A Thermotolerant Yeast Cyberlindnera rhodanensis DK Isolated from Laphet-so Capable of Extracellular Thermostable β-Glucosidase Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Microbial Strains
2.2. Laphet-so Sampling Sites and Sample Collection
2.3. Isolation of Thermotolerant Yeasts
2.4. Characterization of Thermotolerant Yeasts
2.4.1. Evaluation of Tannin-Tolerant Ability
2.4.2. Ethanol Producing Capability
2.4.3. Screening of Polysaccharide Degrading Enzyme Production
2.4.4. Colony and Morphological Characteristics
2.4.5. Molecular Identification
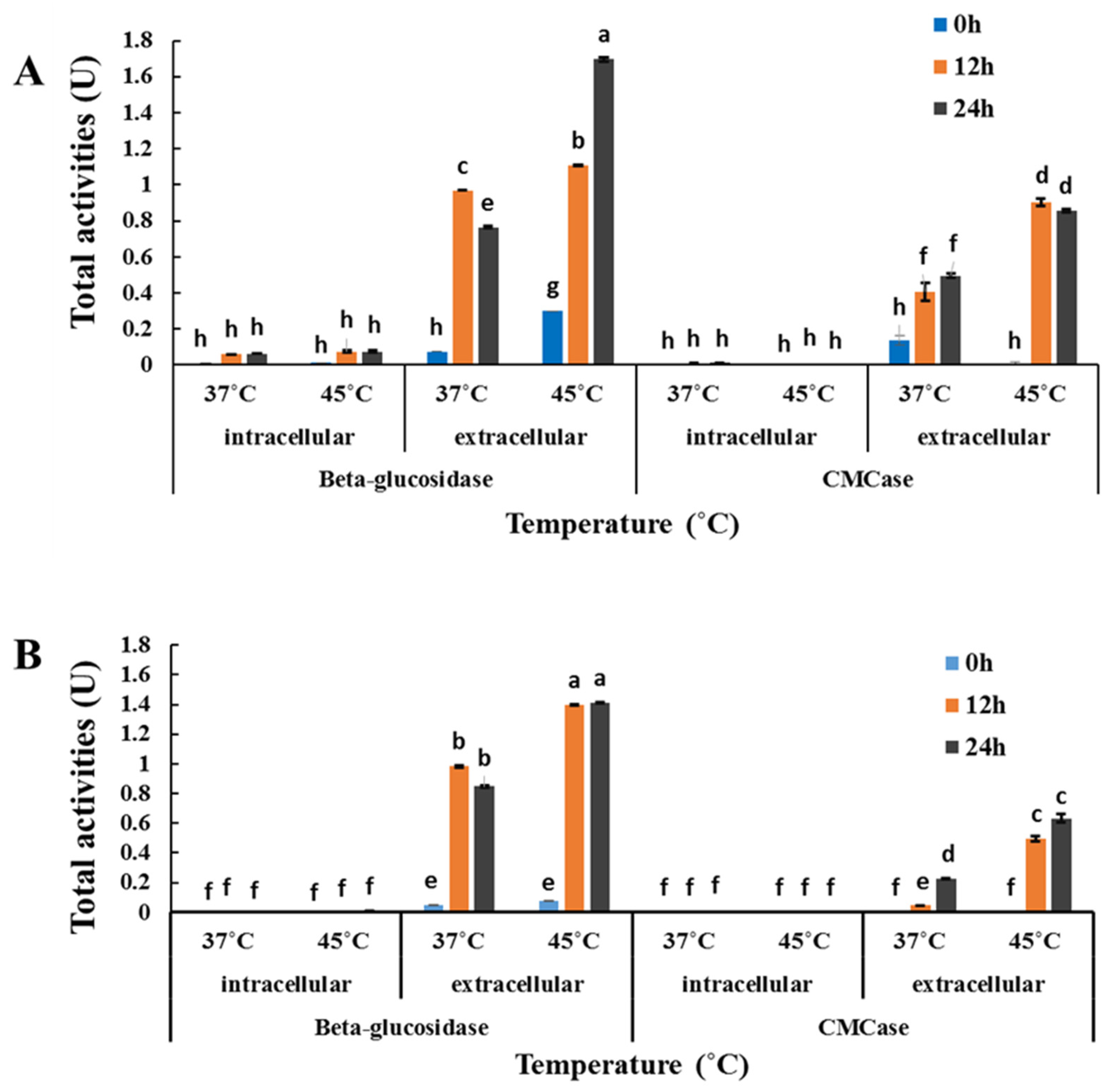
2.5. Investigation of β-Glucosidase and CMCase Production by Cyberlindnera rhodanensis DK
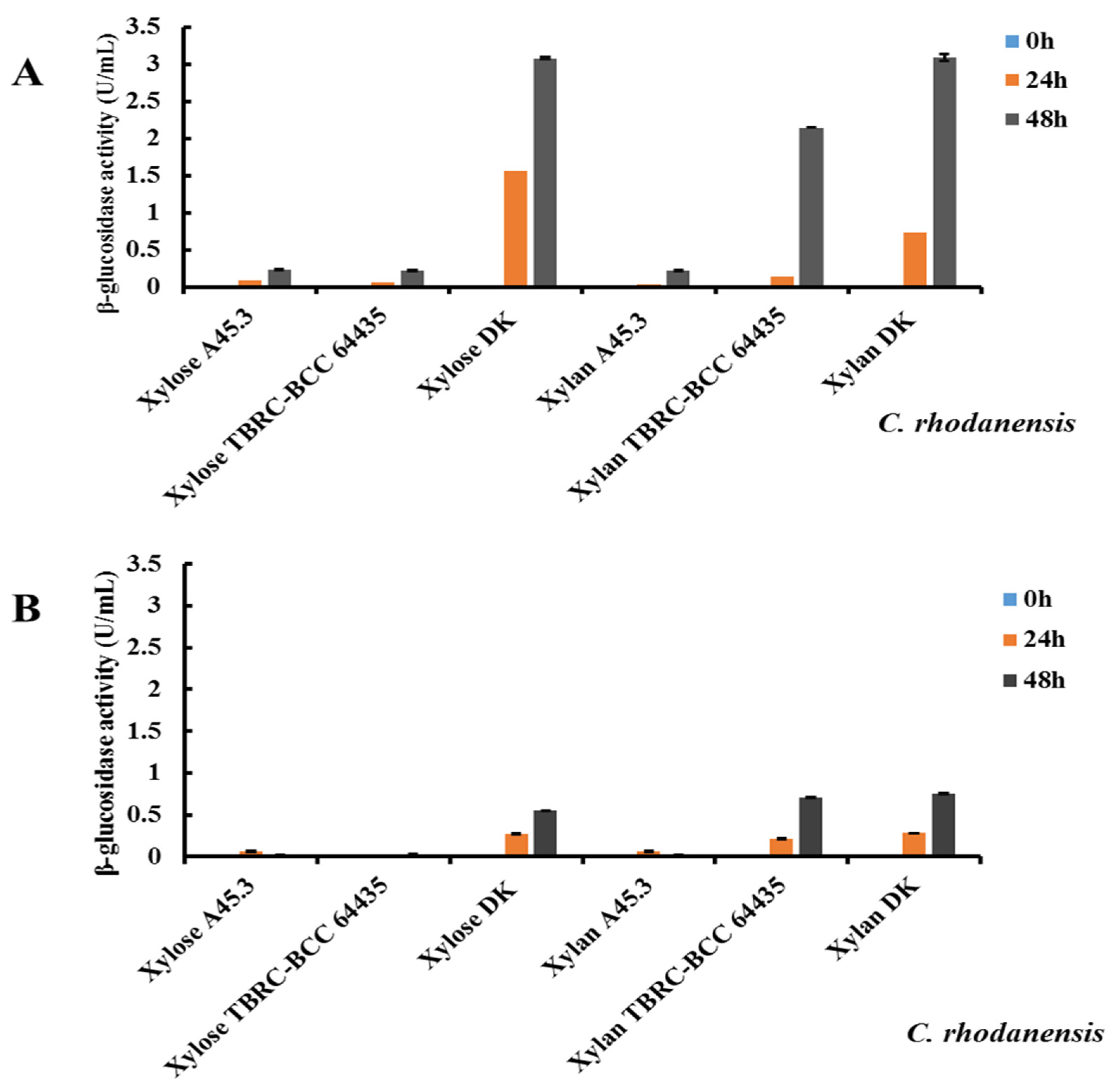
2.6. Screening of Carbon Sources for Extracellular β-Glucosidase Production by C. rhodanensis DK
2.7. Confirmation of the Main Glycosiase Activities in Culture Supernatant of C. rhodanensis DK
2.8. Comparison of Extracellular β-Glucosidase Production of C. rhodanensis DK with the Reference Strains
2.9. Statistical Medium Optimization for Extracellular β-Glucosidase Production Using Xylose and/or Xylan as the Sole Carbon Source
2.9.1. Plackett–Burman Design (PBD)
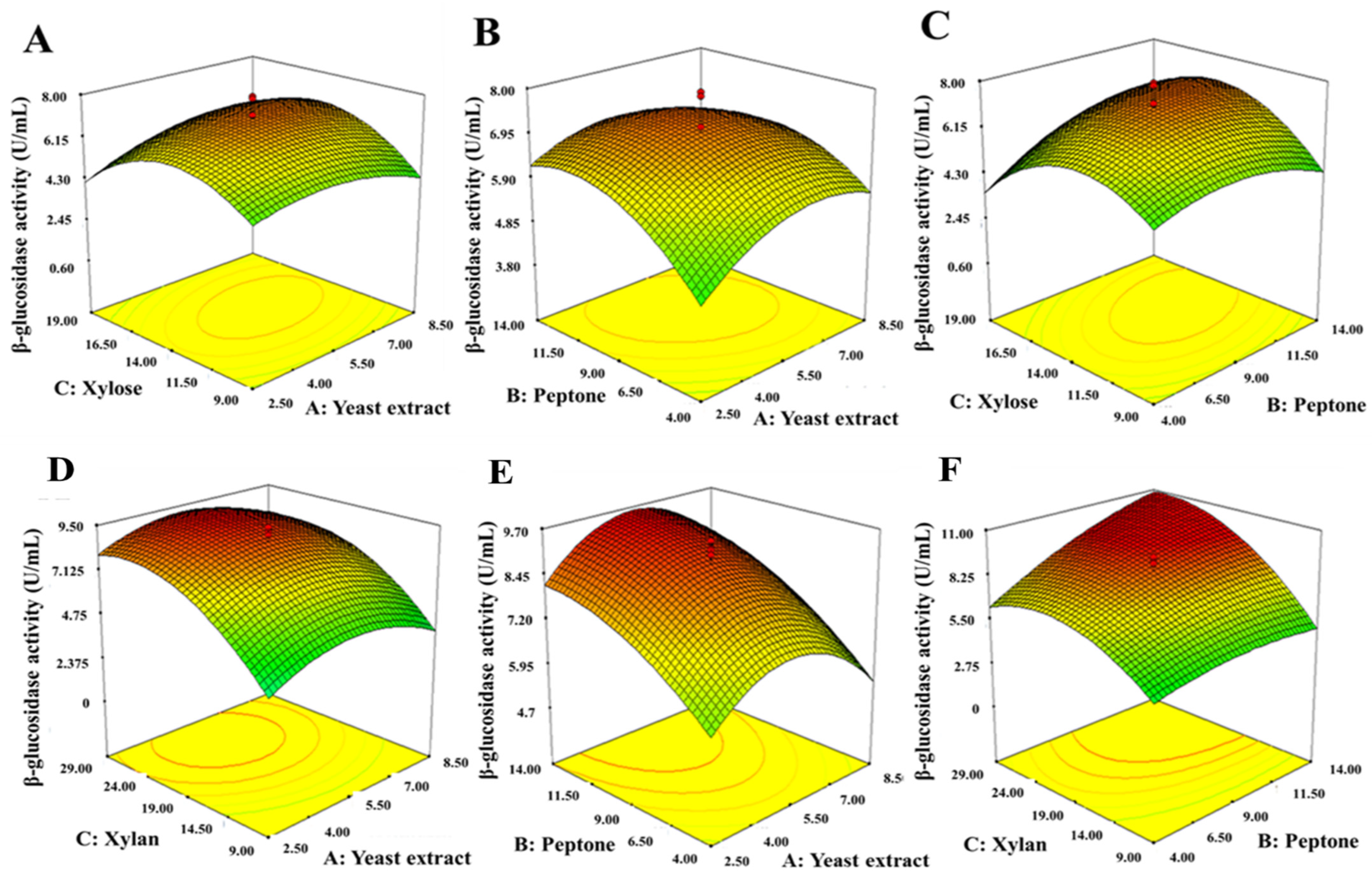
2.9.2. Central Composite Design (CCD) and Response Surface Methodology Analysis (RSM)
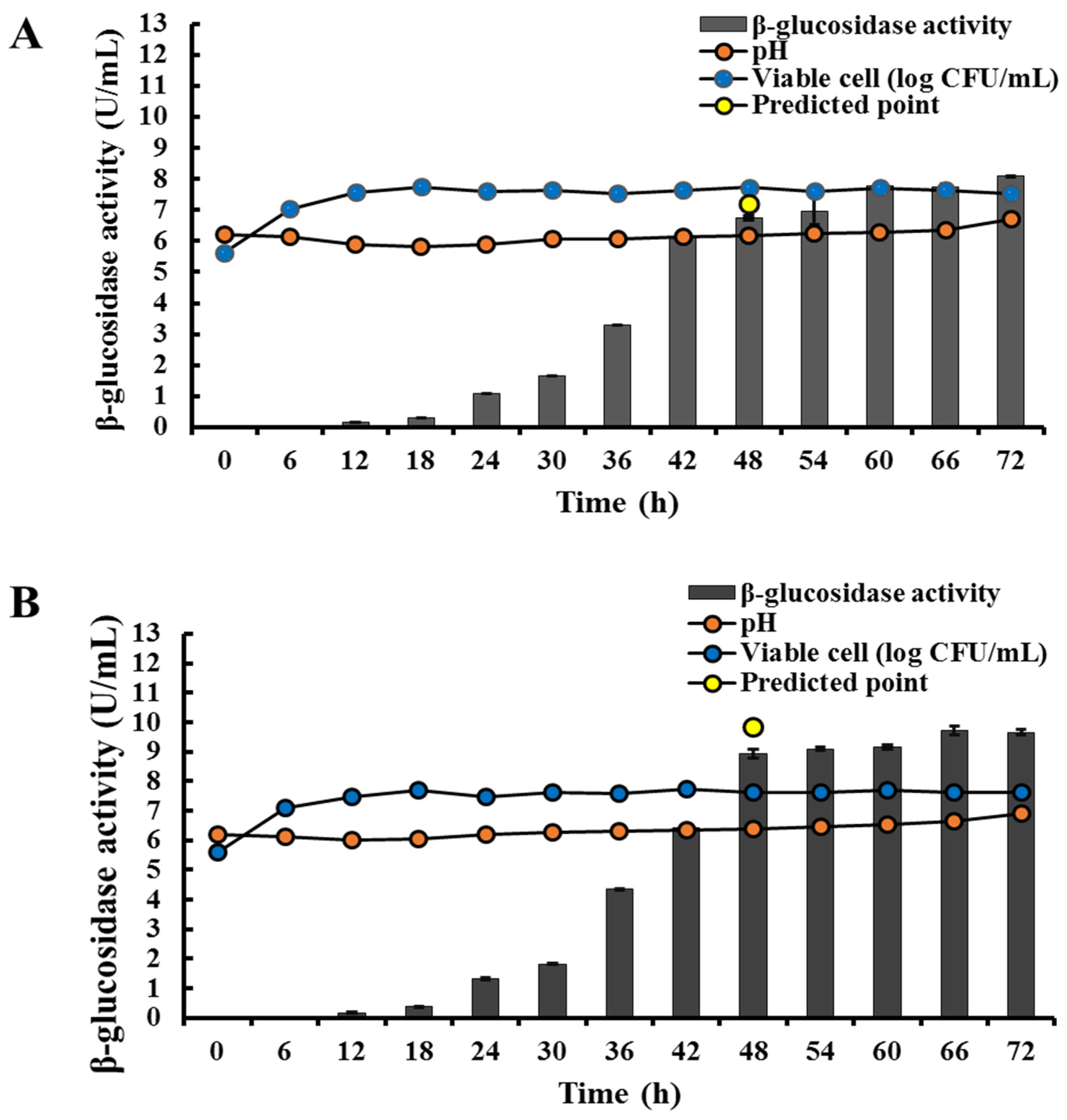
2.10. Thermostability Test
2.11. Statistical Analysis
3. Results and Discussion
3.1. Isolation of Thermotolerant Yeasts
3.2. Characterization of Thermotolerant Yeasts
3.2.1. Evaluation of Tannin-Tolerant Ability
3.2.2. Ethanol Producing Capability
3.2.3. Production of Polysaccharide Degrading Enzymes
3.2.4. Colony and Morphological Characteristics
3.2.5. Molecular Identification
3.3. Investigation of β-Glucosidase and CMCase Production by C. rhodanensis DK
3.4. Screening of Different Carbon Sources for Extracellular β-Glucosidase Production by C. rhodanensis DK
3.5. Confirmation of the Main β-Glucosidase Activities Produced by C. rhodanensis DK
3.6. Comparison of Extracellular β-Glucosidase Production by C. rhodanensis DK with the Reference Strains
3.7. Statistical Medium Optimization for Extracellular β-Glucosidase Production Using Xylose and/or Xylan as the Carbon Sources
3.7.1. Plackett–Burman Design (PBD)
3.7.2. Central Composite Design (CCD) and Response Surface Methodology Analysis (RSM)
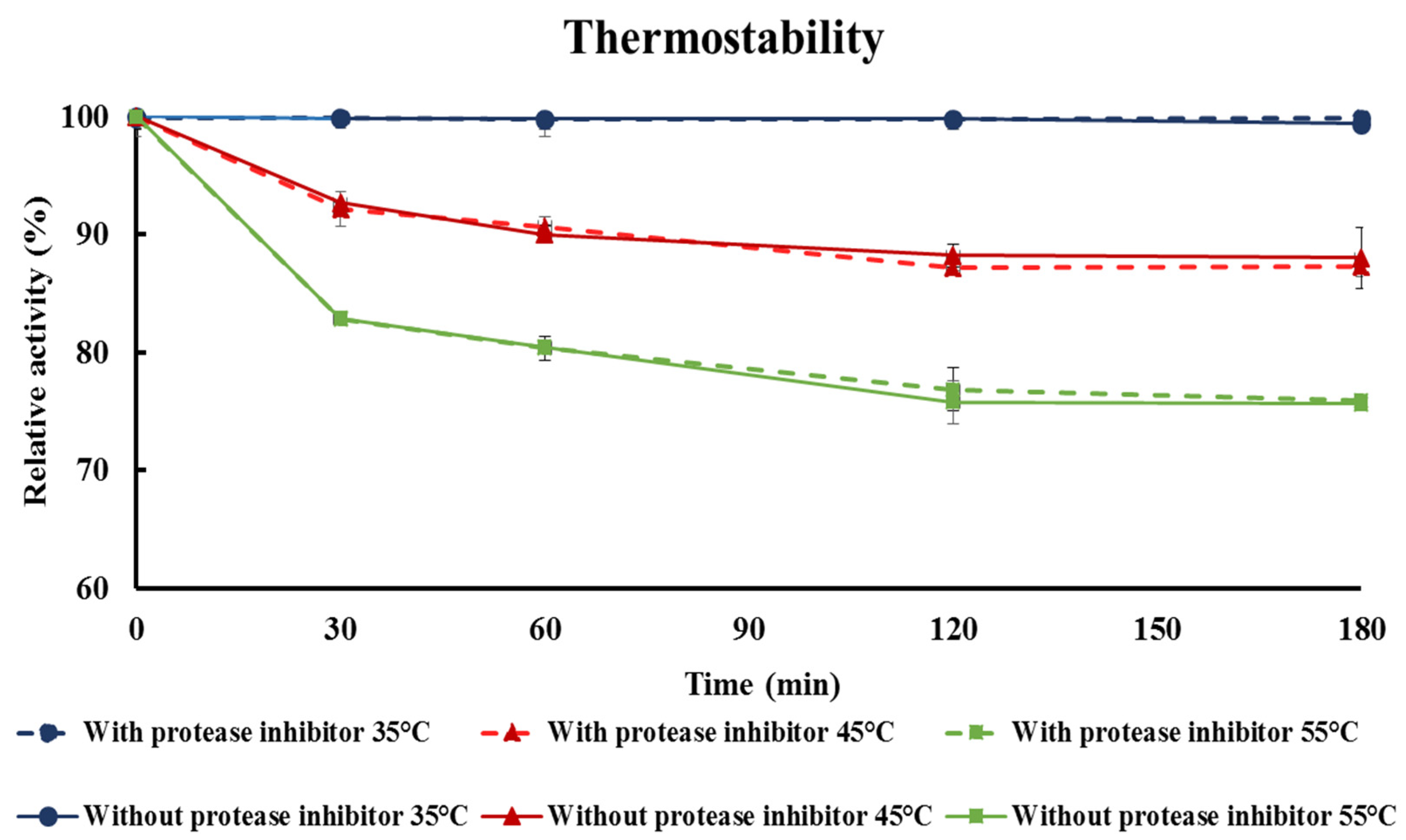
3.8. Thermostability Test
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Htay, H.H.; Kawai, M.; MacNaughton, L.E.; Katsuda, M.; Juneja, L.R. Tea in Myanmar, with special reference to pickled tea. J. Tea Sci. 2006, 5, 11–18. [Google Scholar]
- Han, T.; Aye, K.N. The Legend of Laphet: A Myanmar fermented tea leaf. J. Ethn. Foods 2015, 2, 173–178. [Google Scholar] [CrossRef]
- Maung, P.P.; He, Q.; Chamba, M.V.M. Comparison of polyphenol content between laboratory processed Laphet and China and Myanmar tea (Camellia sinensis) products. Pak. J. Food Sci. 2012, 22, 180–184. [Google Scholar]
- Bo, B.; Kim, S.-A.; Han, N.S. Bacterial and fungal diversity in Laphet, traditional fermented tea leaves in Myanmar, analyzed by culturing, DNA amplicon-based sequencing, and PCR-DGGE methods. Int. J. Food Microbiol. 2020, 320, 108508. [Google Scholar] [CrossRef]
- Khanongnuch, C.; Unban, K.; Kanpiengjai, A.; Saenjum, C. Recent research advances and ethno-botanical history of Miang, a traditional fermented tea (Camellia sinensis var. assamica) of northern Thailand. J. Ethn. Foods 2017, 4, 135–144. [Google Scholar] [CrossRef]
