Abstract
Pyrenophora teres f. teres (Ptt) is a severe pathogen to spring barley in Northern Europe. Ptt with relevant mutations in fungicide target proteins, sterol 14α-demethylase (CYP51A), cytochrome b (Cyt b), and succinate dehydrogenase (SDH) would put efficient disease control at risk. In the growing seasons of 2021 and 2022, 193 Ptt isolates from Estonia were analysed. In this study, mutation detection and in vitro fungicide sensitivity assays of single-spore isolates were carried out. Reduced sensitivity phenotype to mefentrifluconazole was evident in Ptt isolates with a F489L mutation in CYP51A or with 129 bp insert in the Cyp51A gene-promoter region. However, sensitivity to a prothioconazole-desthio remained high regardless of these molecular changes. The Ptt population was mostly sensitive to bixafen, fluxapyroxad, pyraclostrobin, and azoxystrobin. The sensitivity of fluxapyroxad and bixafen has been affected by two mutations, C-S135R and D-H134R, found in SDH subunits. The F129L mutation in Cyt b influenced azoxystrobin but not pyraclostrobin sensitivity. In total, 30 isolates from five fields had relevant mutations in three target protein genes simultaneously. Most of these isolates had a reduced sensitivity phenotype to mefentrifluconazole, fluxapyroxad, and azoxystrobin, while sensitivity to other tested fungicides remained high. Furthermore, possible sexual reproduction may enhance the pathogen’s fitness and help it adapt to fungicides.
Keywords:
net blotch; fungicide sensitivity; CYP51; Cyp51A promoter; SDH subunits; Cyt b; mating type 1. Introduction
Barley is one of the oldest cultivated crops, which is resilient in different growing conditions and under stress factors. It is a valuable crop for multiple uses, e.g., alcohol production, feed for livestock, and human consumption [1]. Spring barley is the second most cultivated crop in Estonia because it fits well in the crop rotation and has satisfactory yields under regional climatic and agricultural conditions with low fertilizer and pesticide input [2].
Net blotch is a common barley disease caused by two forms of Pyrenophora teres: net form is caused by P. teres f. teres (Ptt) and spot form is caused by P. teres f. maculata (Ptm) [3]. The net form net blotch is the most prevalent foliar disease, causing substantial loss of spring barley in Estonia, as well as in other Northern European countries, where the climatic conditions are favourable for the development of the disease and pathogen spread, susceptible hosts are grown, and primary inoculum is not fully diminished [4,5,6,7,8].
The life cycle of Ptt involves both asexual and sexual stages. The asexual lifecycle includes rapid reproduction and conidial spread by wind and rain to neighbouring plants and fields, as well as overwintering of the pathogen as mycelium in seed or in crop debris. For sexual reproduction, the opposite mating types (MAT-1 and MAT-2) of the pathogen need to meet on the host [9]. The sexual fruiting bodies, pseudothecia, are formed on the barley straw after harvest in autumn, enabling pathogen survival through winter [10]. Sexual recombination increases genetic diversity and virulence variation of the pathogen, probably improving the adaptation and fitness of the pathogen [11]. Pathogen populations with mixed reproduction system are expected to quickly adapt to fungicides with a single mode of action [11]. It is probable that sexual reproduction occurs at a high frequency within the worldwide population of P. teres, but the dynamics are influenced by local agronomic and environmental conditions [12].
The primary inoculum sources early in the season are the mature ascospores discharged from the asci in pseudothecia, as well as seed-borne mycelium and conidia released from the barley straws dispersed by wind to nearby fields [10,12,13]. Sowing healthy seeds and practicing longer crop rotation and soil tillage are the main modes of destruction of primary inoculum [13]. Spring barley cultivars bred in Europe are variable in net blotch resistance, but resistance breeding is more relevant than ever [14]. However, reduced and no-tillage cultivation systems are becoming more popular among farmers around the world, making disease management more demanding [15]. Optimizing fertilizer and fungicide applications would increase the crop quality and yield of barley. In Estonia, seed treatment and two foliar applications of fungicide per growing season tend to give the best outcome [7,8]. Optimal disease control with fungicides can give a substantial barley yield increase under high disease pressure [5,6]. However, under continuous selection pressure with fungicide application, a fungal population can evolve toward reduced sensitivity, and the proportion of insensitive phenotypes may reach a level where satisfactory disease control is no longer achieved [16]. It is also evident that selection for fungicide insensitivity in the pathogen population is increased with higher fungicide dosage due to survival and reproduction of the fittest genotypes [16].
Quinone outside inhibitor (QoI), succinate dehydrogenase inhibitor (SDHI), and demethylation inhibitor (DMI) fungicides are widely used in cereal plant protection. The selection of fungicides allowed in Estonia is limited, and the four most popular fungicides (DMIs prothioconazole and tebuconazole, amine fungicide spiroxamine, and QoI pyraclostrobin) constitute over 55% of the total amount of fungicides applied in agriculture [17]. Due to selection pressure, the loss of QoI, SDHI, and DMI sensitivity has spread in Ptt populations in several European countries, Canada, and Australia [18,19,20,21,22,23].
Three amino acid substitutions (e.g., G143A, G137R, F129L) have been detected in the cytochrome b (Cyt b) in plant pathogens with insensitivity to QoI-s. Mutations F129L and G137R have a low to moderate effect on QoI insensitivity in the pathogen´s population, and the field performance of QoI fungicides still remains good in Europe [24]. However, mutation G143A in Cyt b of Pyrenophora tritici-repentis, Blumeria graminis f. sp. tritici, and Zymoseptoria tritici has a strong effect on sensitivity reduction [25,26,27]. Until recently, only the F129L mutation in Cyt b had been reported for P. teres [20,22,23]. In several phytopathogens (e.g., Ptt, Alternaria spp., Puccinia spp.), a nucleotide mutation at a position near the exon/intron junction, such as that which leads to the G143A substitution, would affect the splicing process, and non-functional Cyt b proteins would be produced. It would reduce the fitness and survival of the pathogen markedly [28,29]. There was a rare finding of a Ptt field isolate named ISO-2210 from Denmark in 2019, which was highly insensitive to QoI fungicides due to G143A mutation in Cyt b, which was assumed to be a chimer of P. teres and P. tritici-repentis [27].
Reduced sensitivity to SDHI fungicides in Ptt populations is related to target-site mutations in succinate dehydrogenase (SDH) subunits, the most significant being C-G79R, C-H134R, C-S135R, D-D124E, and D-H134R [21]. The first findings of SDHI insensitive Ptt isolates, which had B-H277Y substitution, were identified in Germany in 2012, and several mutations in SDH subunits have occurred since then [21]. In Eastern Europe, Ptt populations have still high sensitivity to SDHIs, while since 2017, the sensitivity has been reduced in Western Europe [30].
Three primary mechanisms of DMI insensitivity are (i) mutations in the target-encoding sterol 14α-demethylase (Cyp51) gene, (ii) over-expression of the target Cyp51 gene, and (iii) increased efflux caused by the over-expression of genes encoding membrane transporters. Insensitivity levels are often determined by combinations of these mechanisms [31]. The ascomycete species genome has undergone several Cyp51 gene duplication and divergence events; for example, Pyrenophora spp. have two paralogs (Cyp51A and Cyp51B) and Aspergillus spp. and Fusarium spp. have three paralogs (Cyp51A, Cyp51B, and Cyp51C) [19,32]. Species with multiple copies of the Cyp51 gene are intrinsically less sensitive to some DMI fungicides, and mutations conferring acquired insensitivity to effective azoles are usually restricted to one paralogue, most often Cyp51A [33]. Mair et al. [19] showed that Ptt DMI insensitivity was correlated with two genetic modifications, an amino acid change F489L in the CYP51A protein, and overexpression of Cyp51A and Cyp51B genes. Ptm isolates carrying both a 134 bp insertion element in the Cyp51 promoter and an F489L mutation in CYP51A displayed the highly DMI-insensitive phenotype [34].
In Estonia, fungicide sensitivity in the Ptt population is unknown, and testing for fungicide efficacy in field trials is scarce. Being aware of the pathogen sensitivity to fungicides is essential to execute effective and knowledge-based net blotch management strategies and to prolong the effective lifetime of fungicides. Hopefully, it also increases crop yields, additionally taking into account cultivar selection, tillage, optimal fertilization, water supply, climatic conditions, etc.
