Identification and Aggressiveness of Fusarium Species Associated with Onion Bulb (Allium cepa L.) during Storage
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Identification of Pathogens from Diseased Onion Bulbs
2.2. Molecular Analysis
2.2.1. DNA Isolation
2.2.2. DNA Fingerprinting Analysis (BOX-PCR)
2.2.3. Molecular Fungal Species Identification and Phylogenetic Relationships
2.3. Test of Pathogenicity
2.3.1. Preparation of Conidial Suspension of Isolated Fungal Strains
2.3.2. Pathogenicity Assay of Onion Seeds
2.3.3. Pathogenicity Assay of Onion Bulbs
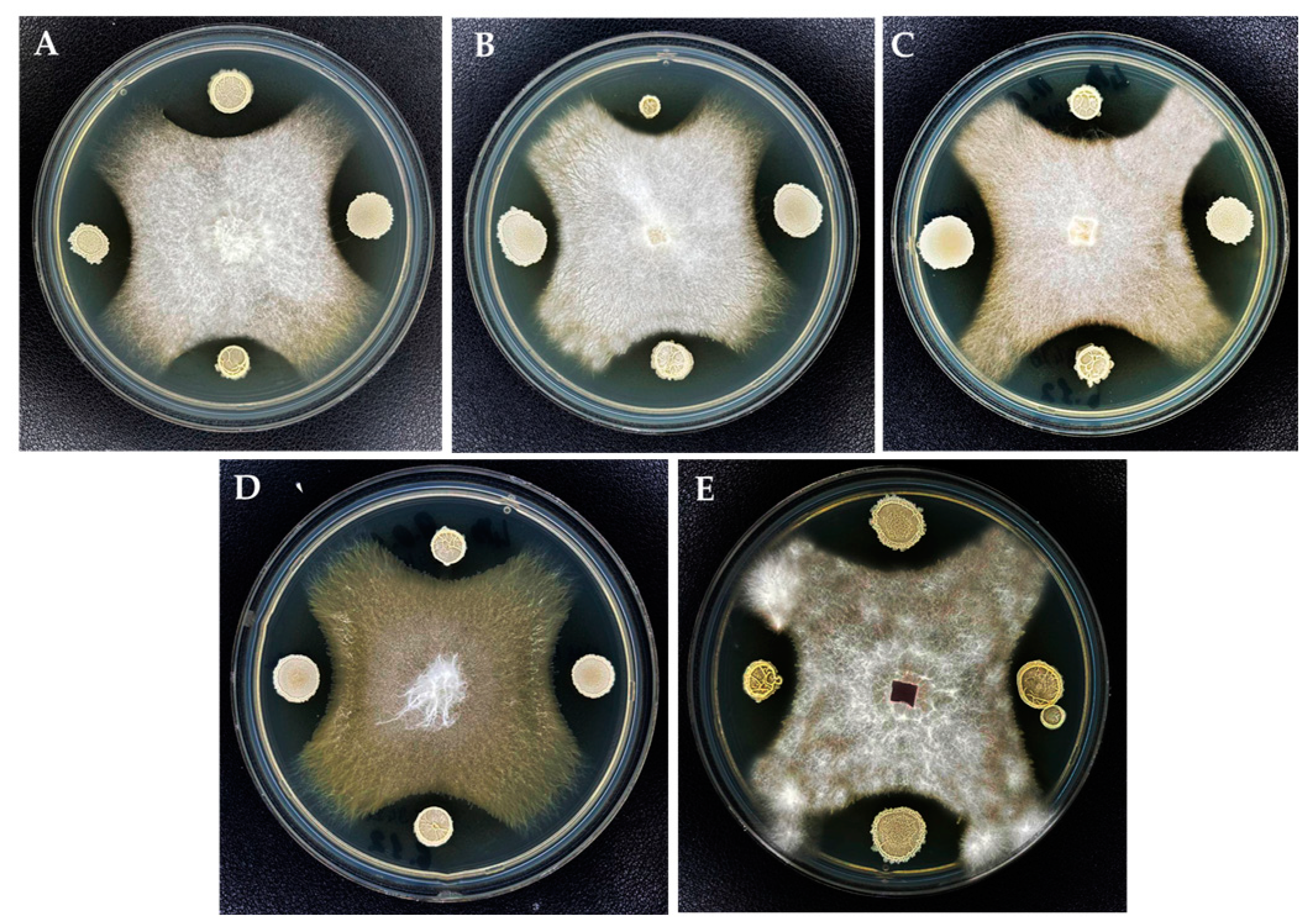