- Chaikaew, S.; Baipong, S.; Sone, T.; Kanpiengjai, A.; Chui-Chai, N.; Asano, K.; Khanongnuch, C. Diversity of lactic acid bacteria from Miang, a traditional fermented tea leaf in northern Thailand and their tannin-tolerant ability in tea extract. J. Microbiol. 2017, 55, 720–729. [Google Scholar] [CrossRef]
- Kanpiengjai, A.; Chui-Chai, N.; Chaikaew, S.; Khanongnuch, C. Distribution of tannin-tolerant yeasts isolated from Miang, a traditional fermented tea leaf (Camellia sinensis var. assamica) in northern Thailand. Int. J. Food Microbiol. 2016, 238, 121–131. [Google Scholar] [CrossRef]
- Akyereko, Y.G.; Yeboah, G.B.; Wireko-Manu, F.D.; Alemawor, F.; Mills-Robertson, F.C.; Odoom, W. Nutritional value and health benefits of cashew apple. JSFA Rep. 2023, 3, 110–118. [Google Scholar] [CrossRef]
- Kodchasee, P.; Nain, K.; Abdullahi, A.D.; Unban, K.; Saenjum, C.; Shetty, K.; Khanongnuch, C. Microbial dynamics-linked properties and functional metabolites during Miang fermentation using the filamentous fungi growth-based process. Food Biosci. 2021, 41, 100998. [Google Scholar] [CrossRef]
- Kodchasee, P.; Pharin, N.; Suwannarach, N.; Unban, K.; Saenjum, C.; Kanpiengjai, A.; Sakar, D.; Shetty, K.; Zarnkow, M.; Khanongnuch, C. Assessment of tannin tolerant non-Saccharomyces yeasts isolated from Miang for production of health-targeted beverage using Miang processing byproducts. J. Fungi 2023, 9, 165. [Google Scholar] [CrossRef]
- Unban, K.; Chaichana, W.; Baipong, S.; Abdullahi, A.D.; Kanpiengjai, A.; Shetty, K.; Khanongnuch, C. Probiotic and antioxidant properties of lactic acid bacteria isolated from indigenous fermented tea leaves (Miang) of north Thailand and promising application in synbiotic formulation. Fermentation 2021, 7, 195. [Google Scholar] [CrossRef]
- Thar, S.P. An Exploratory Value Chain Analysis for Burmese Pickled Tea (LAPHET): A Thesis Presented in Partial Fulfilment of the Requirements for the Degree of Masters of Agricommerce in Agribusiness, Institute of Agriculture and Environment, Massey University, Palmerston North, New Zealand. Ph.D. Thesis, Massey University, Palmerston North, New Zealand, 2016. [Google Scholar]
- Huang, C.-J.; Lu, M.-Y.; Chang, Y.-W.; Li, W.-H. Experimental evolution of yeast for high-temperature tolerance. Mol. Biol. Evol. 2018, 35, 1823–1839. [Google Scholar] [CrossRef] [PubMed]
- Yanase, S.; Hasunuma, T.; Yamada, R.; Tanaka, T.; Ogino, C.; Fukuda, H.; Kondo, A. Direct ethanol production from cellulosic materials at high temperature using the thermotolerant yeast Kluyveromyces marxianus displaying cellulolytic enzymes. Appl. Microbiol. Biotechnol. 2010, 88, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, Y.; Mishra, S.; Bisaria, V.S. Microbial β-glucosidases: Cloning, properties, and applications. Crit. Rev. Biotechnol. 2002, 22, 375–407. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.A.; Mishra, S.; Chaudhuri, T.K. Structural stability and unfolding transition of β-glucosidases: A comparative investigation on isozymes from a thermo-tolerant yeast. Eur. Biophys. J. 2011, 40, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Bibi, A. Fungal cellulase; production and applications: Minireview. LIFE Int. J. Health Life Sci. 2018, 4, 19–36. [Google Scholar] [CrossRef]
