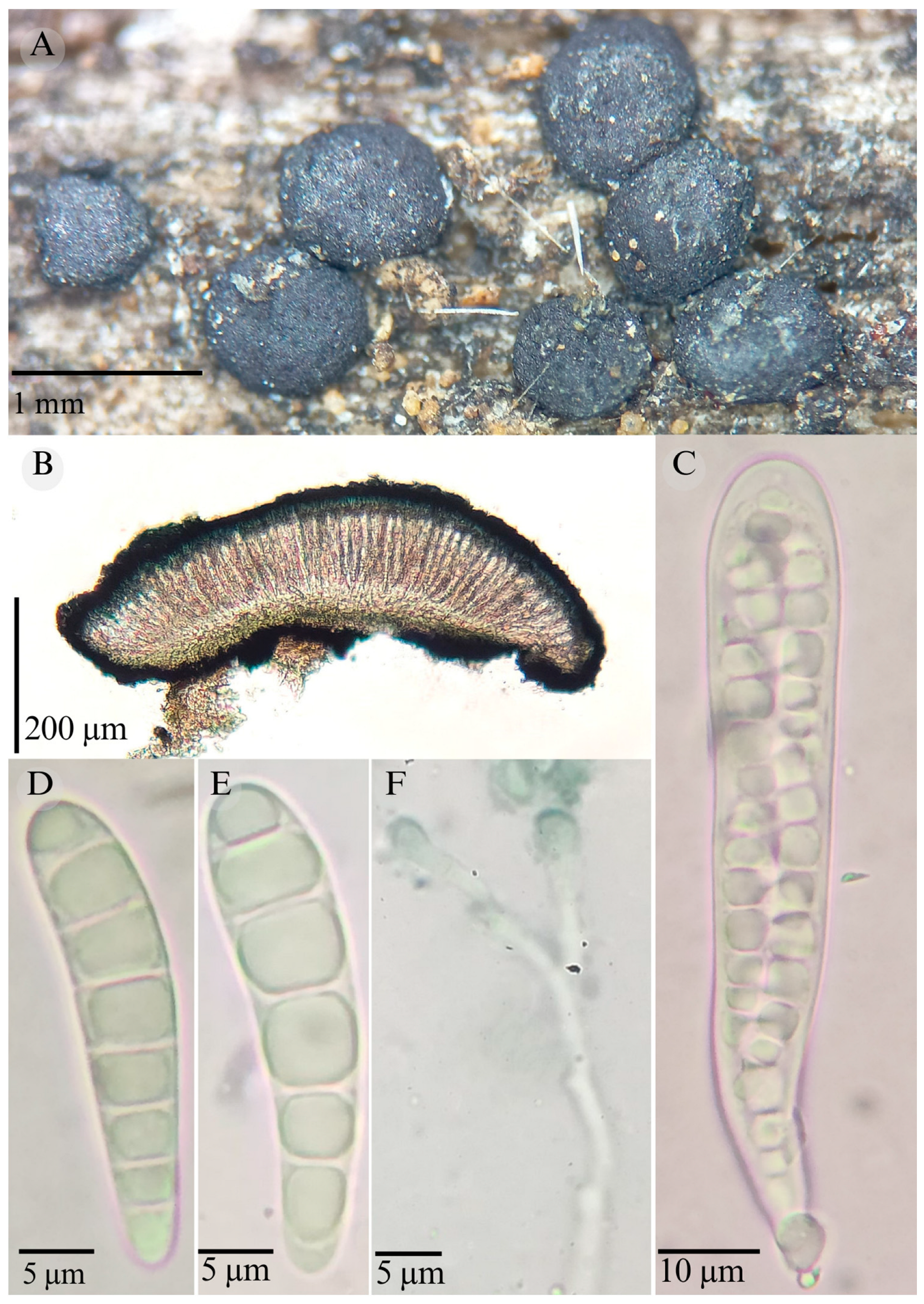
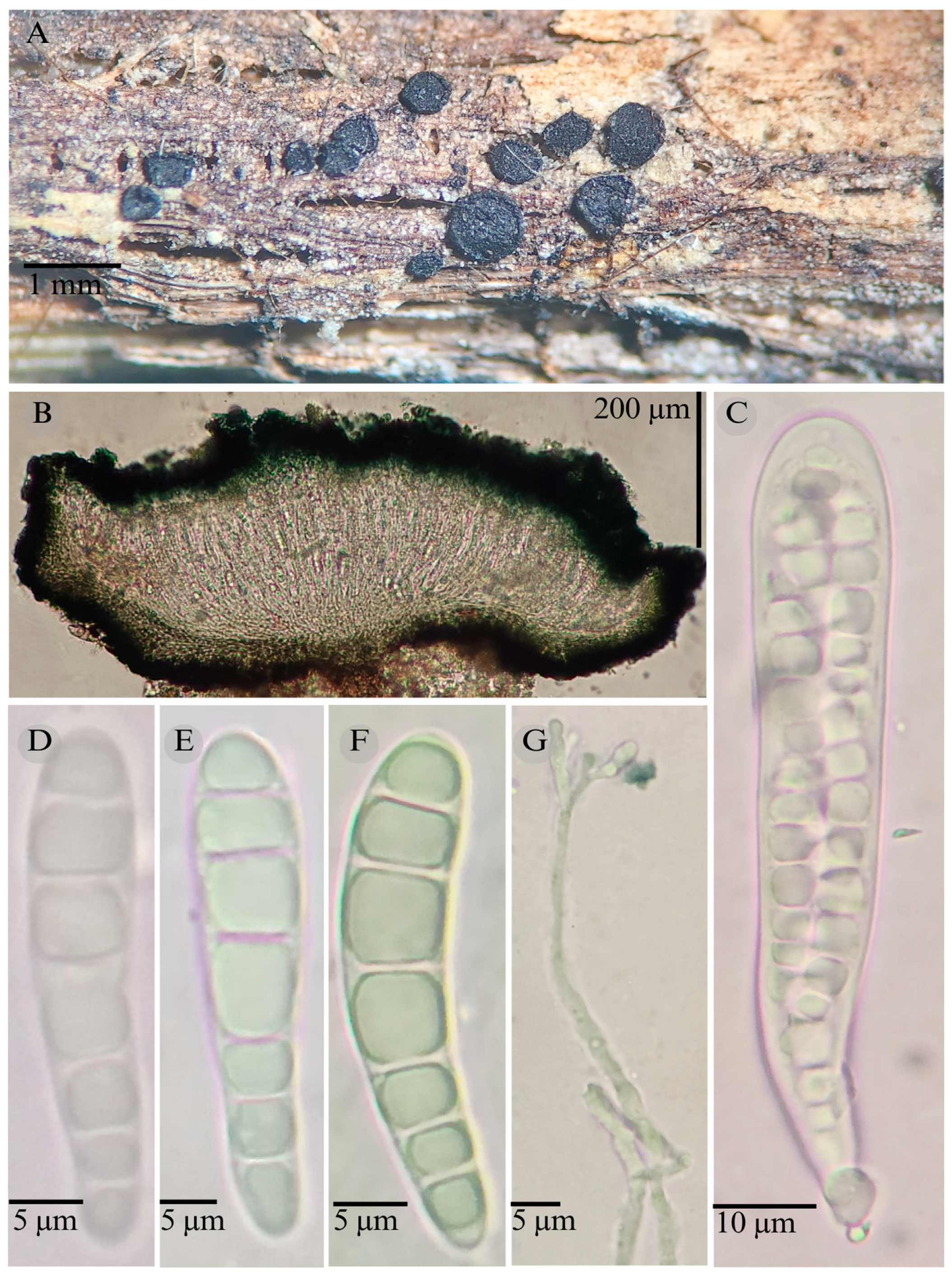
3.2. Taxonomy
- Key to Patellaria species forming ascomata supported by molecular and morphological data
- 1.
- Species with allantoid ascospores and 3–6 septa ……………………….…………………………………………………………2
- 1.
- Species with subfusiform, obclavate, or clavate ascospores; 3–13 septa……………………………………………………………………………3
- 2.
- Ascomata < 700 µm in diam; ascospores 27–34 × 5.2–6 µm, allantoid, 3–7 septa; growing in dead stems of Chromolaenae odorata from Thailand …………………………………………………………………P. chromolaenae
- 2.
- Ascomata > 700 µm in diam; ascospores 21–32 × 4–5 µm, allantoid, 3–6 septa; growing in dead wood of Mexico ……………………………………………………………………P. politecnica
- 3.
- Ascospores < 20 µm long, subfusiform or claviform …………………………………………………………………………………4
- 3.
- Ascospores > 20 µm long, obclavate or claviform …………………………………………………………………………………5
- 4.
- Subfusiform ascospores with three septa growing in the stem of the genus Cyathea ……………………………………………………………………P. esperanzae
- 4.
- Ellipsoid ascospores with eight septa growing in dead wood ……………………………………………………………………P. microspora
- 5.
- Obclavate ascospores, 5–6 septa …………………………………………………………………………………6
- 5.
- Clavate ascospores, 4–13 septa …………………………………………………………………………………7
- 6.
- Discoid ascomata, smooth and thick rim, green jungle coloration in KOH (25F8); unilobed pedicellate asci, ascospores 30–32 × 5–7 µm ( = 31 × 6, n = 30); growing in tropical rain forest ………………………………………………………………………P. esquedii
- 6.
