Ex Situ Conservation, DNA Barcoding and Enzymatic Potential Evaluation of Macrofungi (Basidiomycota, Ascomycota) from Vietnam
Abstract
:1. Introduction
2. Materials and Methods
2.1. Origination and Isolation of Strains
2.2. Preservation of Strains
2.3. Strains Verification
2.3.1. Cultivation and Morphological Methods
2.3.2. Molecular Methods
2.4. Taxonomical Analysis
2.5. Methods for Evaluation of Growth and Enzymatic Activity
3. Results and Discussion
3.1. Ex Situ Conservation of Tropical Macromycetes
3.2. DNA Barcoding of LE-BIN Strains from Vietnam
3.3. Taxonomical Structure of the Vietnamese Fungi in the LE-BIN Collection
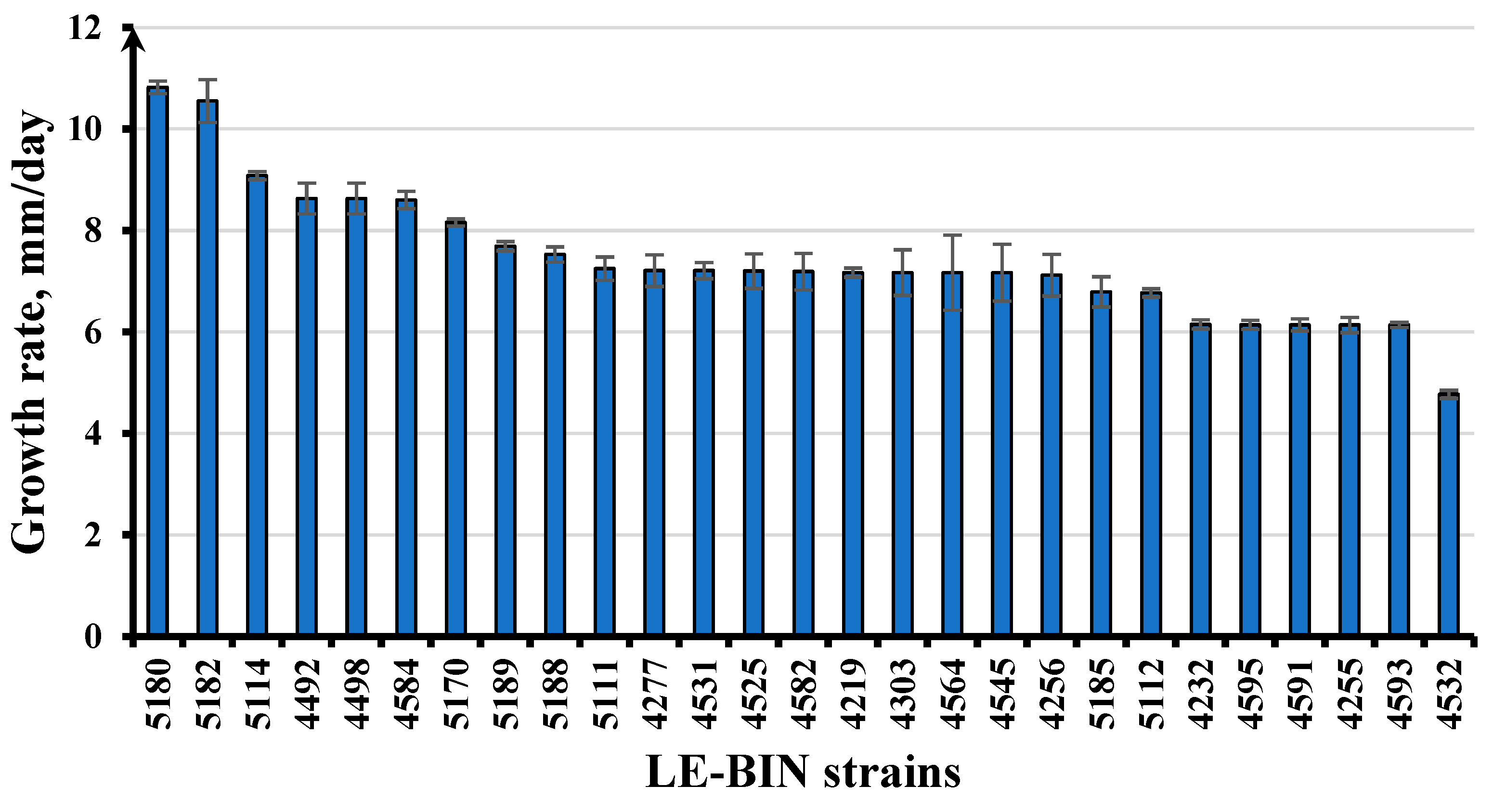
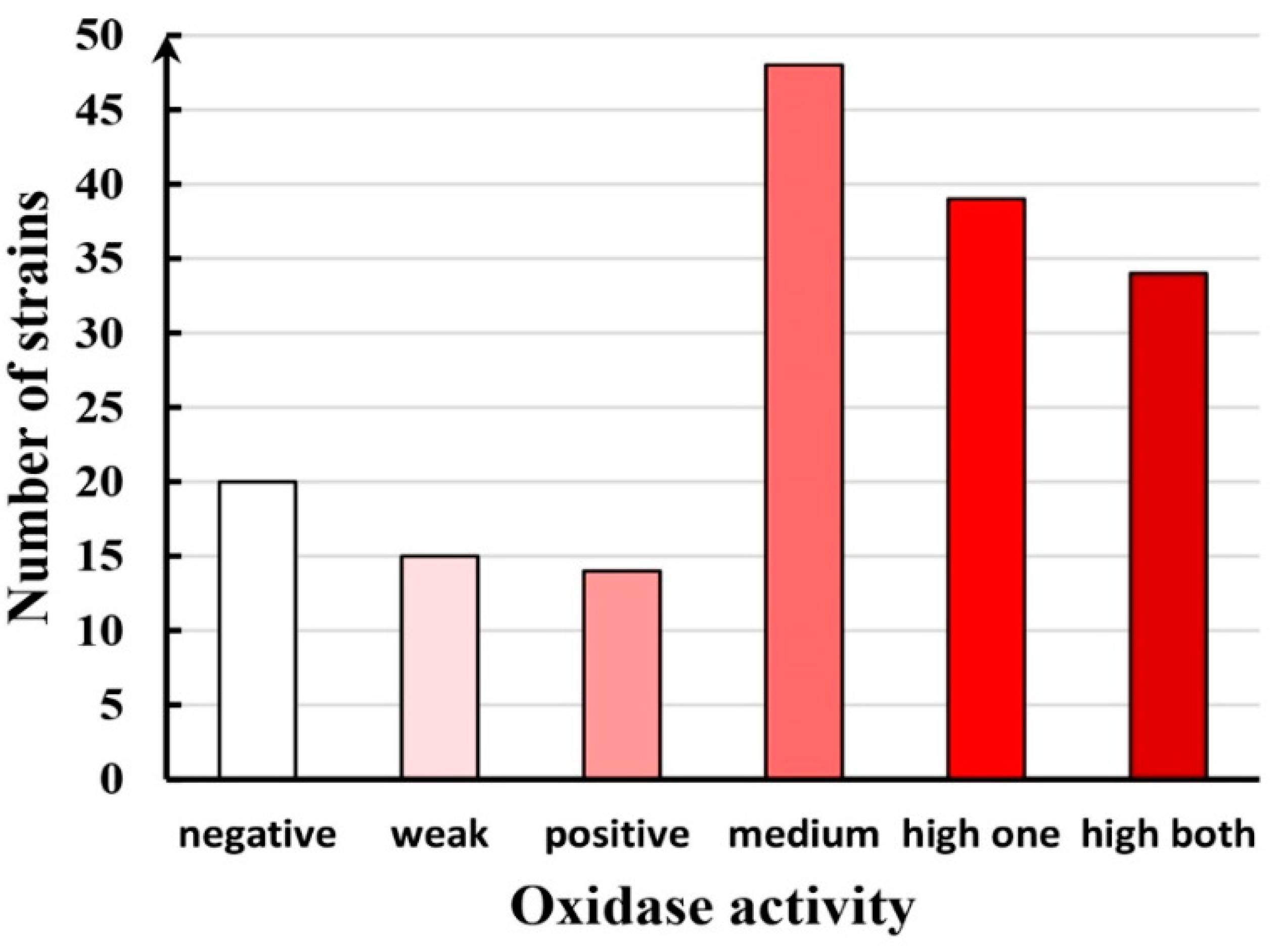
3.4. Growth and Enzymatic Potential of LE-BIN Strains from Vietnam
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jayasekara, S.; Ratnayake, R. Isolation, Identification and studying the potential application of tropical fungi in lignocellulolysis. In Research Advances in the Fungal World—Culture, Isolation, Identification, Classification, Characterization, Properties and Kinetics; Chaurasia, P.K., Lata, S., Eds.; Nova Science Publishers: Hauppauge, NY, USA, 2020; pp. 32–59. [Google Scholar]
- Peraza-Jiménez, K.; De la Rosa-García, S.; Huijara-Vasconselos, J.J.; Reyes-Estebanez, M.; Gómez-Cornelio, S. Enzymatic Bioprospecting of Fungi Isolated from a Tropical Rainforest in Mexico. J. Fungi 2022, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Hyde, K.D.; Xu, J.; Rapior, S. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers. 2019, 97, 1–136. [Google Scholar] [CrossRef]
- Yadav, D.; Negi, P.D. Bioactive components of mushrooms: Processing effects and health benefits. Food Res. Int. 2021, 148, 110599. [Google Scholar] [CrossRef] [PubMed]
- Dudekula, U.T.; Doriya, K.; Devarai, S.K. A critical review on submerged production of mushroom and their bioactive metabolites. 3 Biotech. 2020, 10, 337. [Google Scholar] [CrossRef]
- Niego, A.G.T.; Lambert, C.; Mortimer, P.; Thongklang, N.; Rapior, S.; Grosse, M.; Schrey, H.; Charria-Giron, E.; Walker, A.; Hyde, K.D. The contribution of fungi to the global economy. Fungal Divers. 2023, 121, 95–137. [Google Scholar] [CrossRef]