In this first comprehensive study of the Estonian Ptt population, we focused on the current status of fungicide sensitivity and its association with relevant SNPs in target protein-coding gene sequences linked to fungicide insensitivity [19,21,34,35,36]. Therefore, the objectives of this research were to assess the Estonian Ptt population for (i) fungicide sensitivity level in vitro microtiter plate assays and subsequently associate insensitivity with (ii) Cyt b mutations, (iii) SDH subunit mutations, (iv) molecular changes in the Cyp51A gene and promoter region; and (v) potential sexual recombination by the analysing mating type prevalence rate.
2. Materials and Methods
2.1. Field Sampling and Fungal Isolates
Leaves with net form net blotch symptoms were collected from commercial spring barley fields in the years 2021 and 2022 across Estonia. The crop had been treated once or twice with foliar fungicides (DMIs, DMI, and SDHI mixture or DMI and QoI mixture). The leaves were dried at room temperature (21 °C) and were then surface sterilized with a 70% ethanol solution for 2 min and rinsed with distilled water twice for 2 min. Leaf sections of 10 cm were then placed on 1% water agar Petri dishes (diam. 14 cm) and incubated for 3–5 days (at room temperature in dark). When conidiophores were visible, single conidia were picked with a sterile needle and placed on potato dextrose agar (PDA; VWR International, Leuven, Belgium) plates amended with 0.1 mg/mL streptomycin (AppliChem GmbH, Darmstadt, Germany). The pure culture isolates were incubated at 22 °C in a 12 h white (TL5 54W/840 HO light tubes by Philips, Amsterdam, the Netherlands)/17 °C 12 h dark photoperiod for 1–2 weeks depending on the growth speed of the isolates. A total of 60 and 210 Ptt isolates were acquired in years 2021 and 2022, respectively.
2.2. DNA Extraction and Species Determination
For DNA extraction, the mycelia of each isolate were collected with the sterile inoculation loop from the pure culture isolate plates, lyophilized with a FreeZone 2.5 Liter-50C Benchtop Freeze Dryer (Labconco, Kansas City, MO, USA), and homogenized with a TissueLyzer (Qiagen, Düsseldorf, Germany) in 2 mL tubes with Retsch balls. For further isolation, a QIAcube® HT DNA extractor (Qiagen, Düsseldorf, Germany) and a DNeasy mericon 96 QIAcube HT Kit were used according to the manufacturer’s instructions [37]. After DNA isolation, the Ptt isolates were confirmed with PCR and Sanger sequencing using ITS1 and ITS4 primers according to White et al. [38] and a Ptt form-specific assay with primers according to Knight et al. [35] (Table 1). PCR amplification was performed with a total volume of 20 µL mix consisting of 2 µL of 10× DreamTaq Green PCR buffer (Thermo Fisher Scientific, Waltham, MA, USA), 2 µL 10× GC-rich Enhancer (Solis BioDyne, Tartu, Estonia), 100 µM of each dNTP, 0.4 µM of specific forward and reverse primers (Table 1), 1 unit DreamTaq Hot Start DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, United States), 10 ng of genomic DNA, and the remaining amount of MilliQ water (Merck KGaA, Darmstadt, Germany). The amplification parameters were as follows: an initial denaturation for 3 min at 95 °C, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C (ITS) or 61 °C (Ptt form-specific) for 30 s, and extension at 72 °C for 30 s, with final extension at 72 °C for 5 min. All the amplifications were performed in a Mastercycler nexus (Eppendorf, Hamburg, Germany). The PCR products of the ITS region were sequenced using an Applied Biosystems 3730 DNA Analyzer (Thermo Fisher Scientific, Waltham, MA, USA) at the Institute of Genomics Core Facility, University of Tartu (Estonia). The sequences obtained were analysed using blastn search tools (https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed 21 December 2023)) available at NCBI. The amplicons (213 bp) for the Ptt form-specific control were separated by 1.5% agarose gel electrophoresis in 1xTAE (Tris-acetate-EDTA) buffer (pH 8.0).

Table 1.
Oligonucleotides used for amplification and sequencing to confirm isolate species and detect point mutations in fungicide target genes, CYP51A promoter inserts, and mating type.
2.3. In Vitro Fungicide Sensitivity Assay
A total of 53 Ptt isolates from 2021 and 140 isolates from 2022 were selected randomly to cover a wide selection of commercial fields (12 fields in 2021, 23 fields in 2022) to perform fungicide sensitivity assays and further molecular analysis (Figure 1). The selected isolates were grown on POA medium (20 g of grinded green peanut leaves, 6 g whole-grain oat flour, 6 g agar, filled up to 400 mL of water) amended with 0.1 mg/mL streptomycin on Petri plates (diam. 4.5 cm). The isolates were exposed to periods of white light (12 h at 22 °C) and darkness (12 h at 17 °C) for two weeks. For further preservation, the hyphae and conidia were suspended in 20% sterile glycerol and stored at ultra-low temperature—82 °C. For the sensitivity assay, the fungal mycelium was dislodged from the Petri dishes with a sterile inoculation loop and suspended in 5 mL of 2xYBA medium (8 g Bacto Yeast Extract, 8 g Bacto Tryptone, 16 g anhydrous sodium acetate, 400 mL of distilled water). The Petri dishes were washed with about 2–3 mL of 2xYBA medium and pipetted into the same tube as the mycelium. The tubes were vortexed on a Multi Reax (Heidolph Instruments, Schwabach, Germany) at 2000 rpm for 15 min. The vortexed samples were filtered through pluriStrainer 200 µm mesh (pluriSelect Life Science, Leipzig, Germany) into a sterile 15 mL Falcon tube. Because the spores did not emerge sufficiently, pieces of hyphae were then counted and adjusted to a density of 4 × 103 pcs/mL. Each fungicide technical active ingredient (Dr. Ehrenstorfer GmbH, Augsburg, Germany) was dissolved in 80% ethanol, and the desired dilutions were prepared in sterile deionised water. Approximately 50 µL of fungicide solution and 50 µL of hyphae suspension were mixed in 96-well microtiter plates. The final concentrations of mefentrifluconazole and bixafen were 0, 0.01, 0.04, 0.12, 0.37, 1.11, 3.33, and 10 mg/L; of azoxystrobin, pyraclostrobin, and prothioconazole-desthio were 0, 0.008, 0.025, 0.07, 0.22, 0.67, 2, and 6 mg/L; and of fluxapyroxad were 0, 0.003, 0.01, 0.037, 0.11, 0.33, 1, and 3 mg/L on the microtiter plates. For each isolate, three replicates were used. A negative control with each fungicide solution and 2xYBA medium without hyphae suspension was added to the assay. The microtiter plates were wrapped in aluminium foil and incubated at 20 °C in darkness. After 5 days, the optical density (OD405) was measured with a Tecan Sunrise™ absorbance reader (Tecan, Männedorf, Switzerland). The values were corrected by comparison with the negative controls, and the EC50 was determined by non-linear regression (curve-fit) with a variable slope using GraphPad Prism 9.4.1 (GraphPad Software, San Diego, CA, United States). The Ptt isolates were grouped according to relevant amino acid substitutions in fungicide target molecules and in vitro fungicide sensitivity. According to mefentrifluconazole and prothioconazole-desthio sensitivity results, the Ptt isolates were grouped as follows: EC50 < 1 mg/L as sensitive, EC50 = 1–5 mg/L as reduced sensitivity, EC50 > 5 mg/L as insensitive. The Ptt isolates were grouped based on fluxapyroxad, and the bixafen sensitivity results were as follows: EC50 < 0.1 mg/L as sensitive, EC50 = 0.1–1 mg/L as reduced sensitivity, EC50 > 1 mg/L as insensitive. The Ptt isolates were grouped based on pyraclostrobin, and the azoxystrobin sensitivity results were as follows: EC50 < 0.01 mg/L as sensitive, EC50 = 0.01–1 mg/L as reduced sensitivity, EC50 > 1 mg/L as insensitive.
Figure 1.
Number of Pyrenophora teres f. teres isolates analysed in 2021 (a) and 2022 (b) collected from different Estonian counties (modified maps of administrative units from Estonian Land Board [40]).