- Discoid ascomata, striate and thin rim, coloration nautical blue in KOH (24E7); bilobed pedicel asci, ascospores 30–33 × 6–7 µm ( = 32 × 6, n = 30); growing in Quercus forest “Chaparral” ………………………………………………………………………P. potosina
- 7.
- Clavate ascospores, slightly curved, 4–8 septa, unilobed asci …………………………………………………………………………………8
- 7.
- Clavate ascospores, 8–10 (13) septa, bilobed asci …………………………………………………………………………………15
- 8.
- Ascospores < 30 µm long …………………………………………………………………………………9
- 8.
- Ascospores > 30 µm long …………………………………………………13
- 9.
- Species growing in temperate forests …………………………………………………………………………P. atrata
- 9.
- Species growing in tropical forests or mangroves…………………………………………………………………10
- 10.
- Species growing in mangroves ………………………………………………………………………………11
- 10.
- Species growing in tropical forests; ascospores 26–30 µ ………………………………………………………………………………12
- 11.
- Ascomata with thick rim and turquoise pigmentation in KOH (24F8), ascospores = 26 × 7, 5–6 septa; growing in Rhizophora apiculata ……………………………………………………………………P. apiculate
- 11.
- Ascomata with a thin rim and olive brown pigmentation in KOH (4E8), ascospores = 29 × 7, n = 30, 5–6 septa; growing in Rhizophora mangle ……………………………………………………………………P. mangrovei
- 12.
- Ascomata with thick rim and turquoise pigmentation in KOH (24F8), asci, short pedicellate, ascospores = 28 × 6, n = 30, 4–6 septa; growing in tropical rain forests………………………………………………………………P. ramona
- 12.
- Ascomata with a thin rim with deep green pigmentation in KOH (25D8), asci with long pedicellate, ascospores = 28 × 6, n = 30, 5–7 septa; growing in tropical dry forests ………………………………………………………………P. tropicalis
- 13.
- Ascospores with 5–6 septa ……………………………………………………………P. ghizhuensis
- 13.