- Gafforov, Y.; Rašeta, M.; Rapior, S.; Yarasheva, M.; Wang, X.; Zhou, L.; Wan-Mohtar, W.A.A.Q.I.; Zafar, M.; Lim, Y.W.; Wang, M.; et al. Macrofungi as medicinal resources in Uzbekistan: Biodiversity, ethnomycology, and ethnomedicinal practices. J. Fungi 2023, 9, 922. [Google Scholar] [CrossRef] [PubMed]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; da Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Critical Ecosystem Partnership Fund: Protecting Biodiversity by Supporting People. Available online: https://www.cepf.net/our-work/biodiversity-hotspots/indo-burma (accessed on 11 November 2024).
- Hyde, K.D.; Norphanphoun, C.; Chen, J.; Dissanayake, A.J.; Doilom, M.; Hongsanan, S.; Jayawardena, R.S.; Jeewon, R.; Perera, R.H.; Thongbai, B.; et al. Thailand’s amazing diversity: Up to 96% of fungi in northern Thailand may be novel. Fungal Divers. 2018, 93, 215–239. [Google Scholar] [CrossRef]
- Choudhary, E.; Bithel, N.; Sharma, M. Impact of Climate Change on Ecosystem and its Management Strategies: An Overview. In Climate Change and Water Security: Challenges for Adaptive Water Management; ABS Books Publisher and Exporter: Delhi, India, 2021. [Google Scholar]
- Tischner, Z.; Paldy, A.; Kocsube, S.; Kredics, L.; Dobolyi, C.; Sebok, R.; Kriszt, B.; Szabo, B.; Magyar, D. Survival and growth of microscopic fungi derived from tropical regions under future heat waves in the Pannonian Biogeographical Region. Fungal Biol. 2022, 126, 511–520. [Google Scholar] [CrossRef]
- Singh, S.K. Ex situ Conservation of Fungi: A Review. In Developments in Fungal Biology and Applied Mycology; Satyanarayana, T., Deshmukh, S., Johri, B., Eds.; Springer: Singapore, 2017; pp. 543–562. [Google Scholar] [CrossRef]
- Smith, D.; Ryan, M.J.; Stackebrandt, E. The ex situ conservation of microorganisms: Aiming at a certified quality management. In Encyclopedia of Life Sciences Support; EoLSS: Oxford, UK, 2008; Available online: https://www.eolss.net/sample-chapters/c17/E6-58-13-04.pdf (accessed on 15 September 2023).