2.4. Molecular Analysis of Fungicide Target Site Protein Genes, Cyp51A Promoter Region, and Mating Type
To assess different target site mutations and insertions in the Cyp51A promoter region, PCR amplifications were performed for the Cyp51A, sdhB, sdhC, sdhD, and cyt b gene sequences and the Cyp51A promoter with the selected primers indicated in Table 1. Based on the genome assembly of the Ptt isolate W1-1 (GenBank accession GCA_900232045.3), new primers were designed for detecting G137R mutation in Cyt b using Primer3 software v. 0.4.0 [41,42]. The sequence similarity and specificity were verified using the Primer-BLAST search tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome (accessed 1 June 2023)) of the NCBI GenBank Standard databases. PCR optimisation and specificity assessment was then performed using target (Ptt) and non-target (Z. tritici, Ramularia collo-cygni) isolate DNA. PCR amplification was performed with a total volume of 20 µL mix consisting of 2 µL of 10× DreamTaq Green PCR buffer (Thermo Fisher Scientific, Waltham, MA, USA), 2 µL 10× GC-rich Enhancer (Solis BioDyne, Tartu, Estonia), 100 µM of each dNTP, 0.4 µM of specific forward and reverse primers (Table 1), 1 unit DreamTaq Hot Start DNA Polymerase (Thermo Fisher Scientific, Waltham, MA, United States), 10 ng of genomic DNA, and the remaining amount of MilliQ water. The amplification parameters for the Cyp51A gene and Cyp51A promoter region were as follows: an initial denaturation for 3 min at 95 °C, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C (Cyp51A upstream region), 58 °C (Cyp51A promoter), or 62 °C (Cyp51A) for 30 s, and extension at 72 °C for 2 min, with final extension at 72 °C for 5 min. The amplification parameters for sdhB, sdhC, and sdhD were as follows: an initial denaturation for 3 min at 95 °C, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 60 °C (sdhC and sdhD) or 63 °C (sdhB) for 30 s, and extension at 72 °C for 1 min, with final extension at 72 °C for 5 min. The Cyt b amplification parameters were as follows: an initial denaturation for 3 min at 95 °C, followed by 35 cycles of denaturation at 95 °C for 20 s, annealing at 58 °C for 25 s, and extension at 72 °C for 30 s, with final extension at 72 °C for 5 min. All the amplifications were performed in a Mastercycler nexus (Eppendorf, Hamburg, Germany). The PCR products were sequenced using an Applied Biosystems 3730 DNA Analyzer (Thermo Fisher Scientific, Waltham, MA, USA) at the Institute of Genomics Core Facility, University of Tartu (Estonia). The sequences obtained were analysed, and the target-site mutations were identified using blastn and blastx search tools (https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed 21 December 2023)) available at NCBI. We focused on previously published relevant SNPs in the target protein-coding gene sequences associated with fungicide insensitivity [19,21,34,35,36]. The representative sequences of the fungicide target protein genes of the studied Ptt isolates are deposited in NCBI GenBank with accession numbers OR530176, OR761967 to OR761973, and OR777246 to OR777248 (Table S1).
To determine the mating type of the isolates, PCR amplification was performed in a multiplex of MAT-1 and MAT-2 primers [39] (Table 1) with the following parameters: an initial denaturation for 3 min at 95 °C, followed by 34 cycles of denaturation at 95 °C for 50 s, annealing at 60 °C for 30 s, and extension at 72 °C for 1 min, with final extension at 72 °C for 7 min. The amplicons, 1143 bp for MAT-1 and 1421 bp for MAT-2, were separated by 1% agarose gel electrophoresis in 1xTAE (Tris-acetate-EDTA) buffer (pH 8.0).
2.5. Statistical Analysis
The statistical analysis was performed in GraphPad Prism 9.4.1 (GraphPad Software, San Diego, CA, USA). The EC50 values were log10-transformed prior to the statistical analysis. An unpaired t-test with Welch’s correction was applied to compare the Ptt fungicide sensitivity between the years and between the mutated and wild-type isolates (α = 0.05). A Kruskal–Wallis test with Dunn’s multiple comparison test was performed to compare the three groups (α = 0.05). The mating type distribution within the counties and years was analysed with a binomial test to find if discrepancy is significant (p < 0.05) from the MAT-1 and MAT-2 1:1 distribution. The figures were visualized using Igor Pro 6.36 (WaveMetrics, Portland, OR, USA). The Cyp51A gene-promoter sequence alignment was visualized using Jalview 2.11.3.0 [43].
3. Results
3.1. Pyrenophora teres f. teres Sensitivity to DMI Fungicides
The Ptt population sensitivity to a DMI fungicide mefentrifluconazole was low. Sensitivity to mefentrifluconazole was significantly different between the isolates collected in 2021 and 2022 (p < 0.001, t = 4.135, df = 128.9): the median EC50 were 0.812 mg/L and 1.329 mg/L, respectively (Figure 2). In 2021, 65% of the isolates were sensitive to mefentrifluconazole (EC50 < 1 mg/L) and 35% had a reduced sensitivity phenotype (Figure 3a). Mefentrifluconazole sensitivity reduction was more prominent in 2022, where 4% of the isolates had an insensitive phenotype, 57% had a reduced sensitivity phenotype, and 39% were sensitive (Figure 3a). On the contrary to mefentrifluconazole, the Ptt population was sensitive to another tested DMI prothioconazole derivate, prothioconazole-desthio: the median EC50 was 0.026 mg/L in 2021 and 0.009 mg/L in 2022 (Figure 2). In addition, two exclusive isolates with reduced sensitivity to both DMIs were found.
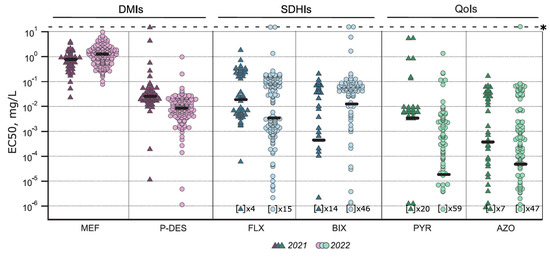
Figure 2.
Fungicide sensitivity of Pyrenophora teres f. teres population in 2021 and 2022. Ptt isolates were tested in microtiter plate sensitivity assay against six fungicides: demethylation inhibitors (DMIs) mefentrifluconazole (MEF) and prothioconazole-desthio (P-DES), succinate dehydrogenase inhibitors (SDHIs) fluxapyroxad (FLX) and bixafen (BIX), quinone outside inhibitors (QoIs) pyraclostrobin (PYR) and azoxystrobin (AZO). Median EC50 values are marked with thick horizontal line and insensitive isolates with asterisk. Note that the EC50 value in the case of number of isolates indicated in brackets remained within the limit of quantification (10−6 mg/L) in the experiments.
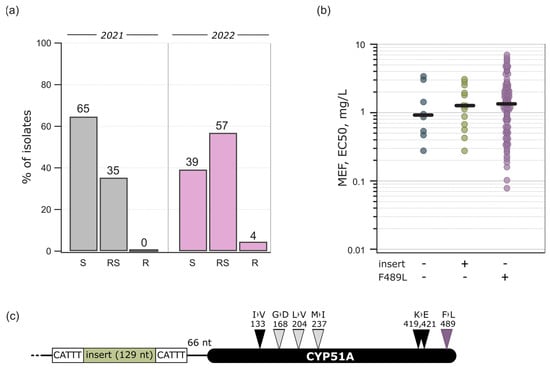
Figure 3.
Mefentrifluconazole sensitivity and variations in its target CYP51A. (a) Percentage of sensitive (S, EC50 < 1 mg/L), reduced sensitivity (RS, EC50 = 1–5 mg/L), and insensitive isolates (R, EC50 > 5 mg/L) in 2021 and 2022; (b) distribution of Ptt isolates according to F489L mutation in CYP51A and 129 bp insert in Cyp51A promoter in 2022; (c) insert position in Cyp51A promoter and CYP51A with the frequent (dark) and rare (light) mutations.
The most frequent mutations in CYP51A were I133V, K419E, K421E, and F489L (Table 2, Figure 3). Two different codons gave rise to the phenylalanine to leucine amino acid change in amino acid position 489 from the wild-type TTC to either CTC or TTA (Table S2). All the Ptt isolates from 2021 (haplotypes PttEE-A3 and 21PttEE-15) and 79% from 2022 had these four mutations in CYP51A (haplotypes PttEE-A3 and PttEE-A4; Table 2). Only a few isolates had other rare mutations, for instance, G168D (haplotypes PttEE-4507 and PttEE-4506), L204V (haplotype 21PttEE-15), and M237I (haplotype PttEE-14).