- Raveendran, S.; Parameswaran, B.; Ummalyma, S.B.; Abraham, A.; Mathew, A.K.; Madhavan, A.; Rebello, S.; Pandey, A. Applications of microbial enzymes in food industry. Food Technol. Biotechnol. 2018, 56, 16–30. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhu, Y.; Lu, M.; Yang, C.; Xie, D.; Tan, J.; Peng, Q.; Zhang, Y.; Ni, D.; Dai, W.; et al. Variation patterns in the content of glycosides during green tea manufacturing by a modification-specific metabolomics approach: Enzymatic reaction promoting an increase in the glycosidically bound volatiles at the pan firing stage. Food Chem. 2019, 279, 80–87. [Google Scholar] [CrossRef]
- Salgado, J.C.S.; Meleiro, L.P.; Carli, S.; Ward, R.J. Glucose tolerant and glucose stimulated β-glucosidases—A review. Bioresour. Technol. 2018, 267, 704–713. [Google Scholar] [CrossRef]
- Singh, G.; Verma, A.K.; Kumar, V. Catalytic properties, functional attributes and industrial applications of β-glucosidases. 3 Biotech 2016, 6, 3. [Google Scholar] [CrossRef]
- Longo, M.; Sanromán, M.A. Production of food aroma compounds: Microbial and enzymatic methodologies. Food Technol. Biotechnol. 2006, 44, 335–353. [Google Scholar]
- Ni, H.; Jiang, Q.; Lin, Q.; Ma, Q.; Wang, L.; Weng, S.; Huang, G.; Li, L.; Chen, F. Enzymatic hydrolysis and auto-isomerization during β-glucosidase treatment improve the aroma of instant white tea infusion. Food Chem. 2021, 342, 128565. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Sun, J.; Lassabliere, B.; Yu, B.; Liu, S.Q. β-Glucosidase activity of Cyberlindnera (Williopsis) saturnus var. mrakii NCYC 2251 and its fermentation effect on green tea aroma compounds. LWT 2021, 151, 112184. [Google Scholar] [CrossRef]
- Sirilun, S.; Chaiyasut, C.; Pengkumsri, N.; Peerajan, S.; Chaiyasut, K.; Suwannalert, P.; Sivamaruthi, B. Screening and characterization of beta-glucosidase production by Saccharomyces cerevisiae. J. Appl. Pharm. Sci. 2016, 6, 029–035. [Google Scholar] [CrossRef][Green Version]
- Ahamed, A.; Vermette, P. Enhanced enzyme production from mixed cultures of Trichoderma reesei RUT-C30 and Aspergillus niger LMA grown as fed batch in a stirred tank bioreactor. Biochem. Eng. J. 2008, 42, 41–46. [Google Scholar] [CrossRef]
- Singhania, R.R.; Patel, A.K.; Pandey, A.; Ganansounou, E. Genetic Modification: A tool for enhancing beta-glucosidase production for biofuel application. Bioresour. Technol. 2017, 245, 1352–1361. [Google Scholar] [CrossRef] [PubMed]
- Uchima, C.A.; Tokuda, G.; Watanabe, H.; Kitamoto, K.; Arioka, M. Heterologous expression in Pichia pastoris and characterization of an endogenous thermostable and high-glucose-tolerant β-glucosidase from the termite Nasutitermes takasagoensis. Appl. Environ. Microbiol. 2012, 78, 4288–4293. [Google Scholar] [CrossRef]
- Keo-oudone, C.; Nitiyon, S.; Sotitham, P.; Tani, A.; Lertwattanasakul, N.; Yuangsaard, N.; Bounphanmy, S.; Limtong, S.; Yamada, M. Isolation and characterization of thermotolerant ethanol-fermenting yeasts from Laos and application of whole-cell matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF/MS) analysis for their quick identification. Afr. J. Biotechnol. 2016, 15, 153–164. [Google Scholar]
- Leangnim, N.; Aisara, J.; Unban, K.; Khanongnuch, C.; Kanpiengjai, A. Acid stable yeast cell-associated tannase with high capability in gallated catechin biotransformation. Microorganisms 2021, 9, 1418. [Google Scholar] [CrossRef]
- Zhang, T.; Fang, K.; Ni, H.; Li, T.; Li, L.J.; Li, Q.B.; Chen, F. Aroma enhancement of instant green tea infusion using β-glucosidase and β-xylosidase. Food Chem. 2020, 315, 126287. [Google Scholar] [CrossRef]
- Phong, H.X.; Klanrit, P.; Dung, N.T.P.; Yamada, M.; Thanonkeo, P. Isolation and characterization of thermotolerant yeasts for the production of second-generation bioethanol. Ann. Microbiol. 2019, 69, 765–776. [Google Scholar] [CrossRef]