- Psurtseva, N.V. Ex situ fungal conservation: The role of culture collections. Mycol. Balc. 2010, 7, 29–35. [Google Scholar]
- Psurtseva, N.V.; Kiyashko, A.A.; Senik, S.V.; Shakhova, N.V.; Belova, N.V. The conservation and study of macromycetes in the Komarov Botanical Institute Basidiomycetes Culture Collection—Their taxonomical diversity and biotechnological prospects. J. Fungi 2023, 9, 1196. [Google Scholar] [CrossRef] [PubMed]
- Vydryakova, G.A.; Van, D.T.; Shoukouhi, P.; Psurtseva, N.V.; Bissett, J. Intergenomic and intragenomic ITS sequence heterogeneity in Neonothopanus nambi (Agaricales) from Vietnam. Mycology 2012, 2, 89–99. [Google Scholar] [CrossRef]
- Psurtseva, N.V.; Zmitrovich, I.V.; Malysheva, V.F. Taxonomy and developmental morphology of Rogersiomyces malaysianus comb. nov. (Cantharellales, Agaricomycetes). Botany 2016, 94, 579–592. [Google Scholar] [CrossRef]
- Senik, S.V.; Psurtseva, N.V.; Shavarda, A.L.; Kotlova, E.R. Role of lipids in the thermal plasticity of basidial fungus Favolaschia manipularis. Can. J. Microbiol. 2019, 65, 870–879. [Google Scholar] [CrossRef] [PubMed]
- Stalpers, J.A. Identification of wood-inhabiting fungi in pure culture. Stud. Mycol. 1978, 16, 1–248. [Google Scholar]
- Clémençon, H. Cytology and plectology of the Hymenomycetes. Bibl. Mycol. 2004, 199, 1–488. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- He, M.Q.; Zhao, R.L.; Hyde, K.D.; Begerow, D.; Kemler, M.; Yurkov, A.; McKenzie, E.H.C.; Raspé, O.; Kakishima, M.; Sánchez-Ramírez, S.; et al. Notes, outline and divergence times of Basidiomycota. Fungal Divers. 2019, 99, 105–367. [Google Scholar] [CrossRef]
- He, M.Q.; Cao, B.; Liu, F.; Boekhout, T.; Denchev, T.T.; Schoutteten, N.; Denchev, C.M.; Kemler, M.; Gorjon, S.P.; Begerow, D.; et al. Phylogenomics, divergence times and notes of orders in Basidiomycota. Fungal Divers. 2024, 126, 127–406. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Dai, D.Q.; Sanchez-Garcia, M.; Goto, B.T.; Magurno, F. Outline of Fungi and Fungus-Like Taxa—2021. Mycosphere 2022, 13, 53–453. [Google Scholar] [CrossRef]
- Kalichman, J.; Kirk, P.M.; Matheny, P.B. A compendium of generic names of agarics and Agaricales. Taxon 2020, 69, 425–447. [Google Scholar] [CrossRef]
- Chen, C.-C.; Chen, C.-Y.; Wu, S.-H. Species diversity, taxonomy and multi-gene phylogeny of phlebioid clade (Phanerochaetaceae, Irpicaceae, Meruliaceae) of Polyporales. Fungal Divers. 2021, 111, 337–442. [Google Scholar] [CrossRef]
- Costa-Rezende, D.-H.; Robledo, G.L.; Drechsler-Santos, E.R.; Glen, M.; Gates, G.; de Madrignac Bonzi, B.R.; Popoff, O.F.; Crespo, E.; Góes-Neto, A. Taxonomy and phylogeny of polypores with ganodermatoid basidiospores (Ganodermataceae). Mycol. Progress 2020, 19, 725–741. [Google Scholar] [CrossRef]
- Justo, A.; Miettinen, O.; Floudas, D.; Ortiz-Santana, B.; Sjökvist, E.; Lindner, D.; Nakasone, K.; Niemelӓ, T.; Larsson, K.-H.; Ryvarden, L.; et al. A revised family-level classification of the Polyporales (Basidiomycota). Fungal Biol. 2017, 121, 798–824. [Google Scholar] [CrossRef]
- Kraisitudomsook, N.; Healy, R.A.; Smith, M.E. Molecular systematics and taxonomic overview of the bird’s nest fungi (Nidulariaceae). Fungal Biol. 2021, 125, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, O.; Vlasák, J.; Larsson, E.; Vlasák, J., Jr.; Seelan, J.S.S.; Hernawati, L.Q.; Larsson, K.-H.; Spirin, V. A revised genus-level classification for Cerrenaceae (Polyporales, Agaricomycetes). Fungal Syst. Evol. 2023, 12, 271–322. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, J.J.S.; Desjardin, D.E.; Jenkinson, T.S.; Margaritescu, S.; Capelari, M.; Moncalvo, J.-M. Taxonomic revision of Marasmius Fr. and Marasmiaceae Roze ex Kühner based on multigene phylogenetics and morphological evidence. Fungal Divers. 2024, 127, 1–54. [Google Scholar] [CrossRef]