Table 2.
The frequency of CYP51A haplotypes with specific amino acid substitutions identified in Pyrenophora teres f. teres population in 2021 and 2022.
The Cyp51A gene-promoter region was mostly wild-type. However, 16 isolates in 2022 had an identical 129 bp insert sequence with 5 bp distinct repeats on both sides of the promoter region (Figure 3c and Figure S1; GenBank accession no. OR530176). None of the isolates had F489L mutation in CYP51A together with an insert in the gene-promoter region. Among the isolates collected in 2022, the mefentrifluconazole sensitivity was highest in the isolates with phenylalanine in position 489 in CYP51A and a wild-type promoter (median EC50 = 0.927 mg/L), followed by isolates with promoter insert (median EC50 = 1.15 mg/L) and F489L-mutated isolates (median EC50 = 1.386 mg/L) with reduced sensitivity (Figure 3b). The difference between these groups of isolates was not significant (p = 0.66). However, the outcome of the prothioconazole-desthio sensitivity analysis was opposite to the mefentrifluconazole analysis. The EC50 of prothioconazole-desthio differentiated significantly between these groups of isolates (p < 0.001, H = 47.3, df = 2), with the sensitivity being the highest in the F489L-mutated isolates (median EC50 = 0.007 mg/L) and comparable between isolates with an unmutated 489 position in CYP51A together with a wild-type promoter and inserted promoter region isolates. The median EC50 values were 0.024 mg/L and 0.025 mg/L, respectively. However, all the isolates except for two were completely sensitive to prothioconazole-desthio, and these molecular changes did not reduce the sensitivity.
3.2. Pyrenophora teres f. teres Sensitivity to SDHI Fungicides
Fluxapyroxad and bixafen sensitivity was analysed in the Ptt population. In 2021, the Ptt population was more sensitive to bixafen than fluxapyroxad and vice versa in 2022 (Figure 2). The fluxapyroxad sensitivity was significantly (p = 0.03) lower in 2021 compared to 2022 (t = 2.105, df = 60.63), and the median EC50 values were 0.02 mg/L and 0.004 mg/L, respectively (Figure 2). In total, nineteen isolates in 2021 (36%) and thirty-one in 2022 (22%) had a reduced sensitivity phenotype (EC50 = 0.1–1 mg/L) to fluxapyroxad. Only 2% of the isolates in both years were insensitive (EC50 > 1 mg/L) (Table 3). The shift in bixafen sensitivity between the years was not significant, and median EC50 values were 4.67 × 10−4 mg/L and 0.011 mg/L in 2021 and 2022, respectively (Figure 2). The frequency of isolates with reduced sensitivity phenotype to bixafen was comparable in 2021 and 2022 (Table 3). Insensitive phenotypes to bixafen occurred only in 2022 in four isolates (3%).

Table 3.
The frequency (%) of amino acid substitutions in SDHI and QoI fungicide target molecules and the frequency (%) of sensitive (S), reduced sensitivity (RS), and insensitive isolates (R) to SDHIs (fluxapyroxad and bixafen) and QoIs (azoxystrobin and pyraclostrobin) in Pyrenophora teres f. teres population in 2021 and 2022.
SDH subunits B, C, and D were sequenced for identifying mutations involved in SDHI sensitivity change. B-H277Y was missing in the Estonian Ptt population (Table 3). C-S135R was present in 16 isolates from 2021 (30%) and 19 isolates from 2022 (14%) (Table 3). It was a surprise to detect 43 isolates with amino acid change D-H134R in 2022 (31%), as no mutations in SDH-D were found in 2021 (Table 3).
The C-S135R and D-H134R mutations in the SDH subunits were mainly responsible for the sensitivity decline of fluxapyroxad and bixafen in the Ptt population. The wild-type and C-S135R-mutated isolates differentiated significantly in fluxapyroxad (p = 0.012, t = 2.615, df = 43.13) and bixafen sensitivity (p < 0.001, t = 4.447, df = 13.04) in 2021 (Figure 4). Additionally, in 2022, the fluxapyroxad (p < 0.001, H = 96.97, df = 2) and bixafen sensitivity (p < 0.001, H = 87.38, df = 2) differentiated significantly between the wild-type and C-S135R/D-H134R-mutated isolates (Figure 4). The fluxapyroxad sensitivity was significantly lower in 2021 among the C-S135R-mutated isolates compared to 2022 (p < 0.001, t = 6.909, df = 23.85), with the EC50 ranging from 0.137 to 0.365 mg/L (median EC50 = 0.238 mg/L) in 2021 and from 0.026 to 0.174 mg/L (median EC50 = 0.098 mg/L) in 2022. However, the fluxapyroxad sensitivity of the D-H134R-mutated isolates was comparable with the C-S135R-mutated isolates in 2022, with the EC50 ranging from 0.016 to 0.28 mg/L (median EC50 = 0.08 mg/L). In general, the bixafen sensitivity was comparable in the SDH-mutated isolates in the two study years, with the EC50 ranging from 0.001 to 0.216 mg/L (median EC50 = 0.058 mg/L) in 2021 and 0.015 to 0.118 mg/L (median EC50 = 0.07 mg/L) in 2022 for the C-S135R-mutated isolates and 0.027 to 0.212 mg/L (median EC50 = 0.067 mg/L) for the D-H134R-mutated isolates in 2022.
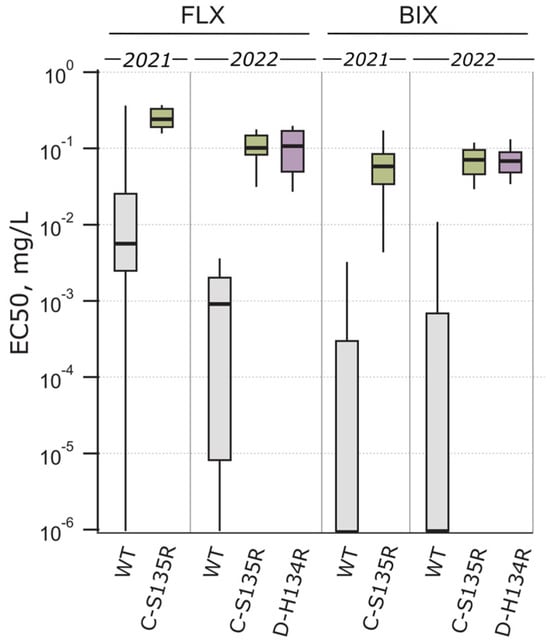
Figure 4.
Fluxapyroxad (FLX) and bixafen (BIX) sensitivity and their dependence on SDH subunit mutations C-S135R and D-H134R. Box plot boundaries were set to 25th percentile and 75th percentile for the box, while lower and upper whiskers extend to 10th and 90th percentile, respectively.
3.3. Pyrenophora teres f. teres Sensitivity to QoI Fungicides
The Ptt population was mostly sensitive to both tested QoI active ingredients, azoxystrobin and pyraclostrobin (Figure 2). The median EC50 for pyraclostrobin were 0.004 mg/L and 1.94 × 10−5 mg/L in 2021 and 2022, respectively. However, there were eight (18%) and nine (7%) reduced sensitivity phenotype isolates (EC50 = 0.01–1 mg/L) in 2021 and 2022, respectively (Table 3). Only two isolates (4%) in 2021 and one isolate (1%) in 2022 were insensitive to pyraclostrobin (EC50 > 1 mg/L) (Table 3). Sensitivity to azoxystrobin was also high, and the median EC50 was less than 0.001 mg/L in both years. However, fifteen isolates (37%) in 2021 and nineteen isolates (14%) in 2022 had a reduced sensitivity phenotype (EC50 = 0.01–1 mg/L), and only one isolate had an insensitive phenotype to azoxystrobin (EC50 > 1 mg/L) in 2022 (Table 3). All these isolates with an insensitive phenotype to pyraclostrobin or azoxystrobin had wild-type target protein Cyt b, so in this case, other mechanisms should be involved.