- Hung, P.Q.; Kumar, S.M.; Govindsamy, V.; Annapurna, K. Isolation and characterization of endophytic bacteria from wild and cultivated soybean varieties. Biol. Fertil. Soils 2007, 44, 155–162. [Google Scholar] [CrossRef]
- Pranay, K.; Padmadeo, S.R.; Prasad, B. Production of amylase from Bacillus subtilis sp. strain KR1 under solid state fermentation on different agrowastes. Biocatal. Agric. Biotechnol. 2019, 21, 101300. [Google Scholar] [CrossRef]
- Kurtzman, C.P.; Robnett, C.J. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26s) ribosomal DNA gene. J. Clin. Microbiol. 1997, 35, 1216–1223. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, A.; Andersen, J.J.; Ahring, B.K.; Teller, P.J.; Lübeck, M. Screening of carbon sources for beta-glucosidase production by Aspergillus saccharolyticus. Int. Biodeterior. Biodegrad. 2014, 93, 78–83. [Google Scholar] [CrossRef]
- Miller, G.L. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Pamueangmun, P.; Abdullahi, A.D.; Kabir, M.H.; Unban, K.; Kanpiengjai, A.; Venus, J.; Shetty, K.; Saenjum, C.; Khanongnuch, C. Lignocellulose degrading Weizmannia coagulans capable of enantiomeric L-lactic acid production via consolidated bioprocessing. Fermentation 2023, 9, 761. [Google Scholar] [CrossRef]
- Yadav, S.; Pandey, A.K.; Dubey, S.K. Molecular modeling, docking and simulation dynamics of β-glucosidase reveals high-efficiency, thermo-stable, glucose tolerant enzyme in Paenibacillus lautus BHU3 strain. Int. J. Biol. Macromol. 2021, 168, 371–382. [Google Scholar] [CrossRef]
- Aouine, M.; Elalami, D.; Koraichi, S.I.; Haggoud, A.; Barakat, A. Exploring natural fermented foods as a source for new efficient thermotolerant yeasts for the production of second-generation bioethanol. Energies 2022, 15, 4954. [Google Scholar] [CrossRef]
- Talukder, A.A.; Easmin, F.; Mahmud, S.A.; Yamada, M. Thermotolerant yeasts capable of producing bioethanol: Isolation from natural fermented sources, identification and characterization. Biotechnol. Biotechnol. Equip. 2016, 30, 1106–1114. [Google Scholar] [CrossRef]
- Choi, D.-H.; Park, E.-H.; Kim, M.-D. Isolation of thermotolerant Yeast Pichia kudriavzevii from Nuruk. Food Sci. Biotechnol. 2017, 26, 1357–1362. [Google Scholar] [CrossRef] [PubMed]
- Kaewkrajay, C.; Dethoup, T.; Limtong, S. Ethanol production from cassava using a newly isolated thermotolerant yeast strain. ScienceAsia 2014, 40, 268–277. [Google Scholar] [CrossRef]
- Khasnabis, J.; Rai, C.; Roy, A. Determination of tannin content by titrimetric method from different types of tea. J. Chem. Pharm. Res. 2015, 7, 238–241. [Google Scholar]
- Kanpiengjai, A.; Kodchasee, P.; Unban, K.; Kumla, J.; Lumyong, S.; Khunnamwong, P.; Sarkar, D.; Shetty, K.; Khanongnuch, C. Three new yeast species from flowers of Camellia sinensis var. assamica collected in northern Thailand and their tannin tolerance characterization. Front. Microbiol. 2023, 14, 1043430. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Wang, N.-N.; Yin, X.-J.; Liang, X.-L.; Wang, Z.-P. Characterization of a robust and pH-stable tannase from mangrove-derived yeast Rhodosporidium diobovatum Q95. Mar. Drugs 2020, 18, 546. [Google Scholar] [CrossRef]
- Song, L.; Wang, X.-C.; Feng, Z.-Q.; Guo, Y.-F.; Meng, G.-Q.; Wang, H.-Y. Biotransformation of gallate esters by a pH-stable tannase of mangrove-derived yeast Debaryomyces hansenii. Front. Molec. Biosci. 2023, 10, 1211621. [Google Scholar] [CrossRef]