- Vizzini, A.; Consiglio, G.; Marchetti, M.; Alvarado, P. Insights into the Tricholomatineae (Agaricales, Agaricomycetes): A new arrangement of Biannulariaceae and Callistosporium, Callistosporiaceae fam. nov., Xerophorus stat. nov., and Pleurocollybia incorporated into Callistosporium. Fungal Divers. 2020, 101, 211–259. [Google Scholar] [CrossRef]
- Vizzini, A.; Consiglio, G.; Marchetti, M.; Borovička, J.; Campo, E.; Cooper, J.; Lebeuf, R.; Ševčíková, H. New data in Porotheleaceae and Cyphellaceae: Epitypification of Prunulus scabripes Murrill, the status of Mycopan Redhead, Moncalvo & Vilgalys and a new combination in Pleurella Horak emend. Mycol. Progress 2022, 21, 44. [Google Scholar] [CrossRef]
- Vizzini, A.; Alvarado, P.; Consiglio, G.; Marchetti, M.; Xu, J. Family matters inside the order Agaricales: Systematic reorganization and classification of incertae sedis clitocyboid, pleurotoid and tricholomatoid taxa based on an updated 6-gene phylogeny. Stud. Mycol. 2024, 107, 67–148. [Google Scholar] [CrossRef]
- Wang, X.W.; Liu, S.L.; Zhou, L.W. An updated taxonomic framework of Hymenochaetales (Agaricomycetes, Basidiomycota). Mycosphere 2023, 14, 452–496. [Google Scholar] [CrossRef]
- Wu, F.; Tohtirjap, A.; Fan, L.-F.; Zhou, L.-W.; Alvarenga, R.L.M.; Gibertoni, T.B.; Dai, Y.-C. Global diversity and updated phylogeny of Auricularia (Auriculariales, Basidiomycota). J. Fungi 2021, 7, 933. [Google Scholar] [CrossRef]
- Muller, G.M.; Bills, G.F.; Foster, M.S. Biodiversity of Fungi, Inventory and Monitoring Methods; Elsevier Academic Press: Watham, UK, 2004; 777p. [Google Scholar]
- Lücking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Ariyawansa, H.A.; Aoki, T.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, D.M.; et al. Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 2020, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chen, Y.Y.; Sun, Y.F.; He, X.L.; Song, C.G.; Si, J.; Liu, D.M.; Gates, G.; Cui, B.K. Systematic classification and phylogenetic relationships of the brown rot fungi within the Polyporales. Fungal Divers. 2023, 118, 1–94. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Xu, J. Fungal DNA barcoding. Genome 2016, 59, 913–932. [Google Scholar] [CrossRef] [PubMed]
- Truong, C.; Mujic, A.B.; Healy, R.; Kuhar, F.; Furci, G.; Torres, D.; Niskanen, T.; Sandoval-Leiva, P.A.; Fernández, N.; Escobar, J.M.; et al. How to know the fungi: Combining field inventories and DNA-barcoding to document fungal diversity. New Phytol. 2017, 214, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Lücking, R.; Truong, B.V.; Huong, D.T.T.; Le, N.H.; Nguyen, Q.D.; Nguyen, V.D.; Raab-Straube, E.v.; Bollendorff, S.; Govers, K.; Di Vincenzo, V. Caveats of fungal barcoding: A case study in Trametes s.lat. (Basidiomycota: Polyporales) in Vietnam reveals multiple issues with mislabelled reference sequences and calls for third-party annotations. Willdenowia 2020, 50, 383–403. [Google Scholar] [CrossRef]
- Corner, E.J.H. A Monograph of Cantharelloid Fungi; Oxford University Press: Oxford, UK, 1966; 255p. [Google Scholar]
- Martínez, A.T.; Ruiz-Dueñas, F.J.; Camarero, S.; Serrano, A.; Linde, D.; Lund, H.; Vind, J.; Tovborg, M.; Herold-Majumdar, O.M.; Hofrichter, M.; et al. Oxidoreductases on their way to industrial biotransformations. Biotechnol. Adv. 2017, 35, 815–831. [Google Scholar] [CrossRef]
- Arregui, L.; Ayala, M.; Gómez-Gil, X.; Gutiérrez-Soto, G.; Hernández-Luna, C.E.; de los Santos, M.H.; Levin, L.; Rojo-Domínguez, A.; Romero-Martínez, D.; Saparrat, M.C.; et al. Laccases: Structure, function, and potential application in water bioremediation. Microb. Cell Fact. 2019, 18, 200. [Google Scholar] [CrossRef] [PubMed]