The Cyt b gene was amplified with two primer pairs to detect F129L and G137R mutations associated with QoI sensitivity in the Ptt isolates. In total, 16 and 17 Ptt isolates had an F129L mutation in Cyt b in 2021 and 2022, respectively (Table 3). At the same time, the G137R mutation was absent in Cyt b. An analysis revealed that all the isolates with the F129L mutation in Cyt b had azoxystrobin EC50 values higher than 0.01 mg/L. There was a significant difference (p < 0.001) between the wild-type and F129L-mutated isolates in azoxystrobin sensitivity in both years (2021: t = 3.646, df = 37.92; 2022: t = 11.49, df = 14.52) (Figure 5). The azoxystrobin sensitivity was significantly higher in 2021 among the F129L-mutated isolates compared to 2022 (p = 0.009, t = 2.802, df = 26.78), with the EC50 ranging from 0.012 to 0.071 mg/L (median EC50 = 0.036 mg/L) in 2021 and 0.029 to 0.088 mg/L (median EC50 = 0.05 mg/L) in 2022. However, there was no difference (p = 0.2) between the Cyt b-mutated and wild-type isolates in pyraclostrobin sensitivity in either year, and the pyraclostrobin sensitivity was highly variable (Figure 5).
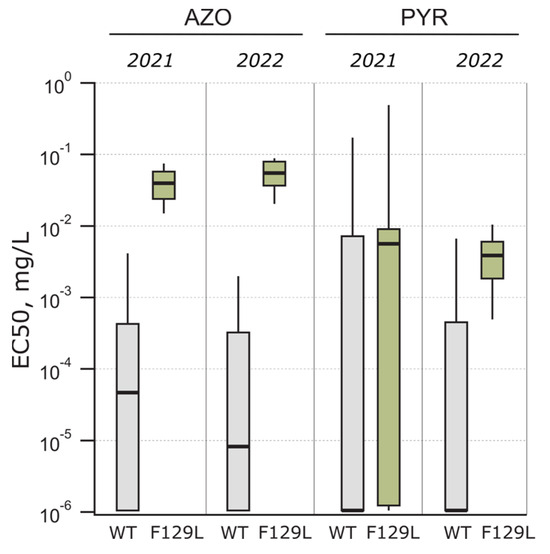
Figure 5.
Azoxystrobin (AZO) and pyraclostrobin (PYR) sensitivity and their dependence on mutation F129L in Cyt b. Box plot boundaries were set to 25th percentile and 75th percentile for the box, while lower and upper whiskers extend to 10th and 90th percentile, respectively.
3.4. Isolates with Relevant Mutations in All Three Target Site Proteins
Furthermore, 16 isolates in 2021 from two fields and 14 isolates in 2022 from three fields had a PttEE-A3 or PttEE-A4 haplotype with I133V, K419E, K421E, and F489L mutations in CYP51A, F129L-mutated Cyt b, and C-S135R mutation in the SDH-C subunit simultaneously (Table S2). Ptt with relevant mutations in all three fungicide target proteins simultaneously would be a high risk for efficient disease control when adapting and spreading in the population. Most of these isolates had a reduced sensitivity phenotype to mefentrifluconazole (median EC50 = 1.319 mg/L), fluxapyroxad (median EC50 = 0.171 mg/L), and azoxystrobin (median EC50 = 0.044 mg/L). However, sensitivity to the other tested fungicides remained high, with the median EC50 being 0.07 mg/L for bixafen, 0.021 mg/L for prothioconazole-desthio, and 0.004 mg/L for pyraclostrobin. Among these high-risk isolates, eight isolates in 2021 and four isolates in 2022 were collected from nearby fields and could have the same origin.
3.5. Pyrenophora teres f. teres Mating Types
The mating type analysis showed that both MAT-1 and MAT-2 are common in the Estonian Ptt population. In 2021, 80% of the isolates were MAT-2, and the population significantly differentiated (p < 0.001) from equal distribution of MAT-1 and MAT-2. However, in 2022, 52% of the isolates were MAT-2 and 48% were MAT-1; the ratio was nearly 1:1 in the population. Only MAT-1 was identified in 8% and 32% of the fields, and MAT-2 prevailed in 33% and 36% of the fields in years 2021 and 2022, respectively. Sexual reproduction can occur when opposite mating types meet on the same field site. Both Ptt mating types occurred together in 59% and 32% of the spring barley fields in 2021 and 2022, respectively.
4. Discussion
This is the first research study characterizing several molecular changes in fungicide target proteins (CYP51A, Cyt b, and SDH subunits C and D) and the Cyp51A gene promoter, which have a potential impact on the fungicide sensitivity of the Ptt field population in Northeastern Europe. Several Ptt isolates had reduced sensitivity to a new DMI active ingredient, mefentrifluconazole, influenced by target site mutations in the DMI target protein CYP51A and insert in the Cyp51A promoter region. The Ptt population was mostly sensitive to prothioconazole-desthio, bixafen, fluxapyroxad, pyraclostrobin, and azoxystrobin. In addition, the mating type analysis revealed that Ptt is expected to reproduce clonally in most of the fields, but possible sexual reproduction events may occur in the fields when both mating types meet.
The Ptt isolates, which had amino acid change phenylalanine to leucine in CYP51A in position 489, dominate in Estonia, 100% in 2021 and 79% in 2022. They are associated with reduced sensitivity to mefentrifluconazole, but not to prothioconazole-desthio. In a study from Australia, the same mutation was assumed to cause a considerable conformational rearrangement within the Ptt CYP51 protein haem cavity, resulting in a low inhibition effect of DMIs, such as tebuconazole, difenoconazole, and prochloraz [19]. In the Estonian Ptt population, two different codons gave rise to the phenylalanine to leucine amino acid change in position 489, from the wild-type TTC to either CTC or TTA (Table 2). In Australia, all the Ptt mutant isolates had an identical codon TTA for leucine [19], and the Ptm mutant isolates had three codons (TTA, TTG, and CTC) for leucine in amino acid position 489 in CYP51A [34]. The mutation F489L has been detected in Australia since 2013 in the Ptt population and since 2016 in the Ptm population [19,34], but in Europe, the Cyp51A mutation prevalence in Ptt populations is so far unknown.
The combination of mutations in Cyp51A with promoter insertions is contributing to the highest levels of DMI resistance in several cereal pathogen (e.g., Z. tritici, Ptm) populations [34,44]. Our finding of the 129 bp insertion element in the promoter of the Cyp51A of Ptt was noticed in isolates with reduced sensitivity to mefentrifluconazole, but their prothioconazole-desthio sensitivity remained high. Direct repeats of five base pairs (CATTT) on both ends of the 129 bp insertion are like 4–6 bp target site duplication sequences (TSDs) characterised in the case of LTR retrotransposons [45]. An identical insert with direct repeats was identified in moderately DMI-resistant Ptm isolates [34], but not in the Ptt isolates from Australia [19]. Moreover, highly DMI-resistant Ptm isolates had both an insertion element in the promoter and mutation F498L in CYP51A [34]. In the Estonian Ptt population, these molecular changes did not occur simultaneously in the same isolates.
However, sensitivity to prothioconazole-desthio was high in the Estonian Ptt population, and molecular changes in the target protein CYP51A and its promoter region did not reduce the sensitivity. A similar observation in the Australian Ptt population revealed high sensitivity to prothioconazole, but reduced sensitivity to other DMIs (tebuconazole, metconazole, and difenoconazole) in the same isolates [19]. Unfortunately, the data on DMI sensitivity of the Ptt population in Europe are not detailed and are limited to annual FRAC reports that show fluctuations in DMI sensitivity in France and Germany, but stable sensitivity in other European countries [46]. The marketed cereal plant protection products in Estonia most often contain prothioconazole as one active ingredient in combination with SDHI, amine, or other ingredients [17]. Although prothioconazole-containing products have been extensively applied to Estonian cereal fields for the last two decades, a significant sensitivity shift has not been noted in other cereal plant pathogen populations (Z. tritici and R. collo-cygni), regardless of the molecular changes in the CYP51 protein [47,48,49]. Furthermore, barley field trials in Northern Europe have shown satisfactory control of net blotch with 1 to 2 foliar applications of prothioconazole fungicide mixtures, even in high disease pressure (for example, in 2019) [6].