- Ascospores with six or more septa……………………………………………………………………14
- 14.
- Ascomata with a thin rim, green jungle pigmentation in KOH (25F8), asci with short pedicellate, ascospores 31–35 × 6–8 µm, = 33 × 7 µm, 6–8 septa; …………………………………………………………P. barronii
- 14.
- Ascomata with thick rim turquoise pigmentation in KOH (24F8), asci long with pedicellate, ascospores 28–30 × 6–7 µm, = 30 × 6 µm, 5–7 septa…………………………………………………………………P. xerofila
- 15.
- Species growing in temperate coniferous forest, ascomata with thin and smooth rims, green jungle pigmentation in KOH(CLA), ascospores with clavate blunt apex ……………………………………………………………………P. garciae
- 15.
- Species in tropical dry forest, ascomata with thick and striate rim, English green pigmentation in KOH, ascospores clavate with conic apex……………………………………………………………………16
- 16.
- Ascospores 32–40 × 6–7 µm, = 36 × 6.2 µm, n = 30, 8–10 (13) septa, asci with long pedicellate, ……………………………………………………………………P. purpurea
- 16.
- Ascospores 38–44 × 6–7 µm, = 40 × 6.6 µm, n = 30, 8–10 septa, asci with short pedicellate………………………………………P. neolonensis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fries, E.M. Systema Mycologicum; Gryphiswaldiae: Sumtibus Emesti, Mauritius, 1822; Volume 2, p. 275. [Google Scholar] [CrossRef]
- Species Fungorum. Available online: http://www.speciesfungorum.org/Index.htm (accessed on 30 September 2024).
- Kutorga, E.; Hawksworth, D.L. A re-assessment of the genera referred to the family Patellariaceae (Ascomcota). Syst. Ascomycetum 1997, 15, 1–110. [Google Scholar]
- Dayarathne, M.C.; Jones, E.B.G.; Maharachchikumbura, S.S.N.; Devadatha, B.; Sarma, V.V.; Khongphinitbunjong, K.; Chonunti, P.; Hyde, K.D. Morpho-molecular characterization of microfungi associated with marine based habitats. Mycosphere 2020, 11, 1–188. [Google Scholar] [CrossRef]
- Yacharoen, S.; Tian, Q.; Chommunti, P.; Boonmee, S.; Chukeatirote, E.; Bhat, J.D.; Hyde, K.D. Patellariaceae revisited. Mycosphere 2015, 6, 290–326. [Google Scholar] [CrossRef]
- Hernandez-Restrepo, M.; Schumacher, R.K.; Wingfield, M.J.; Ishtiaq, A.; Cai, L.; Duong, T.A.; Edwards, J.; Gene, J.; Groenewald, J.Z.; Sana, J.; et al. Fungal systematics and evolution: FUSE 2. Sydowia 2016, 68, 193–230. [Google Scholar]
- Mapook, A.; Hyde, K.D.; McKenzie, E.H.C.; Jones, E.B.G.; Bhat, D.J.; Jeewon, R.; Stadler, M.; Samarakoon, M.C.; Malaithong, M.; Tanunchai, B.; et al. Taxonomic and phylogenetic contributions to fungi associated with the invasive weed Chromolaena odorata (Siam weed). Fungal Divers. 2020, 101, 1–175. [Google Scholar] [CrossRef]
- Hongsanan, S.; Hyde, K.D.; Phookamsak, R.; Wanasinghe, D.N.; McKenzie, E.H.C.; Sarma, V.V.; Lücking, R.; Boonmee, S.; Bhat, J.D.; Liu, N.G.; et al. Refined families of Dothideomycetes: Orders and families incertae sedis in Dothideomycetes. Fungal Divers. 2020, 105, 17–318. [Google Scholar] [CrossRef]
- Zhang, J.F.; Liu, J.K.; Hyde, K.D.; Chen, Y.; Ran, H.; Liu, Z. Ascomycetes from karst landscapes of Guizhou Province, China. Fungal Divers. 2023, 122, 1–160. [Google Scholar] [CrossRef]
- Chacón, S.; Tapia, F. Algunas especies saprobias de dothideomycetes y lecanoromycetes (Pezizomycotina: Ascomycota) en México. Rev. Mex. Biodivers. 2016, 87, 1169–1176. [Google Scholar] [CrossRef]
- Méndez-Mayboca, F.; Checa, J.; Esqueda, M.; Chacón, S. New records of Loculoascomycetes from natural protected areas in Sonora, Mexico. Mycotaxon 2010, 111, 19–30. [Google Scholar] [CrossRef]
- Esqueda, M.; Coronado, M.L.; Gutiérrez, A.; Lizárraga, M.; Raymundo, T.; Valenzuela, R. Hongos de la Reserva de la Biósfera El Pinacate y Gran desierto de Altar, 1st ed.; Centro de Investigación en Alimentación y Desarrollo (CIAD): Hermosillo, Mexico, 2013; p. 23. [Google Scholar]
- García-Martínez, Y.; Heredia Abarca, G.; Guzmán-Guillermo, J.; Valenzuela, R.; Raymundo, T. Hongos asociados al mangle rojo Rhizophora mangle (Rhizophoraceae) en la Reserva de la Biosfera Isla Cozumel, Quintana Roo, México. Acta Bot. Mex. 2021, 128. [Google Scholar] [CrossRef]
- Raymundo, T.; Martínez-Pineda, M.; Reyes, P.H.; Cobos-Villagrán, A.; Martínez, Y.A.G.; Tun, A.A.; Valenzuela, R. Ascomicetos de la Reserva de la Biosfera Isla Cozumel, México. Act. Bot. Mex. 2021, 128. [Google Scholar] [CrossRef]
- Kornerup, A.; Wanscher, J.H. Methuen Handbook of Colour, 3rd ed.; Methuen: London, UK, 1978; pp. 1–254. [Google Scholar]
- Martínez-González, C.R.; Ramírez-Mendoza, R.; Jiménez-Ramírez, J.; Gallegos-Vázquez, C.; Luna-Vega, I. Improved method for genomic DNA extraction for Opuntia Mill. (Cactaceae). Plant Methods 2017, 13, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Zhang, Z.; Schwartz, S.; Wagner, L.; Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000, 7, 203–214. [Google Scholar] [CrossRef]
- Maharachchikumbura, S.S.N.; Wanasinghe, D.N.; Cheewangkoon, R.; Al-Sadi, A.M. Uncovering the hidden taxonomic diversity of fungi in Oman. Fungal Divers. 2021, 106, 229–268. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017, 20, 1160–1166. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Muller, K.; Quandt, D.; Muller, J.; Neinhuis, C. PhyDER-Phylogenetic Data Editor. Program Distributed by the Authors, Ver. 10.0. 2005. Available online: https://www.phyde.de (accessed on 3 September 2024).