- Agustin, M.; Morais de Carvalho, D.; Lahtinen, M.; Hilden, K.; Lundell, T.; Mikkonen, K.S. Laccase as a tool in building advanced lignin-based materials. ChemSusChem 2021, 14, 4615–4635. [Google Scholar] [CrossRef]
- Loi, M.; Glazunova, O.; Fedorova, T.; Logrieco, A.F.; Mulè, G. Fungal laccases: The forefront of enzymes for sustainability. J. Fungi 2021, 7, 1048. [Google Scholar] [CrossRef]
- Singh, A.K.; Bilal, M.; Jesionowski, T.; Iqbal, H.M.N. Deployment of oxidoreductases for sustainable biocatalytic degradation of selected endocrine-disrupting chemicals. Sustain. Chem. Pharm. 2023, 31, 100934. [Google Scholar] [CrossRef]
- Moin, S.F.; Alfarra, H.Y.; Omar, M.N. Azo dyes decolourisation by ABTS-oxidases (Laccases) from a fungus from tropical tree. Biosci. Biotech. Res. Asia 2012, 9, 641–646. Available online: https://www.biotech-asia.org/?p=9997 (accessed on 10 October 2024). [CrossRef]
- Mtui, G.Y.S. Lignocellulolytic enzymes from tropical fungi: Types, substrates and applications. Sci. Res. Essays 2012, 7, 1544–1555. [Google Scholar] [CrossRef]
- Fedorova, T.V.; Shakhova, N.V.; Klyain, O.I.; Glazunova, O.A.; Maloshenok, L.G.; Kulikova, N.A.; Psurtseva, N.V.; Koroleva, O.V. Comparative analysis of the ligninolytic potential of basidiomycetes belonging to different taxonomic and ecological groups. Appl. Biochem. Microbiol. 2013, 49, 570–580. [Google Scholar] [CrossRef]
- Glazunova, O.A.; Shakhova, N.V.; Psurtseva, N.V.; Moiseenko, K.V.; Kleimenov, S.Y.; Fedorova, T.V. White-rot basidiomycetes Junghuhnia nitida and Steccherinum bourdotii: Oxidative potential and laccase properties in comparison with Trametes hirsuta and Coriolopsis caperata. PLoS ONE 2018, 13, e0197667. [Google Scholar] [CrossRef] [PubMed]
- Shakhova, N.; Volobuev, S. Revealing new active and biotechnologically perspective producers of oxidative and cellulolytic enzymes among pure cultures of xylotrophic Agaricomycetes from the Southern Non-Chernozem zone of the European part of Russia. Curr. Res. Environ. Appl. Mycol. J. Fungal Biol. 2020, 10, 113–119. [Google Scholar] [CrossRef]
- Tsiklauri, N.; Jobava, M.; Khvedelidze, R.; Kutateladze, L.Y.; Khokhashvili, I.; Kharatishvili, B. Lignocellulosic Ezymes of Basidial Fungi-Isolated from Different Ecological Niches of Georgia. Eurasia Proc. Sci. Technol. Eng. Math. 2021, 12, 113–118. [Google Scholar] [CrossRef]
- Volobuev, S.; Shakhova, N. Towards the Discovery of Active Lignocellulolytic Enzyme Producers: A Screening Study of Xylotrophic Macrofungi from the Central Russian Upland. Iran. J. Sci. Technol. Trans. A Sci. 2022, 46, 91–100. [Google Scholar] [CrossRef]
- Silva, A.M.; Miranda, A.; Fernandes, E.; Santos, S.; Fraga, I.; Santos, D.L.; Dias, A.A.; Bezerra, R.M. Endopolysaccharides from Ganoderma resinaceum, Phlebia rufa, and Trametes versicolor affect differently the proliferation rate of HepG2 cells. Appl. Biochem. Biotechnol. 2013, 169, 1919–1926. [Google Scholar] [CrossRef] [PubMed]
- Timm, T.G.; Costa, T.M.; Alberton, M.D.; Helm, C.V.; Tavares, L.B.B. Mushroom β-glucans: Application and innovation for food industry and immunotherapy. Appl. Microbiol. Biotechnol. 2023, 107, 5035–5049. [Google Scholar] [CrossRef]