The Estonian Ptt population was mostly sensitive to the tested SDHIs fluxapyroxad and bixafen with a few exceptions. In the wild-type SDH enzyme, histidines at position 134 in subunits SDH-C and SDH-D coordinate the central iron atom of the haem b group. These two amino acid positions have been substituted to arginine in the case of SDHI insensitivity in P. teres [21]. Besides C-H134R and D-H134R, three further amino acid substitutions, C-N75S, C-G79R, and C-S135R, are clustered around haem b [21]. In the current study, out of these only mutations, C-S135R and D-H134R were identified in the Ptt population, and 30% of the Ptt isolates collected in 2021 and 40% in 2022 were mutated. Mutation B-H277Y, which has been found in other countries [21,36], was absent in the Estonian Ptt population. The fluxapyroxad and bixafen sensitivity were both affected in the SDH-mutated Ptt isolates in Estonia. In Rehfus et al.’s [21] study, the field dose of fluxapyroxad showed stable control for P. teres isolates with mutations leading to B-H277Y, C-N75S, C-S135R, D-D124N, and D-D145G. Higher resistance factors have been associated with mutations C-G79R, D-H134R, D-D124E, and C-H134R, with reduced fluxapyroxad efficacy [21,35]. In Argentina, the detection of Ptt isolates carrying double mutation C-N75S + D-D145G raises concerns in fluxapyroxad field efficacy [36]. As fluxapyroxad-containing products (e.g., Priaxor and Revytrex by BASF) are often used in cereal disease control and the spread of SDH-mutated Ptt isolates within the Estonian population is remarkable, the field efficacy should be monitored.
In general, the Ptt in Estonia was highly sensitive to QoI fungicides, and mutation F129L in Cyt b was identified in around 17% of the isolates and none had the G137R mutation. The target site mutation impacted azoxystrobin, but not pyraclostrobin sensitivity. The same effect was noted previously in another study, where pyraclostrobin was the most effective QoI fungicide, regardless of F129L mutation in the Ptt isolates [20]. Mutation F129L in Ptt Cyt b protein in Western Europe is frequent, and since the first notice of it in 2003, it rose to the frequency of 25% in 2014 [21]. Ptt population studies in Europe, Northern Africa, and Canada have shown variability but mostly high sensitivity to pyraclostrobin, azoxystrobin, and picoxystrobin [18,20,50]. Positively for disease control, it has been noted that mutation F129L does not reduce QoI sensitivity significantly, and these Ptt isolates can be controlled by recommended field rates of QoIs in vivo to almost the same extent as wild-type isolates [22,23]. In addition, Ptt cannot have mutation G143A in Cyt b, which has a high resistance factor and causes substantial disease control failure in several other cereal pathogen populations, for instance, P. tritici-repentis and Z. tritici [23,51,52].
Laboratory assays on new isolates with reduced sensitivity can be used as possible indicators of future changes in field performance [51]. The new less-sensitive isolates have a fitness advantage over wild-type strains under the selection pressure of fungicide [51]. In this study, 30 Ptt isolates had target site mutations in all three investigated fungicide target proteins, CYP51A, Cyt b, and SDH, and most of them had a reduced sensitivity phenotype to mefentrifluconazole, azoxystrobin, and fluxapyroxad. Although these isolates were mostly sensitive to other tested fungicides, it is critical to monitor and establish the spread of these mutated isolates prior to disease control failures.
In 2021, plant protection products Revystar XL, Revytrex (mefentrifluconazole, fluxapyroxad; BASF), and Balaya (mefentrifluconazole, pyraclostrobin; BASF) were marketed in Estonia for the first time, and farmers adopted these promptly into their cereal plant protection plans [17]. In Estonia, farmers are advised to apply these products once or twice in the active growing phases (BBCH 30-69) to protect spring barley against several plant diseases (net blotch, scald, ramularia leaf spot, powdery mildew, and leaf rust) and provide high quality and yield of the crop. Though mefentrifluconazole was not specifically developed to control Ptt, the active use of products containing mefentrifluconazole is a selection pressure for resistance development and needs further research if it selects for specific alterations in the CYP51 protein. As long as the Estonian Ptt population remains sensitive to SDHI agent fluxapyroxad and QoI agent pyraclostrobin, these products with a combination of two modes of action are efficient and provide sufficient control against net blotch, among other barley diseases.
Ptt is a seedborne pathogen, and gene flow or introduction of isolates with new traits (for example, target site mutations in fungicide target proteins) is possible through seed trade, then evolving and adapting to local environments, and through selection pressure of host cultivars and fungicide applications [12]. Farmers in Estonia import new batches of barley seed of various cultivars from Finnish, Danish, German, and other breeding companies, in addition to propagating seeds themselves. For instance, in Denmark and Germany, Ptt Cyt b protein F129L mutation levels are medium or medium to high [24]. SDH mutations C-G79R, C-H134R, C-N75S, and C-S135R occur most often in European populations [30]. Mutations in the SDH protein are absent in Finland but occur at low frequency in Denmark and high levels in Western Europe, including Germany [30].
Ptt has a mixed reproduction system, which contributes to the evolution of the pathogen [12]. Equal distribution of the two mating types is assumed in the absence of segregation bias and selection of one mating type [9]. In the present study, within the Estonian Ptt population in 2022, the MAT-1/MAT-2 ratio was 1:1, as expected in sexually reproducing population. Although several fields had only one mating type and sexual reproduction within these fields would be limited. In 2022, the sampling was more widespread compared to 2021 when MAT-1/MAT-2 ratio was 1:4 and one mating type dominated. Ptt population studies from Finland, Hungary, Italy, Australia, and Canada reported a 1:1 mating type ratio and possible sexual reproduction contributing to genetic diversity [9,53,54,55,56]. However, evidence from Dahanayaka et al. [54] showed deviation from the expected 1:1 ratio in population clusters from South Africa and Australia.
5. Conclusions
As this is the first detailed study of the current fungicide sensitivity of a widely distributed barley pathogen Ptt population in Estonia, it will serve as a baseline for future assessments. We identified multiple relevant molecular changes—F489L mutation in CYP51A, 129 bp insert in Cyp51A promoter, mutations C-S135R and D-H134R in SDH protein, and F129L mutation in Cyt b protein—affecting the sensitivity of the Ptt field population to fungicides of different modes of action. To prolong the effective lifetime of fungicides, they should be applied in alternation, as well as in mixtures where available, preferably including other modes of action. However, the selection of fungicide products is limited, and cereals as the main crops in Estonian commercial farms are generally protected with the same products. Thus, the selection pressure for fungicide resistance development in pathogen populations is strong. Barley seed trade may introduce new fungicide resistance-related mutations, and probable sexual reproduction in the Ptt population in Estonia could increase the genetic diversity and fitness of the pathogen. Therefore, it is crucial to continue with monitoring of the pathogen and its adaptation to fungicides to prevent disease management failures.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof10040260/s1, Figure S1: Alignment of Cyp51A gene-promoter sequences. Insert-containing CYP51A promoter sequence from representative Estonian Pyrenophora teres f. teres isolate 22PTEE-0701 (Ptt_07-01: GenBank accession OR530176) compared to Ptt wild-type (Ptt_W1: GenBank accession HG992977) and P. teres f. maculata respective sequences (Ptm_SG1…PTM_P7: GenBank accessions OCTF02000001 and MT499783–MT499787) previously described in Mair et al. [34]. Sequence alignment was visualized using Jalview [43]; Table S1: Summary of Estonian Pyrenophora teres f. teres representative isolates and their GenBank accession numbers; Table S2: Collection of Estonian Pyrenopora teres f. teres isolates with relevant data of fungicide target gene sequences and GenBank accession number of representative isolates.
Author Contributions
Conceptualization, R.K., R.P. and A.M.; methodology, R.K. and R.P.; validation, R.K., R.P. and L.A.; formal analysis, R.K., R.P. and L.A.; investigation, all authors; resources, A.M. and R.K.; data curation, R.K., R.P. and L.A.; writing—original draft preparation, R.K. and R.P.; writing—review and editing, all authors; visualization, R.P. and L.A.; supervision, A.M. and R.K.; project administration, A.M. and R.K.; funding acquisition, A.M. and R.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Ministry of Regional Affairs and Agriculture of the Republic of Estonia with project nos. 10.1-2/177 and 10.1-2/256, by the Ministry of Education and Research of the Republic of Estonia with project “Sordiaretus” no. 2014-2020.4.01.16-0037, and by the Estonian Research Council, grant no. PSG827.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The DNA sequences of the Ptt fungicide target molecule genes with relevant single nucleotide mutations are made publicly available in the NCBI GenBank database with accession numbers OR530176, OR761967 to OR761973, and OR777246 to OR777248. Other raw data that support the findings of this study are available from the corresponding author upon reasonable request.