- Maddison, W.P.; Maddison, D.R. Mesquite: A Modular System for Evolutionary Analysis. Version 3.31. 2017. Available online: http://mesquiteproject.org (accessed on 3 September 2024).
- Swofford, D.L. Phylogenetic Analysis Using Parsimony (PAUP*) 4.0; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MrBayes: Bayesian inference of phylogeny. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Frandsen, P.B.; Calcott, B.; Mayer, C.; Lanfear, R. Automatic selection of partitioning schemes for phylogenetic analyses using iterative k-means clustering of site rates. BMC Evol. Biol. 2015, 15, 13. [Google Scholar] [CrossRef]
- Lanfear, R.; Calcott, B.; Kainer, D.; Mayer, C.; Stamatakis, A. Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol. Biol. 2014, 14, 82. [Google Scholar] [CrossRef] [PubMed]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. FigTree v.1.4.2. A Graphical Viewer of Phylogenetic Trees. 2014. Available online: https://tree.bio.ed.ac.uk/software/figtree (accessed on 3 September 2024).
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 135–322. [Google Scholar] [CrossRef]
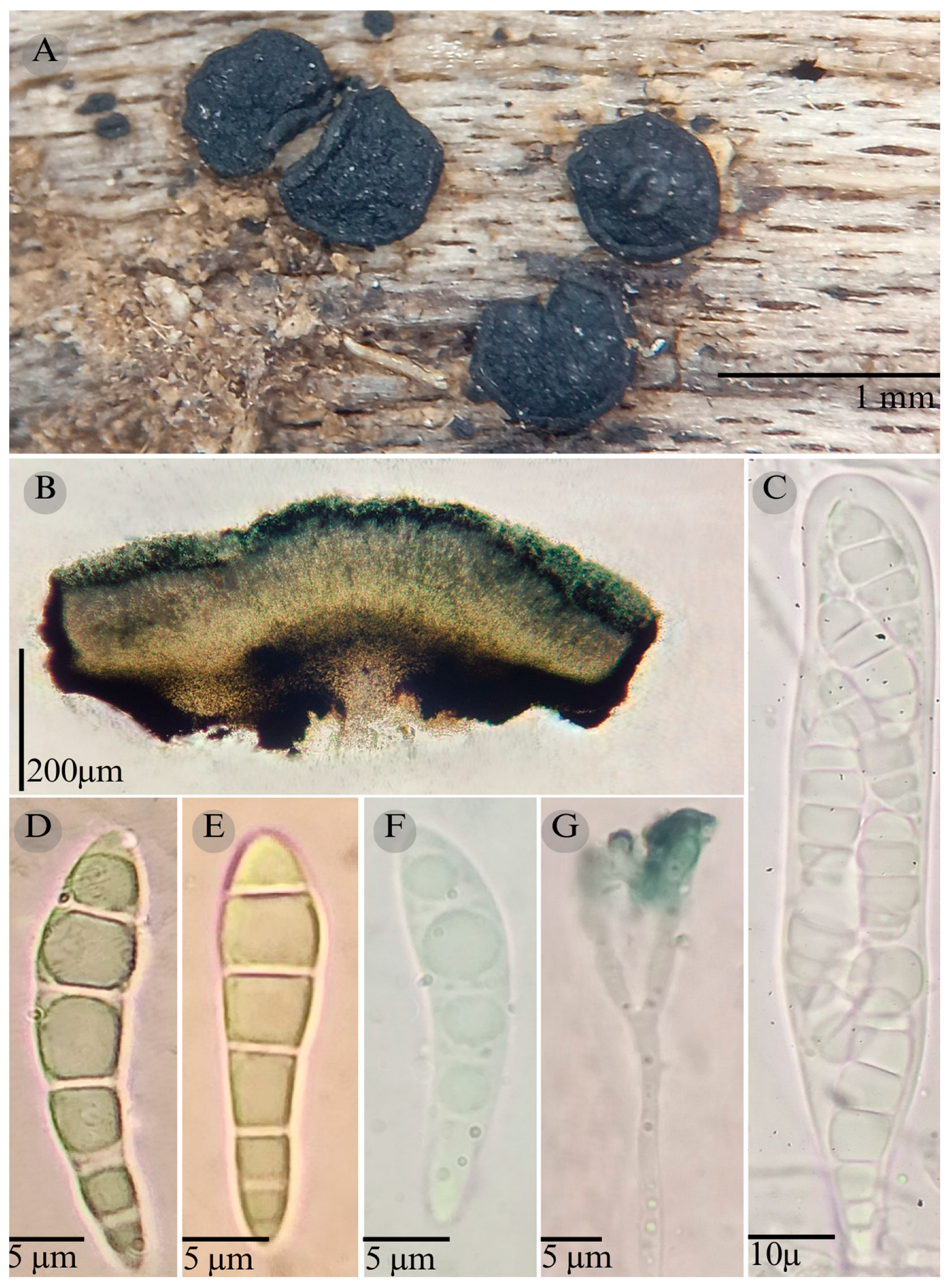
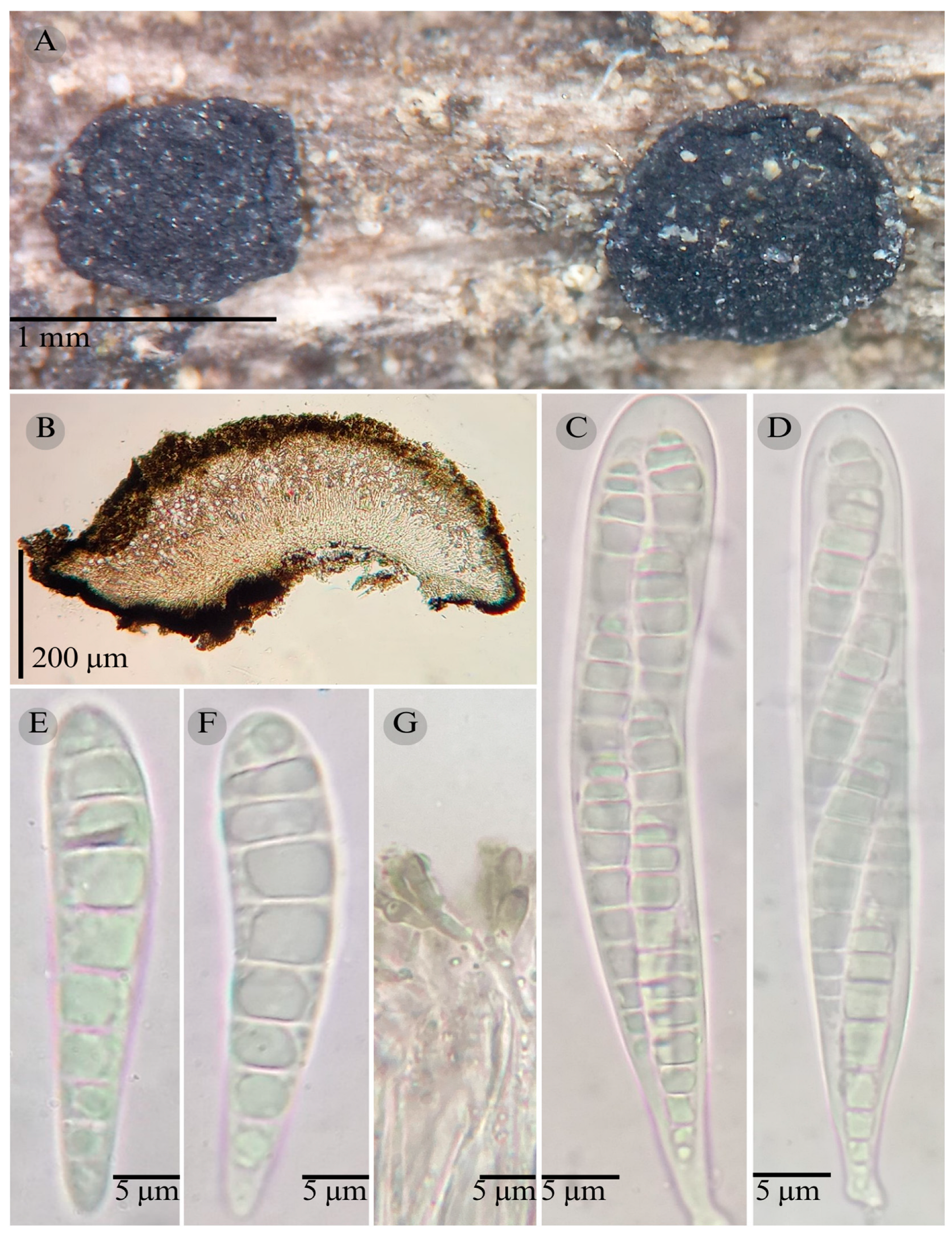
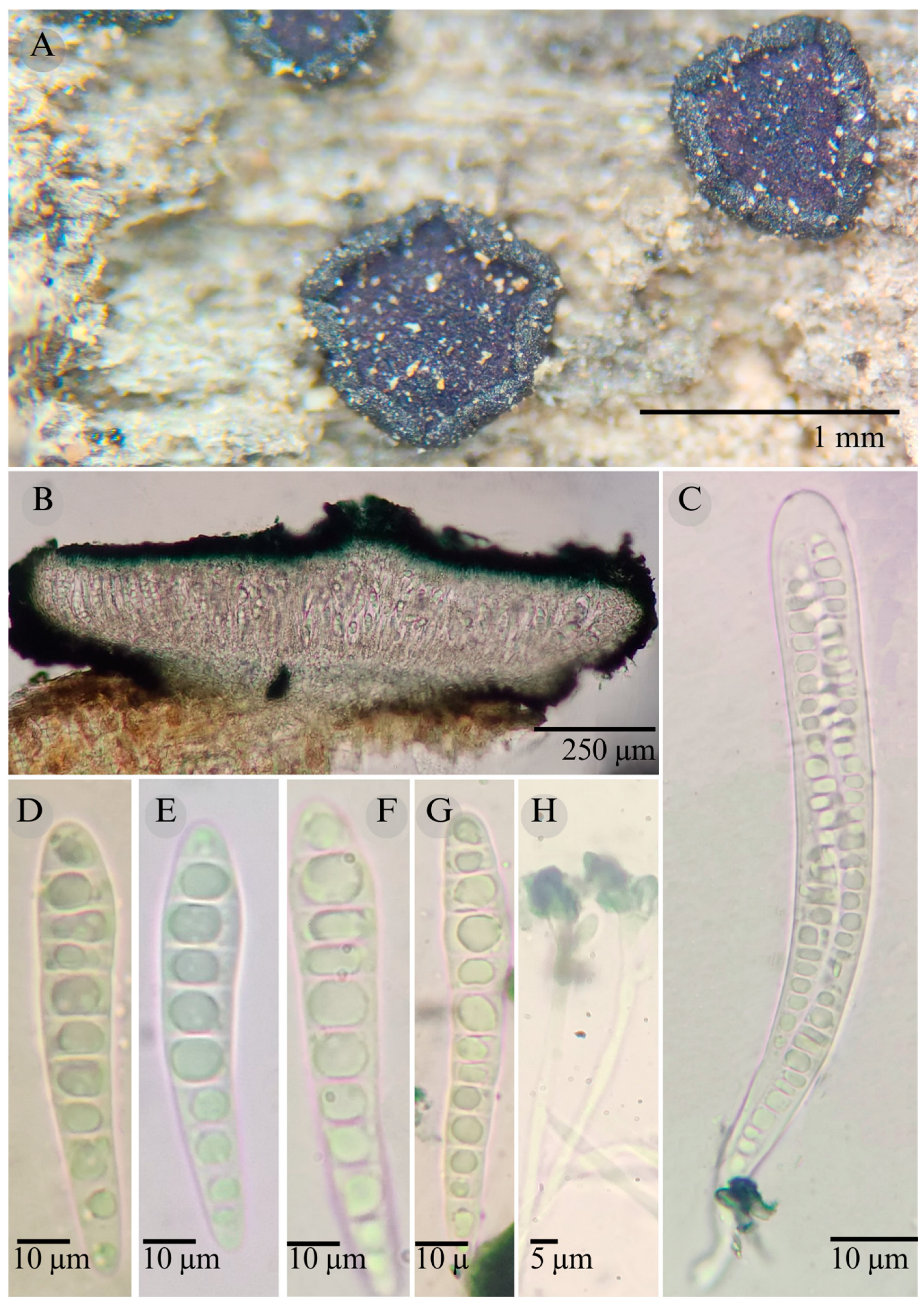
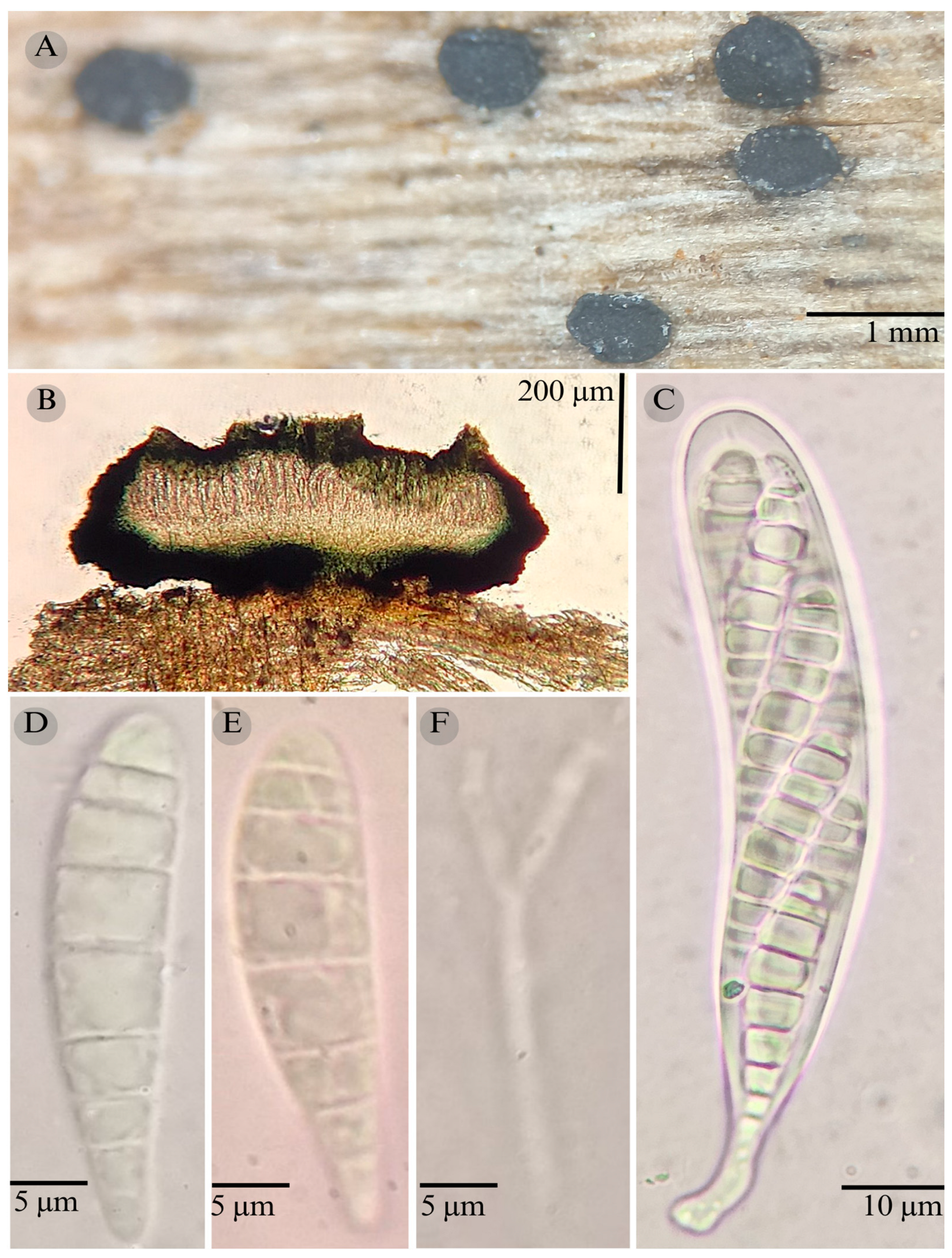

| Species | Strain No. | GenBank Accession No. | ||
|---|---|---|---|---|
| nLSU | ITS | SSU | ||
| Glyphium elatum | EB 0329 | KM220937 | KM220943 | KM220951 |
| Glyphium elatum | EB 0342 | KM220938 | KM220944 | KM220952 |
| Glyphium elatum | EB 0388 | KM220940 | KM220946 | ----- |
| Hysteropatella clavispora | CBS 247.34 | AY541493 | ----- | ----- |
| Hysteropatella prostii | H.B. 9934b | KT876980 | KT876980 | ----- |
| Hysteropatella prostii | G.M. 2014-05-20 | KM220949 | KM220949 | ----- |