- Vetter, J. The Mushroom Glucans: Molecules of High Biological and Medicinal Importance. Foods 2023, 12, 1009. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xu, Y.; Xie, L.; Shu, X.; Zhang, S.; Wang, Y.; Wang, H.; Dong, Q.; Peng, W. The function and application of edible fungal polysaccharides. Adv. Appl. Microbiol. 2024, 127, 45–142. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Shen, R.; Jiao, Z.; Chen, W.; Peng, D.; Wang, L.; Yu, N.; Peng, C.; Cai, B.; Song, H.; et al. Current Advancements in Antitumor Properties and Mechanisms of Medicinal Components in Edible Mushrooms. Nutrients 2022, 14, 2622. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, F.; Chopra, H.; Baig, A.A.; Avula, S.K.; Kumari, S.; Mohanta, T.K.; Saravanan, M.; Mishra, A.K.; Sharma, N.; Mohanta, Y.K. Edible Mushrooms as Novel Myco-Therapeutics: Effects on Lipid Level, Obesity and BMI. J. fungi 2022, 8, 211. [Google Scholar] [CrossRef] [PubMed]
- Venturella, G.; Ferraro, V.; Cirlincione, F.; Gargano, M.L. Medicinal Mushrooms: Bioactive Compounds, Use, and Clinical Trials. IJMS 2021, 22, 634. [Google Scholar] [CrossRef]
- Téllez-Téllez, M.; Diaz-Godinez, G. Mushrooms and Fungi and Their Biological Compounds with Antidiabetic Activity: A Review. Int. J. Med. Mushrooms 2024, 26, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Wasser, S.P. Medicinal mushrooms as a source of antitumor and immunomodulating polysaccharides. Appl. Microbiol. Biotechnol. 2002, 60, 258–274. [Google Scholar]
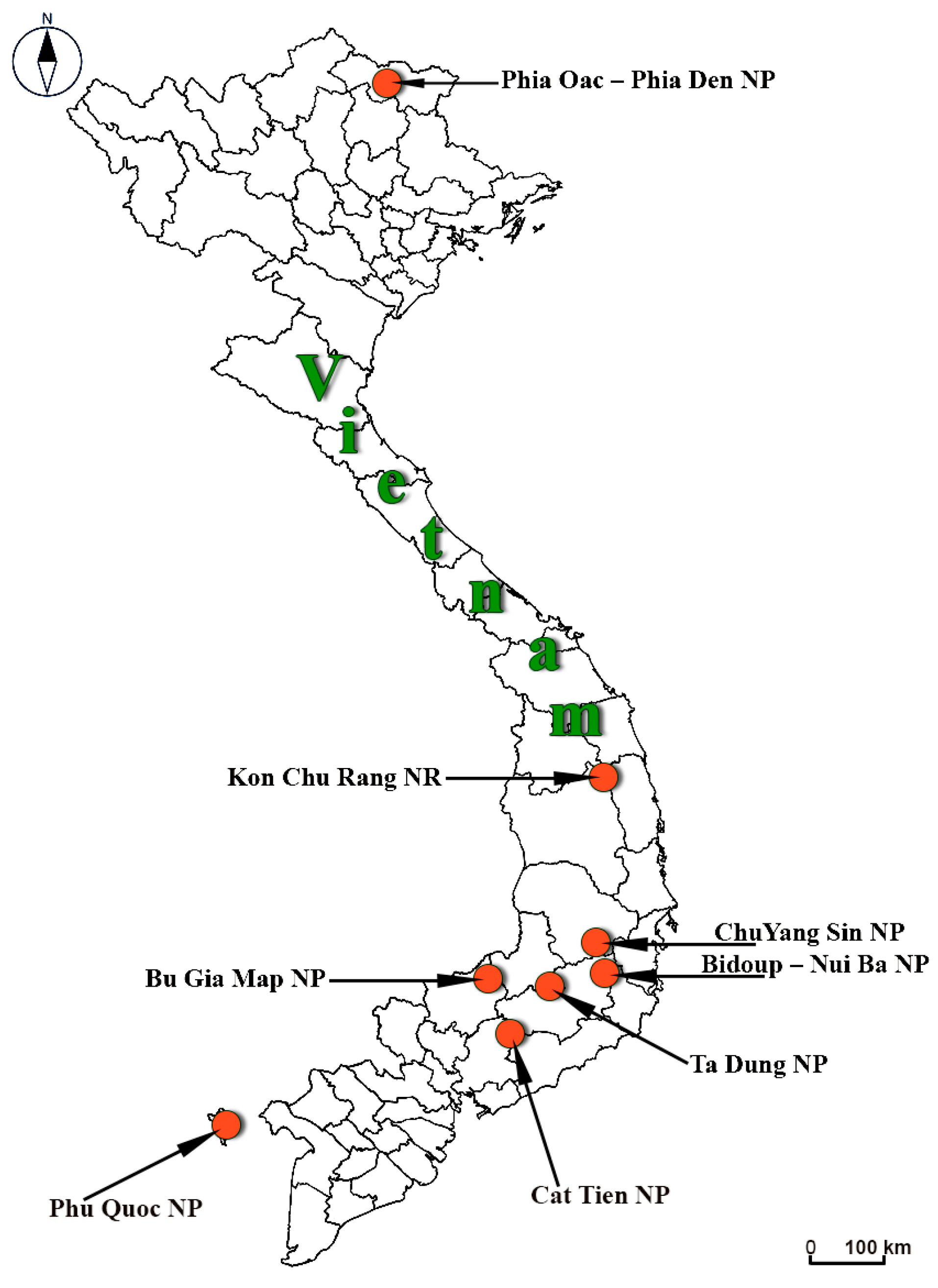

| Vietnam Protected Territories | Collection Dates |
|---|---|
| NP Phu Quoc | 18–24 October 2012 |
| NP Cat Tien | 28–31 October 2012; 5–12 May 2014; 6–20 February 2021; 20–21 November 2022 |
| NP Bidoup—Nui Ba | 2–8 November 2012; 23–29 May 2014; 29 October–6 November 2019; 3–11 July 2023; 19–30 October 2023 |
| NP Bu Gia Map | 13–18 May 2014; 25 October–2 November 2018; 21–24 October 2019 |
| NR Kon Chu Rang | 25 February–3 March 2021; 23–28 October 2022; 14–20 July 2023; 4–6 November 2023 |
| NP Chu Yang Sin | 16–27 May 2019 |
| NP Ta Dung | 3–19 October 2022 |
| NP Phia Oac—Phia Den | 7–17 November 2018; 4–13 June 2019; 4–12 October 2019 |
| Vietnam Protected Territories | Number of Specimens | Number of LE-BIN Strains | |
|---|---|---|---|
| Collected | Isolates | ||
| NP Phu Quoc | 88 | 50 | 41 |
| NP Cat Tien | 189 | 127 | 92 |
| NP Bidoup—Nui Ba | 445 | 279 | 184 |
| NP Bu Gia Map | 224 | 137 | 127 |
| NR Kon Chu Rang | 191 | 103 | 80 |
| NP ChuYang Sin | 58 | 33 | 14 |
| NP Ta Dung | 165 | 115 | 104 |
| NP Phia Oac—Phia Den | 247 | 160 | 143 |
| Total: | 1519 | 1004 | 785 |
| Class | Order | Family | Genus |
|---|---|---|---|
| ASCOMYCOTA | |||
| Leotiomycetes | Helotiales | Chlorociboriaceae | Chlorociboria 1 (1) |
| Chlorospleniaceae | Chlorosplenium 1 (1) | ||
| Gelatinodiscaceae | Ascocoryne 1 (1) | ||
| Helotiaceae | Dicephalospora ≥ 1 (3), Hymenoscyphus 1 (1), Tatraea ≥ 1 (3) | ||
| Orbiliomycetes | Orbiliales | Orbiliaceae | Orbilia 1 (1) |
| Pezizomycetes | Pezizales | Sarcoscyphaceae | Cookeina 2 (2) |
| Sarcosomataceae | Plectania ≥ 1 (2) | ||
| Sordariomycetes | Hypocreales | Cordycipitaceae | Cordyceps 1 (1), Isaria 1 (1) |
| Xylariales | Xylariaceae | Durotheca 1 (1), Nemania 1 (1), Xylaria ≥ 6 (14) | |
| BASIDIOMYCOTA | |||
| Agaricomycetes | Agaricales | Agaricaceae | Agaricus ≥ 1 (5), Leucoagaricus 1 (1), Leucocoprinus 1 (1), Micropsalliota ≥ 1 (2) |
| Bolbitiaceae | Conocybe 1 (1) | ||
| Callistosporiaceae | Callistosporium 1 (1) | ||
| Campanellaceae | Campanella ≥ 1 (5), Tetrapyrgos ≥ 1 (4) | ||
| Clitocybaceae | Clitocybe 1 (1), Collybia 1 (1) | ||
| Crepidotaceae | Crepidotus 1 (1) | ||
| Cystostereaceae | Crustomyces 1 (1) | ||
| Entolomataceae | Clitopilus 2 (2) | ||
| Fayodiaceae | Conchomyces ≥ 1 (2) | ||
| Hymenogastraceae | Gymnopilus ≥ 2 (7), Psilocybe ≥ 1 (4) | ||
| Lyophyllaceae | Tephrocybe 1 (1), Termitomyces (1) | ||
| Marasmiaceae | Chaetocalathus 1 (1), Crinipellis ≥ 1 (3), Marasmius ≥ 2 (25), Paramarasmius 2 (2) | ||