Acknowledgments
The authors are grateful to Pille Sooväli and Kersti Lilleväli from The Centre of Estonian Rural Research and Knowledge for providing professional advice and technical support. We acknowledge Meelis Värnik and Tiiu Annuk from the Estonian Farmers’ Cooperative Kevili for their collaboration. This work is part of a PhD thesis of Regina Pütsepp in partial fulfilment for the requirements for a doctorate degree at Estonian University of Life Sciences.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Newton, A.C.; Flavell, A.J.; George, T.S.; Leat, P.; Mullholland, B.; Ramsay, L.; Revoredo-Giha, C.; Russell, J.; Steffenson, B.J.; Swanston, J.S.; et al. Crops that feed the world 4. Barley: A resilient crop? Strengths and weaknesses in the context of food security. Food Secur. 2011, 3, 141–178. [Google Scholar] [CrossRef]
- Sooväli, P. Integrated Plant Disease Management in Spring Barley and Oat Production. Ph.D. Thesis, Estonian University of Life Sciences, Tartu, Estonia, 21 December 2011. [Google Scholar]
- Smedegård-Petersen, V. Pyrenophora teres f. maculata f. nov. and Pyrenophora teres f. teres on Barley in Denmark; Agricultural University of Copenhagen: Copenhagen, Denmark, 1971; pp. 124–144. [Google Scholar]
- Jalli, M.; Laitinen, P.; Latvala, S. The emergence of cereal fungal diseases and the incidence of leaf spot diseases in Finland. Agric. Food Sci. 2011, 20, 62–73. [Google Scholar] [CrossRef]
- Jalli, M.; Kaseva, J.; Andersson, B.; Ficke, A.; Jørgensen, L.N.; Ronis, A.; Kaukoranta, T.; Ørum, J.-E.; Djurle, A. Yield increases due to fungicide control of leaf blotch diseases in wheat and barley as a basis for IPM decision-making in the Nordic-Baltic region. Eur. J. Plant Pathol. 2020, 158, 315–333. [Google Scholar] [CrossRef]
- Jørgensen, L.N.; Matzen, N.; Ficke, A.; Andersson, B.; Jalli, M.; Ronis, A.; Nielsen, G.C.; Erlund, P.; Djurle, A. Using risk models for control of leaf blotch diseases in barley minimises fungicide use—Experiences from the Nordic and Baltic countries. Acta Agric. Scand. Sect. B Soil Plant Sci. 2021, 71, 247–260. [Google Scholar] [CrossRef]
- Kangor, T.; Sooväli, P.; Tamm, Ü.; Tamm, I.; Koppel, M. Malting barley diseases, yield and quality—Responses to using various agro-technology regimes. Proc. Latv. Acad. Sci. Sect. B. Nat. Exact Appl. Sci. 2017, 71, 57–62. [Google Scholar] [CrossRef]
- Sooväli, P.; Koppel, M. Efficacy of fungicide tebuconazole in barley varieties with different resistance level. Agric. Food Sci. 2010, 19, 34–42. [Google Scholar] [CrossRef]
- Rau, D.; Maier, F.J.; Papa, R.; Brown, A.H.D.; Balmas, V.; Saba, E.; Schaefer, W.; Attene, G. Isolation and characterization of the mating-type locus of the barley pathogen Pyrenophora teres and frequencies of mating-type idiomorphs within and among fungal populations collected from barley landraces. Genome 2005, 48, 855–869. [Google Scholar] [CrossRef] [PubMed][Green Version]
- McLean, M.S.; Howlett, B.J.; Hollaway, G.J. Epidemiology and control of spot form of net blotch (Pyrenophora teres f. maculata) of barley: A review. Crop Pasture Sci. 2009, 60, 303–315. [Google Scholar] [CrossRef]
- McDonald, B.A.; Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 2002, 40, 349–379. [Google Scholar] [CrossRef]
- Liu, Z.; Ellwood, S.R.; Oliver, R.P.; Friesen, T.L. Pyrenophora teres: Profile of an increasingly damaging barley pathogen. Mol. Plant Pathol. 2011, 12, 1–19. [Google Scholar] [CrossRef]
- Jordan, V.W.L.; Allen, E.C. Barley net blotch: Influence of straw disposal and cultivation methods on inoculum potential, and on incidence and severity of autumn disease. Plant Pathol. 1984, 33, 547–559. [Google Scholar] [CrossRef]
- Jørgensen, J.H.; Bech, C.; Jensen, J. Reaction of European spring barley varieties to a population of the net blotch fungus. Plant Breed. 2000, 119, 43–46. [Google Scholar] [CrossRef]
- Rusu, T.; Gus, P.; Bogdan, I.; Moraru, P.I.; Pop, A.I.; Clapa, D.; Marin, D.I.; Oroian, I.; Pop, L.I. Implications of minimum tillage systems on sustainability of agricultural production and soil conservation. J. Food Agric. Environ. 2009, 8, 335–338. [Google Scholar]
- van den Bosch, F.; Paveley, N.; Shaw, M.; Hobbelen, P.; Oliver, R. The dose rate debate: Does the risk of fungicide resistance increase or decrease with dose? Plant Pathol. 2011, 60, 597–606. [Google Scholar] [CrossRef]
- Statistics Estonia. Statistical Database. Sales of Pesticides by Active Substance. Available online: https://andmed.stat.ee/et/stat/keskkond__pollumajanduskeskkond/KK2085/table/tableViewLayout2 (accessed on 2 February 2023).
- Akhavan, A.; Strelkov, S.E.; Askarian, H.; Kher, S.V.; Fraser, M.; Kutcher, H.R.; Turkington, T.K. Sensitivity of western Canadian Pyrenophora teres f. teres and P. teres f. maculata isolates to propiconazole and pyraclostrobin. Can. J. Plant Pathol. 2017, 39, 11–24. [Google Scholar] [CrossRef]
- Mair, W.J.; Deng, W.; Mullins, J.G.L.; West, S.; Wang, P.; Besharat, N.; Ellwood, S.R.; Oliver, R.P.; Lopez-Ruiz, F.J. Demethylase inhibitor fungicide resistance in Pyrenophora teres f. sp. teres associated with target site modification and inducible overexpression of Cyp51. Front. Microbiol. 2016, 7, 1279. [Google Scholar] [CrossRef]
- Marzani, Q.A.; Swarbrick, P.; Rossall, S. Correlation of the F129L mutation in Pyrenophora teres, the pathogen of net blotch of barley, with the efficacy of QoI fungicides. IOSR J. Agric. Vet. Sci. 2013, 3, 66–72. [Google Scholar] [CrossRef]
- Rehfus, A.; Miessner, S.; Achenbach, J.; Strobel, D.; Bryson, R.; Stammler, G. Emergence of succinate dehydrogenase inhibitor resistance of Pyrenophora teres in Europe. Pest Manag. Sci. 2016, 72, 1977–1988. [Google Scholar] [CrossRef] [PubMed]
- Semar, M.; Strobel, D.; Koch, A.; Klappach, K.; Stammler, G. Field efficacy of pyraclostrobin against populations of Pyrenophora teres containing the F129L mutation in the cytochrome b gene. J. Plant Dis. Prot. 2007, 114, 117–119. [Google Scholar] [CrossRef]
- Sierotzki, H.; Frey, R.; Wullschleger, J.; Palermo, S.; Karlin, S.; Godwin, J.; Gisi, U. Cytochrome b gene sequence and structure of Pyrenophora teres and P. tritici-repentis and implications for QoI resistance. Pest Manag. Sci. 2007, 63, 225–233. [Google Scholar] [CrossRef]
- QoI Working Group of FRAC. Fungicide Resistance Action Committee Monitoring Results and Use Recommendations for QoI Fungicides. Available online: https://www.frac.info/frac-teams/working-groups/qol-fungicides/recommendations-for-qoi (accessed on 2 April 2023).