| Hysteropatella prostii | G.M. 2016-02-20.2 | MT341324 | MT341324 | MT341324 |
| Neohelicosporium acrogenisporum | MFLUCC 17-2019 | MH558871 | NR160376 | ----- |
| Neohelicosporium thailandicum | MFLUCC 16-0221 | MF467941 | MF467928 | MF467928 |
| Patellaria andina | G.M. 2000-06-08.222.173-6 | MT273251 | ----- | ----- |
| Patellaria andina | G.M. 2003-05-30.716-5 | MT293616 | MT293616 | ----- |
| Patellaria andina | G.M. 2003-05-30.715.195-2 | MT273250 | MT273250 | ----- |
| Patellaria andina | G.M. 2003-05-28.712-5 | MT293619 | MT293619 | ----- |
| Patellaria andina | G.M. 2003-05-28.710-5 | MT293624 | MT293624 | ----- |
| Patellaria apiculatae | MFLU 19-1236 | MN017860 | MN047094 | MN017925 |
| Patellaria atrata | JAC11495 | MK431438 | MK432702 | MK432702 |
| Patellaria atrata | CBS 958.97 | GU301855 | ----- | GU296181 |
| Patellaria atrata | G.M. 2013-03-19.1 | MN565888 | MN565888 | ----- |
| Patellaria atrata | SQUCC 15117 | MW077152 | MW077143 | ----- |
| Patellaria barronii | S. García 1 ENCB | PQ766174 | PQ817147 | PQ773270 |
| Patellaria cf. atrata | BCC 28877 | GU371829 | ----- | GU371837 |
| Patellaria cf. atrata | BCC 28876 | GU371828 | ----- | GU371836 |
| Patellaria chromolaenae | MFLUCC 16-0578 | MW142387 | MW136695 | MW127179 |
| Patellaria chromolaenae | MFLUCC 17-1482 | MT214475 | MT214381 | MT214426 |
| Patellaria chromolaenae | MFLUCC 17-1479 | MT214474 | MT214380 | ----- |
| Patellaria esperanzae | T. Raymundo 5700 ENCB | PQ766175 | PQ817148 | PQ773271 |
| Patellaria esquedii | T. Raymundo 7255 ENCB | PQ766176 | PQ817149 | PQ773272 |
| Patellaria garciae | T. Raymundo 7425 ENCB | PQ766177 | PQ817150 | PQ773273 |
| Patellaria guizhouensis | GZCC 19-0222 | OR209665 | ----- | OR134437 |
| Patellaria mangrovei | T. Raymundo 8326 ENCB | PQ766178 | PQ817151 | PQ773274 |