- Zhang, L.-L.; Li, J.; Wang, Y.-L.; Liu, S.; Wang, Z.-P.; Yu, X.-J. Integrated approaches to reveal genes crucial for tannin degradation in Aureobasidium melanogenum T9. Biomolecules 2019, 9, 439. [Google Scholar] [CrossRef]
- Choudhary, J.; Singh, S.; Nain, L. Thermotolerant fermenting yeasts for simultaneous saccharification fermentation of lignocellulosic biomass. Electron. J. Biotechnol. 2016, 21, 82–92. [Google Scholar] [CrossRef]
- Techaparin, A.; Thanonkeo, P.; Klanrit, P. High-temperature ethanol production using thermotolerant yeast newly isolated from greater Mekong subregion. Braz. J. Microbiol. 2017, 48, 461–475. [Google Scholar] [CrossRef]
- Waghmare, P.R.; Kshirsagar, S.D.; Saratale, R.G.; Govindwar, S.P.; Saratale, G.D. Production and characterization of cellulolytic enzymes by isolated Klebsiella sp. prw-1 using agricultural waste biomass. Emir. J. Food Agric. 2014, 26, 44–59. [Google Scholar] [CrossRef]
- Delgado-Ospina, J.; Triboletti, S.; Alessandria, V.; Serio, A.; Sergi, M.; Paparella, A.; Rantsiou, K.; Chaves-López, C. Functional biodiversity of yeasts isolated from Colombian fermented and dry cocoa beans. Microorganisms 2020, 8, 1086. [Google Scholar] [CrossRef]
- Rai, P.; Tiwari, S.; Gaur, R. Optimization of process parameters for cellulase production by novel thermotolerant yeast. BioResources 2012, 7, 5401–5414. [Google Scholar] [CrossRef]
- Adelabu, B.; Kareem, S.O.; Adeogun, A.I.; Wakil, S.M. Optimization of cellulase enzyme from sorghum straw by yeasts isolated from plant feeding-termite Zonocerus Variegatus: Production of cellulase enzyme. Food Appl. Biosci. J. 2019, 7, 81–101. [Google Scholar]
- Sousa, A.M.; Machado, I.; Nicolau, A.; Pereira, M.O. Improvements on colony morphology identification towards bacterial profiling. J. Microbiol. Methods 2013, 95, 327–335. [Google Scholar] [CrossRef]
- Bourdichon, F.; Casaregola, S.; Farrokh, C.; Frisvad, J.C.; Gerds, M.L.; Hammes, W.P.; Harnett, J.; Huys, G.; Laulund, S.; Ouwehand, A.; et al. Food fermentations: Microorganisms with technological beneficial use. Int. J. Food Microbiol. 2012, 154, 87–97. [Google Scholar] [CrossRef]
- Burns, R.G.; Dick, R.P. Enzymes in the Environment: Activity, Ecology, and Applications; CRC Press: Boca Raton, FL, USA, 2002; ISBN 978-0-203-90403-9. [Google Scholar]
- Saritha Mohanram, V.R. Beta-Glucosidase: Key enzyme in determining efficiency of cellulase and biomass hydrolysis. J Bioprocess Biotech. 2015, 5, 197. [Google Scholar] [CrossRef]
- Chuengcharoenphanich, N.; Watsuntorn, W.; Qi, W.; Wang, Z.; Hu, Y.; Chulalaksananukul, W. The potential of biodiesel production from grasses in Thailand through consolidated bioprocessing using a cellulolytic oleaginous yeast, Cyberlindnera rhodanensis CU-CV7. Energy 2023, 263, 125759. [Google Scholar] [CrossRef]
- He, Y.; Wang, C.; Jiao, R.; Ni, Q.; Wang, Y.; Gao, Q.; Zhang, Y.; Xu, G. Biochemical characterization of a novel glucose-tolerant GH3 β-glucosidase (Bgl1973) from Leifsonia sp. ZF2019. Appl. Microbiol. Biotechnol. 2022, 106, 5063–5079. [Google Scholar] [CrossRef]
- Eisenmesser, E.Z.; Bosco, D.A.; Akke, M.; Kern, D. Enzyme dynamics during catalysis. Science 2002, 295, 1520–1523. [Google Scholar] [CrossRef]
- Somero, G.N. Adaptation of enzymes to temperature: Searching for basic “Strategies”. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2004, 139, 321–333. [Google Scholar] [CrossRef]