- Fraaije, B.A.; Buttersb, J.A.; Coelhoa, J.M.; Jonesc, D.R.; Hollomonb, D.W. Following the dynamics of strobilurin resistance in Blumeria graminis f.sp. tritici using quantitative allele-specific real-time PCR measurements with the fluorescent dye SYBR Green I. Plant Pathol. 2002, 51, 45–54. [Google Scholar] [CrossRef]
- Fraaije, B.A.; Cools, H.J.; Fountaine, J.; Lovell, D.J.; Motteram, J.; West, J.S.; Lucas, J.A. Role of ascospores in further spread of QoI-resistant cytochrome b alleles (G143A) in field populations of Mycosphaerella graminicola. Phytopathology 2005, 95, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Hoffmeister, M.; Mehl, A.; Hinson, A.; Siepe, I.; Taufferner, T.; Stammler, G. Acquired QoI resistance in Pyrenophora teres through an interspecific partial gene transfer by Pyrenophora tritici repentis? J. Plant Dis. Prot. 2022, 129, 1073–1086. [Google Scholar] [CrossRef]
- Grasso, V.; Palermo, S.; Sierotzki, H.; Garibaldi, A.; Gisi, U. Cytochrome b gene structure and consequences for resistance to Qo inhibitor fungicides in plant pathogens. Pest Manag. Sci. 2006, 62, 465–472. [Google Scholar] [CrossRef] [PubMed]
- Vallieres, C.; Trouillard, M.; Dujardin, G.; Meunier, B. Deleterious effect of the Qo inhibitor compound resistance-conferring mutation G143A in the intron-containing cytochrome b gene and mechanisms for bypassing it. Appl. Environ. Microbiol. 2011, 77, 2088–2093. [Google Scholar] [CrossRef] [PubMed]
- SDHI Working Group of FRAC. Fungicide Resistance Action Committee Monitoring Results and Use Recommendations for SDHI Fungicides. Available online: https://www.frac.info/frac-teams/working-groups/sdhi-fungicides/recommendations-for-sdhi (accessed on 2 April 2023).
- Cools, H.J.; Fraajie, B.A. Update on mechanisms of azole resistance in Mycosphaerella graminicola and implications for future control. Pest Manag. Sci. 2013, 69, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Celia-Sanchez, B.N.; Mangum, B.; Brewer, M.; Momany, M. Analysis of Cyp51 protein sequences shows 4 major Cyp51 gene family groups across fungi. G3 Genes Genomes Genet. 2022, 12, jkac249. [Google Scholar] [CrossRef] [PubMed]
- Becher, R.; Wirsel, S.G. Fungal cytochrome P450 sterol 14α-demethylase (CYP51) and azole resistance in plant and human pathogens. Appl. Microbiol. Biotechnol. 2012, 95, 825–840. [Google Scholar] [CrossRef] [PubMed]
- Mair, W.J.; Thomas, G.J.; Dodhia, K.; Hills, A.L.; Jayasena, K.W.; Ellwood, S.R.; Oliver, R.P.; Lopez-Ruiz, F.J. Parallel evolution of multiple mechanisms for demethylase inhibitor fungicide resistance in the barley pathogen Pyrenophora teres f. sp. maculata. Fungal Genet. Biol. 2020, 145, 103475. [Google Scholar] [CrossRef]
- Knight, N.L.; Adhikari, K.C.; Dodhia, K.N.; Mair, W.J.; Lopez-Ruiz, F.J. Workflows for detecting fungicide resistance in net form and spot form net blotch pathogens. Pest Manag. Sci. 2024, 80, 2131–2140. [Google Scholar] [CrossRef]
- Sautua, F.J.; Carmona, M.A. SDHI resistance in Pyrenophora teres f. teres and molecular detection of novel double mutations in sdh genes conferring high resistance. Pest Manag. Sci. 2023, 79, 3300–3311. [Google Scholar] [CrossRef] [PubMed]
- Qiagen—Kits Handbooks. DNeasy Mericon 96 QIAcube HT Handbook. Available online: https://www.qiagen.com/us/resources/resourcedetail?id=6c38af78-01df-4640-af5e-c208737f96b1&lang=en (accessed on 11 January 2023).
- White, T.J.; Bruns, T.D.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Eds.; Academic Press: London, UK, 1990; pp. 315–322. [Google Scholar]
- Lu, S.; Platz, G.J.; Edwards, M.C.; Friesen, T.L. Mating type locus-specific polymerase chain reaction markers for differentiation of Pyrenophora teres f. teres and P. teres f. maculata, the causal agents of barley net blotch. Phytopathology 2010, 100, 1298–1306. [Google Scholar] [CrossRef]
- Estonian Land Board. Spatial Data. Administrative and Settlement Division. Available online: https://geoportaal.maaamet.ee/eng/spatial-data/administrative-and-settlement-division-p312.html (accessed on 1 November 2020).
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program Primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Cools, H.J.; Bayon, C.; Atkins, S.; Lucas, J.A.; Fraaije, B.A. Overexpression of the sterol 14α-demethylase gene (MgCYP51) in Mycosphaerella graminicola isolates confers a novel azole fungicide sensitivity phenotype. Pest Manag. Sci. 2012, 68, 1034–1040. [Google Scholar] [CrossRef] [PubMed]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef] [PubMed]
- SBI Working Group of FRAC. Fungicide Resistance Action Committee Monitoring Results and Use Recommendations for SBI Fungicides. Available online: https://www.frac.info/frac-teams/working-groups/sbi-fungicides/recommendations-for-sbi (accessed on 2 April 2023).
- Kiiker, R.; Juurik, M.; Heick, T.M.; Mäe, A. Changes in DMI, SDHI, and QoI fungicide sensitivity in the Estonian Zymoseptoria tritici population between 2019 and 2020. Microorganisms 2021, 9, 814. [Google Scholar] [CrossRef]
- Kiiker, R.; Juurik, M.; Mäe, A. Fungicide resistance evolving in Ramularia collo-cygni population in Estonia. Microorganisms 2021, 9, 1514. [Google Scholar] [CrossRef]
- Mäe, A.; Fillinger, S.; Sooväli, P.; Heick, T.M. Fungicide sensitivity shifting of Zymoseptoria tritici in Finnish-Baltic region and a novel insertion in the MFS1 promoter. Front. Plant Sci. 2020, 11, 385. [Google Scholar] [CrossRef]
- Lammari, H.-I.; Rehfus, A.; Stammler, G.; Benslimane, H. Sensitivity of the Pyrenophora teres population in Algeria to quinone outside inhibitors, succinate dehydrogenase inhibitors and demethylation inhibitors. Plant Pathol. J. 2020, 36, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Blake, J.J.; Gosling, P.; Fraaije, B.A.; Burnett, F.J.; Knight, S.M.; Kildea, S.; Paveley, N.D. Changes in field dose–response curves for demethylation inhibitor (DMI) and quinone outside inhibitor (QoI) fungicides against Zymoseptoria tritici, related to laboratory sensitivity phenotyping and genotyping assays. Pest Manag. Sci. 2018, 74, 302–313. [Google Scholar] [CrossRef] [PubMed]
- Kildea, S.; Bucar, D.E.; Hutton, F.; de la Rosa, S.; Welch, T.E.; Phelan, S. Prevalence of QoI resistance and mtDNA diversity in the Irish Zymoseptoria tritici population. Ir. J. Agric. Food Res. 2019, 58, 27–33. [Google Scholar] [CrossRef]
- Akhavan, A.; Turkington, T.K.; Kebede, B.; Xi, K.; Kumar, K.; Tekauz, A.; Kutcher, H.R.; Tucker, J.R.; Strelkov, S.E. Genetic structure of Pyrenophora teres f. teres and P. teres f. maculata populations from western Canada. Eur. J. Plant Pathol. 2016, 146, 325–335. [Google Scholar] [CrossRef]
- Dahanayaka, B.A.; Vaghefi, N.; Knight, N.L.; Bakonyi, J.; Prins, R.; Seress, D.; Snyman, L.; Martin, A. Population structure of Pyrenophora teres f. teres barley pathogens from different continents. Phytopathology 2021, 111, 2118–2129. [Google Scholar] [CrossRef] [PubMed]
- Linde, C.C.; Smith, L.M. Host specialisation and disparate evolution of Pyrenophora teres f. teres on barley and barley grass. BMC Evol. Biol. 2019, 19, 139. [Google Scholar] [CrossRef]
- Serenius, M.; Mironenko, N.; Manninen, O. Genetic variation, occurrence of mating types and different forms of Pyrenophora teres causing net blotch of barley in Finland. Mycol. Res. 2005, 109, 809–817. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).