| Mycenaceae | Cruentomycena 1 (1), Cynema 1 (3), Favolaschia ≥ 4 (16), Filoboletus ≥ 1 (2), Mycena ≥ 4 (25), Panellus ≥ 3 (19), Resinomycena 1 (1) | ||
| Nidulariaceae | Cyathus 1 (1), Nidula 1 (1), Nidularia 1 (1) | ||
| Omphalotaceae | Anthracophyllum ≥ 1 (3), Collybiopsis ≥ 3 (24), Gymnopus ≥ 2 (10), Lentinula 3 (5), “Marasmiellus” 2 (4), Mycetinis ≥ 1 (8), Neonothopanus 1 (10), Omphalotus ≥ 1 (2), Pseudomarasmius 1 (2) | ||
| Phyllotopsidaceae | Phyllotopsis 1 (1) | ||
| Physalacriaceae | Cyptotrama ≥ 2 (6), Flammulina 1 (1), Gloeocephala 1 (1), Hymenopellis ≥ 1 (4), Mucidula ≥ 1 (7), Oudemansiella ≥ 1 (2), Xerula ≥ 1 (7) | ||
| Pleurotaceae | Hohenbuehelia ≥ 1 (2), Pleurotus ≥ 4 (12) | ||
| Porotheleaceae | Clitocybula ≥ 1 (2), Gerronema ≥ 1 (3), Hydropus ≥ 1 (8), Pseudohydropus 1 (1), Trogia ≥ 1 (4), Vanromburghia 1 (1) | ||
| Psathyrellaceae | Candolleomyces ≥ 1 (5), Coprinellus ≥ 1 (2), Coprinopsis ≥ 1 (3), Psathyrella ≥ 1 (4) | ||
| Pterulaceae | Pterulicium 1 (1), | ||
| Radulomycetaceae | Radulomyces ≥ 1 (2) | ||
| Resupinataceae | Resupinatus 1 (1) | ||
| Schizophyllaceae | Schizophyllum 1 (1) | ||
| Strophariaceae | Deconica 1 (1), Hypholoma ≥ 2 (12), Kuehneromyces 1 (1), Melanotus ≥ 1 (4), Pholiota ≥ 1 (5) | ||
| Xeromphalinaceae | Heimiomyces 1 (1), Xeromphalina ≥ 1 (4) | ||
| Amylocorticiales | Amylocorticiaceae | Irpicodon 1 (1) | |
| Atheliales | Atheliaceae | Athelia 1 (1) | |
| Auriculariales | Auriculariaceae | Auricularia ≥ 1 (13), Eichleriella 1 (1), Elmerina 2 (2), Protodaedalea 1 (1) | |
| Cantharellales | Hydnaceae | Rogersiomyces 1 (1) | |
| Gloeophyllales | Gloeophyllaceae | Hispidaedalea 1 (1) | |
| Hymenochaetales | Hirschioporaceae | Pallidohirschioporus ≥ 1 (3) | |
| Hymenochaetaceae | Erythromyces 1 (1), Fulvifomes ≥ 1 (2), Fuscoporia ≥ 2 (4), Hymenochaete ≥ 5 (8), Phellinus ≥ 1 (6), Pyrrhoderma 2 (2) | ||
| Hyphodontiaceae | Hyphodontia 1 (1) | ||
| Rigidoporaceae | Rigidoporus ≥ 1 (2) | ||
| unknown | Trichaptum s. l. (4) | ||
| Phallales | Phallaceae | Phallus 1 (1) | |
| Polyporales | Cerrenaceae | Cerrena ≥ 1 (5) | |
| Fomitopsidaceae | Antrodia 1 (1), Fomitopsis ≥ 1 (4), Rhodofomitopsis 1 (2) | ||
| Ganodermataceae | Ganoderma ≥ 6 (23), Sanguinoderma 1(1) | ||
| Hypochniciaceae | Hypochnicium 1 (1) | ||
| Incrustoporiaceae | Incrustoporia 1 (1), Skeletocutis 1 (1) | ||
| Irpicaceae | Efibula 1 (1), Flavodon 1 (5), Gloeoporus 1 (1), Irpex 1 (1) | ||
| Laetiporaceae | Laetiporus 1 (1) | ||
| Meripilaceae | Physisporinus ≥ 2 (6) | ||
| Meruliaceae | Climacodon 1 (1), Crustodontia (1), Merulius 1 (1), Mycoacia ≥ 1 (2), Mycoaciella 1 (1), Phlebia 1 (3), Scopuloides 1 (1) | ||
| Panaceae | Cymatoderma 2 (4), Panus ≥ 2 (8) | ||
| Phaeolaceae | Phaeolus 1 (1) | ||
| Phanerochaetaceae | Bjerkandera 1 (3), Oxychaete 1 (1), Phlebiopsis ≥ 1 (2), Terana 1(1) | ||
| Polyporaceae | Abundisporus 1 (1), Cellulariella 1 (5), Coriolopsis 1 (2), Cubamyces 3 (4), Earliella 1 (2), Favolus ≥ 4 (9), Fomes 1 (1), Fomitella 1 (1), Funalia 2 (2), Grammothele 1 (2), Hexagonia ≥ 1 (3), Lentinus ≥ 6 (19), Lopharia ≥ 1 (2), Megasporoporiella 1 (1), Microporellus 1 (1), Microporus ≥ 2 (25), Neofomitella 2 (3), Perenniporiopsis 1 (1), Picipes ≥ 1 (3), Podofomes 1 (1), Polyporus ≥ 1 (5), Porogramme 1 (1), Theleporus 1 (1), Tinctoporellus 1 (2), Trametes ≥ 11 (36), Truncospora 1 (2) | ||
| Steccherinaceae | Junghuhnia 1 (1), Nigroporus 1 (7), Steccherinum ≥ 3 (6) | ||
| Russulales | Auriscalpiaceae | Artomyces ≥ 1 (2), Auriscalpium 1 (1) | |
| Bondarzewiaceae | Bondarzewia 1 (1) | ||
| Hericiaceae | Dentipellis ≥ 1 (2) | ||
| Peniophoraceae | Duportella 1 (1), Vararia ≥ 1 (2) | ||
| Stereaceae | Stereum ≥ 3 (23), Xylobolus ≥ 3 (10), Megalocystidium 1 (1) | ||