| Patellaria microspora | MFLU 16-0567 | MW142388 | NR175661 | |
| Patellaria neoleonensis | R. Valenzuela 17594 ENCB | PQ766179 | PQ817152 | PQ773275 |
| Patellaria politecnica | T. Raymundo 7979 ENCB | PQ766180 | PQ817153 | PQ773276 |
| Patellaria potosina | T. Raymundo 5307 ENCB | PQ766181 | PQ817154 | PQ773277 |
| Patellaria magenta | T. Raymundo 5427 ENCB | PQ766182 | PQ817155 | PQ773278 |
| Patellaria magenta | R. Valenzuela 15818 ENCB | PQ766183 | PQ817156 | PQ773279 |
| Patellaria magenta | T. Raymundo 9218 ENCB | PQ766184 | PQ817157 | PQ773280 |
| Patellaria quercus | CPC 27232 | NG059696 | NR152540 | |
| Patellaria ramona | T. Raymundo 8444 ENCB | PQ766185 | PQ817158 | PQ773281 |
| Patellaria tropicalis | T. Raymundo 8042 ENCB | PQ766186 | PQ817159 | PQ773282 |
| Patellaria tropicalis | R. Valenzuela 19131 ENCB | PQ766187 | PQ817160 | PQ773283 |
| Patellaria xerofila | R. Valenzuela 15286 ENCB | PQ766188 | PQ817161 | PQ773284 |
| Species | Ascomata | Rim | Exciple | Sub Hymenium | Asci | Pedicel | Epithecium Color & Key | Colour KOH 10% | Ascospores | Septa | Vegetation |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patellaria barronii | Discoidal 500–730 µm | Thin striate | 33–40 µm = 37 | 35–40 µm = 37 | 101.3 × 14.8–15.3 µm ( = 101 × 15 µm) | Short unilobed | = 20 µm Color: 25E7 Green Jungle |  | 28–36 × 5–8 = 35 × 7 µm Clavate | 6–8 | XS |
| Patellaria esperanzae | Discoidal 400–600 µm | Thick smooth | 45–52 µm = 47.5 µm | 25–40 µm = 30 µm | 70–95 × 10 µm ( = 83 × 10) | - | = 25 µm Color: 27E5 Olive green |  | 16–20 × 4–5 = 18 × 4 µm Subfusiform | 3 | TMCF |
| Patellaria esquedii | Discoidal to irregular 600–1000 µm | Thick smooth | 35–40 µm = 37.3 µm | 35–40 µm = 37 µm | 85–98 × 13–17 µm ( = 91 × 13.83 µm) | Short unilobed | = 22.3 µm Color: 25E7 Green jungle |  | 27–32 × 6 = 30 × 6 µm Obclavate | 5–6 | TRF |