- Corrêa, T.L.R.; Franco Cairo, J.P.L.; Cota, J.; Damasio, A.; Oliveira, L.C.; Squina, F.M. A novel mechanism of β-glucosidase stimulation through a monosaccharide binding-induced conformational change. Int. J. Biol. Macromol. 2021, 166, 1188–1196. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, S.; Demain, A.L. Metabolic regulation and overproduction of primary metabolites. Microb. Biotechnol. 2008, 1, 283–319. [Google Scholar] [CrossRef] [PubMed]
- Zanoelo, F.F.; Polizeli, M.d.L.T.d.M.; Terenzi, H.F.; Jorge, J.A. Beta-glucosidase activity from the thermophilic fungus Scytalidium thermophilum is stimulated by glucose and xylose. FEMS Microbiol. Lett. 2004, 240, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Bonfá, E.C.; De Souza Moretti, M.M.; Gomes, E.; Bonilla-Rodriguez, G.O. Biochemical characterization of an isolated 50 kda beta-glucosidase from the thermophilic fungus Myceliophthora thermophila M.7.7. Biocatal. Agric. Biotechnol. 2018, 13, 311–318. [Google Scholar] [CrossRef]
- Bhalla, A.; Bischoff, K.M.; Sani, R.K. Highly thermostable GH39 β-xylosidase from a Geobacillus sp. Strain WSUCF1. BMC Biotechnol. 2014, 14, 963. [Google Scholar] [CrossRef][Green Version]
- Masoud, W.; Jespersen, L. Pectin degrading enzymes in yeasts involved in fermentation of Coffea arabica in east Africa. Int. J. Food Microbiol. 2006, 110, 291–296. [Google Scholar] [CrossRef]
- Pietikäinen, J.; Pettersson, M.; Bååth, E. Comparison of temperature effects on soil respiration and bacterial and fungal growth rates. FEMS Microbiol. Ecol. 2005, 52, 49–58. [Google Scholar] [CrossRef]
- Fendt, S.-M.; Sauer, U. Transcriptional regulation of respiration in yeast metabolizing differently repressive carbon substrates. BMC Syst. Biology 2010, 4, 12. [Google Scholar] [CrossRef]
- Landry, C.R.; Oh, J.; Hartl, D.L.; Cavalieri, D. Genome-wide scan reveals that genetic variation for transcriptional plasticity in yeast is biased towards multi-copy and dispensable genes. Gene 2006, 366, 343–351. [Google Scholar] [CrossRef]
- Unban, K.; Puangkhankham, N.; Kanpiengjai, A.; Govindarajan, R.K.; Kalaimurugan, D.; Khanongnuch, C. Improvement of polymer grade L-lactic acid production using Lactobacillus rhamnosus SCJ9 from low-grade cassava chips by simultaneous saccharification and fermentation. Processes 2020, 8, 1143. [Google Scholar] [CrossRef]
- Analytical Methods Committee. AMCTB No 55. Experimental design and optimisation (4): Plackett–Burman designs. Anal. Methods 2013, 5, 1901–1903. [Google Scholar] [CrossRef]
- Mahapatra, S.; Manian, R. Enhancement, production, and immobilization of beta-glucosidase from Zobellella denitrificans VIT SB117 and its utilization in bioethanol production from lignocellulosic feedstock. Biomass Conv. Bioref. 2022, 12, 447–458. [Google Scholar] [CrossRef]
- Huang, C.; Feng, Y.; Patel, G.; Xu, X.-Q.; Qian, J.; Liu, Q.; Kai, G.-Y. Production, immobilization and characterization of beta-glucosidase for application in cellulose degradation from a novel Aspergillus versicolor. Int. J. Biol. Macromol. 2021, 177, 437–446. [Google Scholar] [CrossRef]
- Dutta, S.G.; Shaik, A.B.; Ganesh Kumar, C.; Kamal, A. Statistical optimization of production conditions of β-glucosidase from Bacillus stratosphericus strain SG9. 3 Biotech 2017, 7, 221. [Google Scholar] [CrossRef] [PubMed]
- Nanjundaswamy, A.; Okeke, B.C. Comprehensive optimization of culture conditions for production of biomass-hydrolyzing enzymes of trichoderma SG2 in submerged and solid-state fermentation. Appl. Biochem. Biotechnol. 2020, 191, 444–462. [Google Scholar] [CrossRef]