| Wrightoporiaceae | Wrightoporia 1 (1) | ||
| Dacrymycetes | Dacrymycetales | Dacrymycetaceae | Dacrymyces 1 (2) |
| Tremellomycetes | Tremellales | Phaeotremellaceae | Phaeotremella 1 (1) |
| Species | Strain Number |
|---|---|
| Neonothopanus nambi | 10 |
| Microporus xanthopus | 9 |
| Favolaschia manipularis | 8 |
| Trametes vernicipes | 8 |
| Nigroporus vinosus | 7 |
| Stereum ostrea | 7 |
| Auricularia delicata | 6 |
| Microporus affinis | 6 |
| Pleurotus pulmonarius s. l. | 6 |
| Trametes versicolor | 6 |
| Candolleomyces candolleanus s. l. | 5 |
| Cellulariella acuta | 5 |
| Flavodon flavus | 5 |
| Lentinus arcularius s. l. | 5 |
| Mucidula mucida | 5 |
| Panellus luxfilamentus | 5 |
| Xylobolus princeps | 5 |
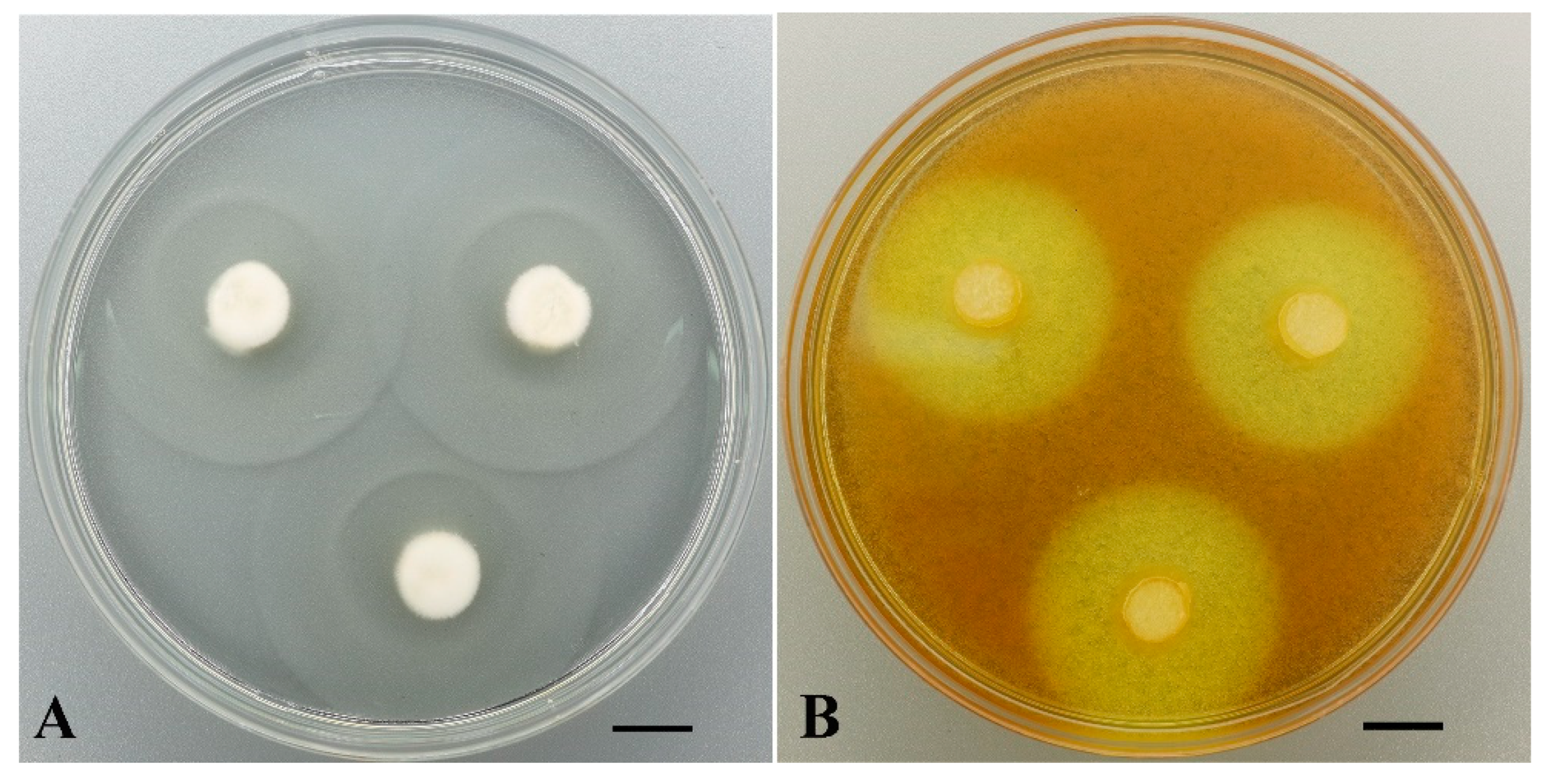
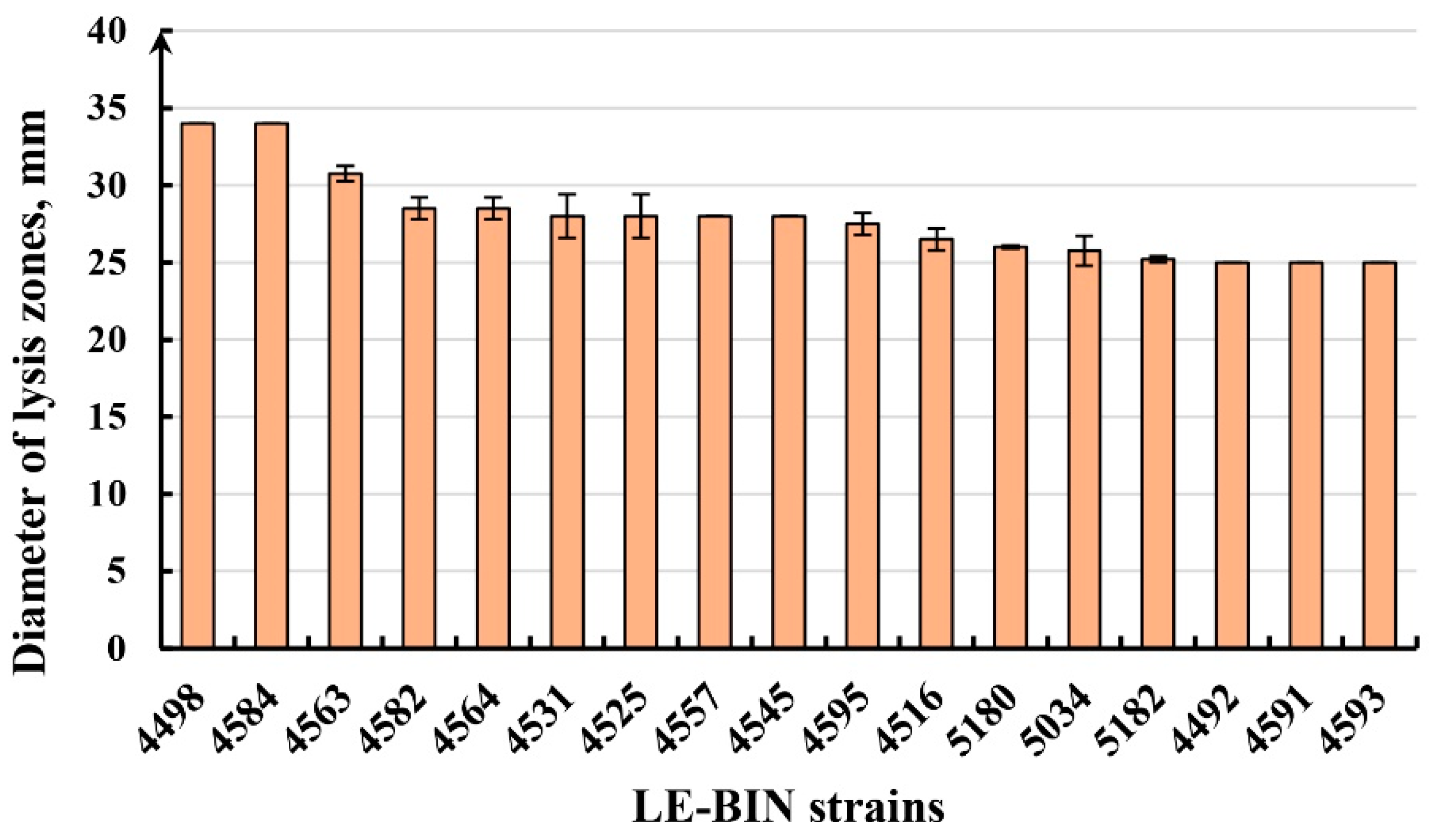
| Taxon, Strain Num. | Growth Rate (mm/d) | Oxidases (SG) | Oxidases (GU) | Proteinases (GA, mm) | Cellulases (CA, mm) |
|---|---|---|---|---|---|
| Phlebia acerina LE-BIN 4498 | 8.63 ± 0.30 | ++ * | ++ | — | 34 ± 0 |
| Phlebia acerina LE-BIN 4584 | 8.6 ± 0.17 | +++ | ++ | — | 34 ± 0 |
| Xylobolus spectabilis LE-BIN 4563 | 7.88 ± 1.01 | + | +++ | — | 30.75 ± 0.5 |
| Fomitopsis sp. LE-BIN 5106 | 4.33 ± 0.57 | − | − | 31.9 ± 0.7 | 29.3 ± 0.4 |
| Fulvifomes fastuosus LE-BIN 4531 | 7.21 ± 0.16 | ++ | +++ | — | 28.00 ± 1.41 |
| Trametes versicolor LE-BIN 4557 | 6.18 ± 0.38 | +++ | ++ | — | 28 ± 0 |
| Cellulariella acuta LE-BIN 4545 | 7.17 ± 0.56 | +++ | +++ | — | 28 ± 0 |
| Trametes sanguinea LE-BIN 4595 | 6.14 ± 0.09 | +++ | + | — | 27.5 ± 0.71 |
| Trametes versicolor LE-BIN 4516 | 8.6 ± 0.21 | +++ | ++ | — | 26.5 ± 0.71 |
| Trametes meyenii LE-BIN 5182 | 10.55 ± 0.42 | — | — | 25 ± 0.1 | 25.2 ± 0.2 |
| Xylaria grammica LE-BIN 5102 | 2.42 ± 0.26 | + | +++ | 45 ± 0.7 | 24.5 ± 0.3 |
| Truncospora tephropora LE-BIN 5184 | 6.24 ± 0.05 | — | — | Full lysis | 23.4 ± 0.3 |
| Candolleomyces aff. candolleanus LE-BIN 5112 | 6.77 ± 0.08 | ++ | − | 44.2 ± 0.6 | 23 ± 0.5 |
| Lentinus squarrosulus LE-BIN 5189 | 7.69 ± 0.09 | − | +/− | 27.2 ± 0.2 | 20.5 ± 0.1 |
| Nemania sp. LE-BIN 5155 | 4.64 ± 0.04 | +/− | − | 26 ± 1.1 | 18.7 ± 0.2 |
| Auricularia delicata LE-BIN 5164 | 3.25 ± 0.07 | ++ | ++ | 25.4 ± 1.4 | 24.7 ± 0.4 |
| Tinctoporellus epimiltinus LE-BIN 5180 | 10.82 ± 0.12 | — | — | 23.7 ± 0.3 | 26 ± 0.1 |
| Theleporus membranaceus LE-BIN 5170 | 8.16 ± 0.07 | +++ | +++ | 18.4 ± 0.6 | 22.9 ± 0.6 |
| Coriolopsis brunneoleuca LE-BIN 5131 | 3.97 ± 0.55 | ++ | +++ | 16.6 ± 0.9 | 27.8 ± 0.6 |
| Cerrena zonata LE-BIN 4492 | 8.6 ± 0.34 | ++ | +++ | — | 25 ± 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Psurtseva, N.V.; Kiyashko, A.A.; Senik, S.V.; Pham, T.H.G. Ex Situ Conservation, DNA Barcoding and Enzymatic Potential Evaluation of Macrofungi (Basidiomycota, Ascomycota) from Vietnam. J. Fungi 2025, 11, 34. https://doi.org/10.3390/jof11010034
Psurtseva NV, Kiyashko AA, Senik SV, Pham THG. Ex Situ Conservation, DNA Barcoding and Enzymatic Potential Evaluation of Macrofungi (Basidiomycota, Ascomycota) from Vietnam. Journal of Fungi. 2025; 11(1):34. https://doi.org/10.3390/jof11010034
Chicago/Turabian StylePsurtseva, Nadezhda V., Anna A. Kiyashko, Svetlana V. Senik, and Thi Ha Giang Pham. 2025. "Ex Situ Conservation, DNA Barcoding and Enzymatic Potential Evaluation of Macrofungi (Basidiomycota, Ascomycota) from Vietnam" Journal of Fungi 11, no. 1: 34. https://doi.org/10.3390/jof11010034
APA StylePsurtseva, N. V., Kiyashko, A. A., Senik, S. V., & Pham, T. H. G. (2025). Ex Situ Conservation, DNA Barcoding and Enzymatic Potential Evaluation of Macrofungi (Basidiomycota, Ascomycota) from Vietnam. Journal of Fungi, 11(1), 34. https://doi.org/10.3390/jof11010034