| Patellaria garciae | Discoidal 700–800 µm | Thin smooth | 50 µm | 30 µm | 100–115 × 15 µm ( = 108 × 15 µm) | Short bilobuled | = 30 µm Color: 25F3 Spruce green |  | 35–42 × 68 = 40 × 7 µm Clavate | 8–10 | CF |
| Patellaria magenta | Discoidal 600–1000 µm | Thick inovulte striate | 40–56 µm = 49.6 µm | 25–37 µm = 30.5 µm | 95–145 × 12–20 µm ( = 120.6 × 13.1 µm) | Long bilobed | = 25 µm Color: 25E7 Green jungle |  | 30–54 × 6–8 µm = 36 × 6 Clavate | 7–13 | TDF, XS |
| Patellaria mangrovei | Discoidal to ovoid 500–700 µm | Thin smooth | 40–43 µm = 42 | 60 µm | 75–90 × 14–16 µm ( = 83 × 15 µm) | Short unilobed | = 20 µm Color: 4E8 Olive brown |  | 25–35 × 6–9 = 30 × 7 µm Clavate | 5–8 | M |
| Patellaria neoleonensis | Discoidal 830–1000 µm | Thick involute striate | 45–62 µm = 55 µm | 20–35 µm = 28 | 93–154 × 12–20 ( = 109.7 × 14.7 µm) | Long bilobed | = 21 µm Color: 24F8 Dark turquoise |  | 32–50 × 6–8 = 40 × 6 µm Clavate | 8–10 | XS |
| Patellaria politecnica | Discoidal 700–860 µm | Thin smooth | 25–30 µm = 28.3 | 10 µm | 55–76 × 10–15 µm ( = 65.18 × 12.60 µm) | Short unilobed | = 18 µm Color: 24E7 Nautic bue |  | 21–32 × 4–5, = 25 × 4 µm Allantoid | 3–6 | TRF |
| Patellaria potosina | Discoidal 550–750 µm | Thin striate | 35–45 µm = 40 µm | 40–55 µm = 45 µm | 75–116 × 11–15 µm ( = 91.42 × 12.56 µm) | Short bilobed | = 22.5 µm Color: 24E7 Nautic bue |  | 31–38 × 5–7 = 31 × 6 µm Obclavate | 5–8 | XS |
| Patellaria ramona | Irregular 450–700 µm | Thin smooth | 25–30 µm | 35–40 µm = 37.5 µm | 75–96 × 15–17 µm ( = 88.5 × 15.8 µm) | Long bilobed | = 15 µm Color: 24F8 Dark turquoise |  | 22–29 × 6 = 2 × 6 µm Clavate | 4–6 | TRF |