- Tasharrofi, N.; Adrangi, S.; Fazeli, M.; Rastegar, H.; Khoshayand, M.R.; Faramarzi, M.A. Optimization of chitinase production by Bacillus pumilus using Plackett-Burman Design and Response Surface Methodology. Iran J. Pharm. Res. 2011, 10, 759–768. [Google Scholar] [PubMed]
- Javed, M.M.; Siddiq, Z.; Saleem, T. Triggering of β-glucosidase production in Trichoderma viride with nutritional and environmental control. J. Appl. Sci. Res. 2006, 2, 884–889. [Google Scholar]
- El-Naggar, N.E.-A.; Haroun, S.A.; Owis, E.A.; Sherief, A.A. Optimization of β-glucosidase production by Aspergillus terreus strain EMOO 6-4 using Response Surface Methodology under solid-state fermentation. Prep. Biochem. Biotechnol. 2015, 45, 568–587. [Google Scholar] [CrossRef] [PubMed]
- Fawzi, E. Production and Purification of β-glucosidase and protease by Fusarium proliferatum NRRL 26517 grown on Ficus nitida wastes. Ann. Microbiol. 2003, 53, 463–476. [Google Scholar]
- Gong, G.; Zheng, Z.; Liu, H.; Wang, L.; Diao, J.; Wang, P.; Zhao, G. Purification and characterization of a β-glucosidase from Aspergillus niger and its application in the hydrolysis of geniposide to genipin. J. Microbiol. Biotechnol. 2014, 24, 788–794. [Google Scholar] [CrossRef]
- Jaenicke, R. Protein stability and molecular adaptation to extreme conditions. Eur. J. Biochem. 1991, 202, 715–728. [Google Scholar] [CrossRef] [PubMed]
- García-Carreño, F.L. Protease inhibition in theory and practice. Biotechnol. Educ. 1992, 3, 145–150. [Google Scholar]


| Isolates | Tannin- Tolerant Ability | Cellulases | β- Mannanase | Pectinase | Xylanase | Amylase | Ethanol Production |
|---|---|---|---|---|---|---|---|
| DK | ++ | + | − | − | − | − | + |
| MD1 | ++ | + | − | − | − | − | − |
| MD2 | ++ | + | − | − | − | − | − |
| MD3 | +++ | + | − | − | − | − | − |
| MD6 | +++ | + | − | − | − | − | − |
| MD7 | ++ | + | − | − | − | − | − |
| MD8 | ++ | + | − | − | − | − | − |
| MD12 | +++ | + | − | − | − | − | − |
| MD14 | +++ | + | − | − | − | − | − |
| MD20 | +++ | + | − | − | − | − | − |
| MD21 | ++ | + | − | − | − | − | − |
| MD23 | ++ | + | − | − | − | − | − |
| MD24 | ++ | + | − | − | − | − | − |
| TN1 | ++ | + | − | − | − | − | − |
| TN2 | ++ | + | − | − | − | − | − |
| TN4 | ++ | + | − | − | − | − | − |
| TN5 | +++ | + | − | − | − | − | − |
| TN7 | ++ | + | − | − | − | − | − |
| Run | A: Yeast Extract (g/L) | B: Peptone (g/L) | C: Malt Extract (g/L) | D: KH2PO4 (g/L) | E: MgSO4 (g/L) | F: Xylose or Xylan (g/L) | β-Glucosidase (Xylose) (U/mL) | β-Glucosidase (Xylan) (U/mL) |
|---|---|---|---|---|---|---|---|---|
| 1 | 5.5 | 9 | 0.5 | 1.5 | 0.9 | 19 | 7.76 | 4.87 |
| 2 | 0.5 | 9 | 5.5 | 0.5 | 0.9 | 19 | 1.00 | 1.85 |
| 3 | 5.5 | 1 | 5.5 | 1.5 | 0.1 | 19 | 0.62 | 2.91 |
| 4 | 0.5 | 9 | 0.5 | 1.5 | 0.9 | 1 | 0.02 | 0.02 |
| 5 | 0.5 | 1 | 5.5 | 0.5 | 0.9 | 19 | 0.11 | 1.51 |
| 6 | 0.5 | 1 | 0.5 | 1.5 | 0.1 | 19 | 0.05 | 0.09 |
| 7 | 5.5 | 1 | 0.5 | 0.5 | 0.9 | 1 | 0.03 | 0.05 |
| 8 | 5.5 | 9 | 0.5 | 0.5 | 0.1 | 19 | 7.56 | 6.57 |
| 9 | 5.5 | 9 | 5.5 | 0.5 | 0.1 | 1 | 0.98 | 0.35 |
| 10 | 0.5 | 9 | 5.5 | 1.5 | 0.1 | 1 | 1.57 | 0.17 |
| 11 | 5.5 | 1 | 5.5 | 1.5 | 0.9 | 1 | 0.05 | 0.06 |
| 12 | 0.5 | 1 | 0.5 | 0.5 | 0.1 | 1 | 0.03 | 0.06 |
| 13 | 3 | 5 | 3 | 1 | 0.5 | 10 | 2.43 | 3.67 |
| 14 | 3 | 5 | 3 | 1 | 0.5 | 10 | 2.89 | 3.76 |
| 15 | 3 | 5 | 3 | 1 | 0.5 | 10 | 3.39 | 3.36 |
| Variable | Xylose | Xylan | ||
|---|---|---|---|---|
| Coefficient | p-Value | Coefficient | p-Value | |
| Intercept | 1.69 | 0.0308 | 1.54 | 0.0066 |
| Yeast extract | 1.14 | 0.0417 | 0.93 | 0.0095 |
| Peptone | 1.46 | 0.0155 | 0.70 | 0.0212 |
| Malt extract | −0.97 | 0.0729 | −0.40 | 0.01557 |
| MgSO4 | −0.19 | 0.6851 | −0.19 | 0.5659 |
| KH2PO4 | 0.071 | 0.8814 | −0.15 | 0.4725 |
| Xylose | 1.24 | 0.0302 | - | - |
| Xylan | - | - | 1.42 | 0.0012 |
| R2 = 0.8016 | R2 = 0.9188 | |||
| Run | A: Yeast Extract (g/L) | B: Peptone (g/L) | C: Xylose (g/L) | β-Glucosidase (U/mL) |
|---|---|---|---|---|
| 1 | 2.5(−1) | 4(−1) | 9(−1) | 4.44 |
| 2 | 8.5(+1) | 4(−1) | 9(−1) | 4.83 |
| 3 | 2.5(−1) | 14(+1) | 9(−1) | 4.95 |
| 4 | 8.5(+1) | 14(+1) | 9(−1) | 4.64 |
| 5 | 2.5(−1) | 4(−1) | 29(+1) | 1.32 |
| 6 | 8.5(+1) | 4(−1) | 29(+1) | 4.01 |
| 7 | 2.5(−1) | 14(+1) | 29(+1) | 5.06 |
| 8 | 8.5(+1) | 14(+1) | 29(+1) | 6.04 |
| 9 | 0.45(−α) | 9(0) | 19(0) | 4.69 |
| 10 | 10.55(+α) | 9(0) | 19(0) | 4.17 |
| 11 | 5.5(0) | 0.59(−α) | 19(0) | 3.86 |
| 12 | 5.5(0) | 17.41(+α) | 19(0) | 4.52 |
| 13 | 5.5(0) | 9(0) | 2.18(−α) | 0.62 |
| 14 | 5.5(0) | 9(0) | 35(+α) | 2.90 |
| 15 | 5.5(0) | 9(0) | 19(0) | 6.89 |
| 16 | 5.5(0) | 9(0) | 19(0) | 6.25 |
| 17 | 5.5(0) | 9(0) | 19(0) | 7.91 |
| 18 | 5.5(0) | 9(0) | 19(0) | 7.82 |
| 19 | 5.5(0) | 9(0) | 19(0) | 7.12 |
| 20 | 5.5(0) | 9(0) | 19(0) | 6.79 |
| Run | A: Yeast Extract (g/L) | B: Peptone (g/L) | C: Xylan (g/L) | β-Glucosidase (U/mL) |
|---|---|---|---|---|
| 1 | 2.5(−1) | 4(−1) | 9(−1) | 3.30 |
| 2 | 8.5(+1) | 4(−1) | 9(−1) | 3.24 |
| 3 | 2.5(−1) | 14(+1) | 9(−1) | 4.11 |
| 4 | 8.5(+1) | 14(+1) | 9(−1) | 4.23 |
| 5 | 2.5(−1) | 4(−1) | 29(+1) | 4.99 |
| 6 | 8.5(+1) | 4(−1) | 29(+1) | 4.11 |
| 7 | 2.5(−1) | 14(+1) | 29(+1) | 9.45 |
| 8 | 8.5(+1) | 14(+1) | 29(+1) | 9.34 |
| 9 | 0.45(−α) | 9(0) | 19(0) | 4.92 |
| 10 | 10.55(+α) | 9(0) | 19(0) | 4.76 |
| 11 | 5.5(0) | 0.59(−α) | 19(0) | 5.53 |
| 12 | 5.5(0) | 17.41(+α) | 19(0) | 8.94 |
| 13 | 5.5(0) | 9(0) | 2.18(−α) | 0.07 |
| 14 | 5.5(0) | 9(0) | 35(+α) | 7.99 |
| 15 | 5.5(0) | 9(0) | 19(0) | 9.18 |
| 16 | 5.5(0) | 9(0) | 19(0) | 8.29 |
| 17 | 5.5(0) | 9(0) | 19(0) | 9.13 |
| 18 | 5.5(0) | 9(0) | 19(0) | 8.31 |
| 19 | 5.5(0) | 9(0) | 19(0) | 9.37 |
| 20 | 5.5(0) | 9(0) | 19(0) | 8.98 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kham, N.N.N.; Phovisay, S.; Unban, K.; Kanpiengjai, A.; Saenjum, C.; Lumyong, S.; Shetty, K.; Khanongnuch, C. A Thermotolerant Yeast Cyberlindnera rhodanensis DK Isolated from Laphet-so Capable of Extracellular Thermostable β-Glucosidase Production. J. Fungi 2024, 10, 243. https://doi.org/10.3390/jof10040243
Kham NNN, Phovisay S, Unban K, Kanpiengjai A, Saenjum C, Lumyong S, Shetty K, Khanongnuch C. A Thermotolerant Yeast Cyberlindnera rhodanensis DK Isolated from Laphet-so Capable of Extracellular Thermostable β-Glucosidase Production. Journal of Fungi. 2024; 10(4):243. https://doi.org/10.3390/jof10040243
Chicago/Turabian StyleKham, Nang Nwet Noon, Somsay Phovisay, Kridsada Unban, Apinun Kanpiengjai, Chalermpong Saenjum, Saisamorn Lumyong, Kalidas Shetty, and Chartchai Khanongnuch. 2024. "A Thermotolerant Yeast Cyberlindnera rhodanensis DK Isolated from Laphet-so Capable of Extracellular Thermostable β-Glucosidase Production" Journal of Fungi 10, no. 4: 243. https://doi.org/10.3390/jof10040243
APA StyleKham, N. N. N., Phovisay, S., Unban, K., Kanpiengjai, A., Saenjum, C., Lumyong, S., Shetty, K., & Khanongnuch, C. (2024). A Thermotolerant Yeast Cyberlindnera rhodanensis DK Isolated from Laphet-so Capable of Extracellular Thermostable β-Glucosidase Production. Journal of Fungi, 10(4), 243. https://doi.org/10.3390/jof10040243