| Patellaria tropicalis | Discoidal irregular 530–800 µm | Thin smooth | 25–40 µm = 39.6 | 17–30 µm = 20.28 | 72–110 × 12–15 µm ( = 85.12 × 14.15 µm) | Short unilobed | = 20 µm Color: 25D8 Deep green |  | Obclavate 24–37 × 6–7 = 28 × 6 µm Clavate | 5–7 | TDF |
| Patellaria xerófila | Discoidal irregular 510–660 µm | Thin striate | 14–45 µm = 36 | 14–45 µm = 25 | 82–110 × 11–15 ( = 92.9 × 13.03 µm) | Short unilobed | = 17 µm Color: 24F8 Dark turquoise |  | 26–39 × 5–7 = 30 × 6 µm Clavate | 5–7 | XS |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Jacobo, I.; Raymundo, T.; Martínez-González, C.R.; Martínez-Pineda, M.; Valenzuela, R. Phylogenetic and Morphological Analyses Reveal Twelve New Species of the Genus Patellaria (Dothideomycetes, Ascomycota) from Mexico. J. Fungi 2025, 11, 44. https://doi.org/10.3390/jof11010044
García-Jacobo I, Raymundo T, Martínez-González CR, Martínez-Pineda M, Valenzuela R. Phylogenetic and Morphological Analyses Reveal Twelve New Species of the Genus Patellaria (Dothideomycetes, Ascomycota) from Mexico. Journal of Fungi. 2025; 11(1):44. https://doi.org/10.3390/jof11010044
Chicago/Turabian StyleGarcía-Jacobo, Ilian, Tania Raymundo, Cesar R. Martínez-González, Michelle Martínez-Pineda, and Ricardo Valenzuela. 2025. "Phylogenetic and Morphological Analyses Reveal Twelve New Species of the Genus Patellaria (Dothideomycetes, Ascomycota) from Mexico" Journal of Fungi 11, no. 1: 44. https://doi.org/10.3390/jof11010044
APA StyleGarcía-Jacobo, I., Raymundo, T., Martínez-González, C. R., Martínez-Pineda, M., & Valenzuela, R. (2025). Phylogenetic and Morphological Analyses Reveal Twelve New Species of the Genus Patellaria (Dothideomycetes, Ascomycota) from Mexico. Journal of Fungi, 11(1), 44. https://doi.org/10.3390/jof11010044