2.3.4. In Vitro Control of Fusarium Isolates Using Bacillus velezensis KS04-AU and Streptomyces albidoflavus MGMM6
2.4. In Planta Control of Fusarium Isolates through Bacillus velezensis KS04-AU and Streptomyces albidoflavus MGMM6
2.5. Statistical Analysis
3. Results
3.1. Characterization and Identification of Fungal Isolates
3.2. BOX PCR for Genetic Diversity Analyses
3.3. Molecular Identification
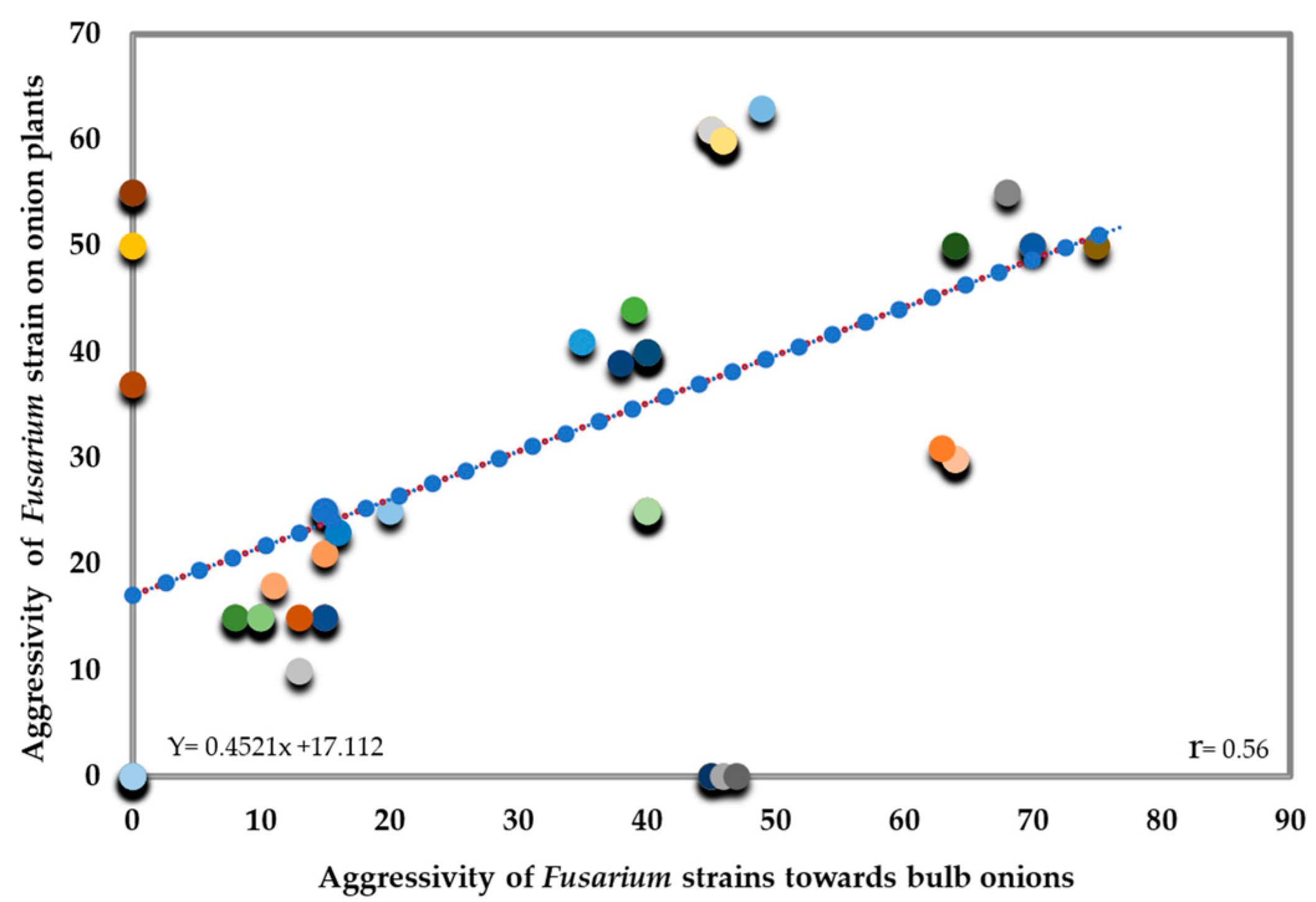
3.4. Pathogenicity Assay of Onion Seeds and Planta Control Using Bacillus velezensis KS04-AU
3.4.1. Pathogenicity Assay of Onion Seeds
3.4.2. Pathogenicity Assay of Onion Bulbs
3.5. In Vitro Control of Fusarium Isolates Using Bacillus velezensis KS04-AU and S. albidoflavus MGMM6
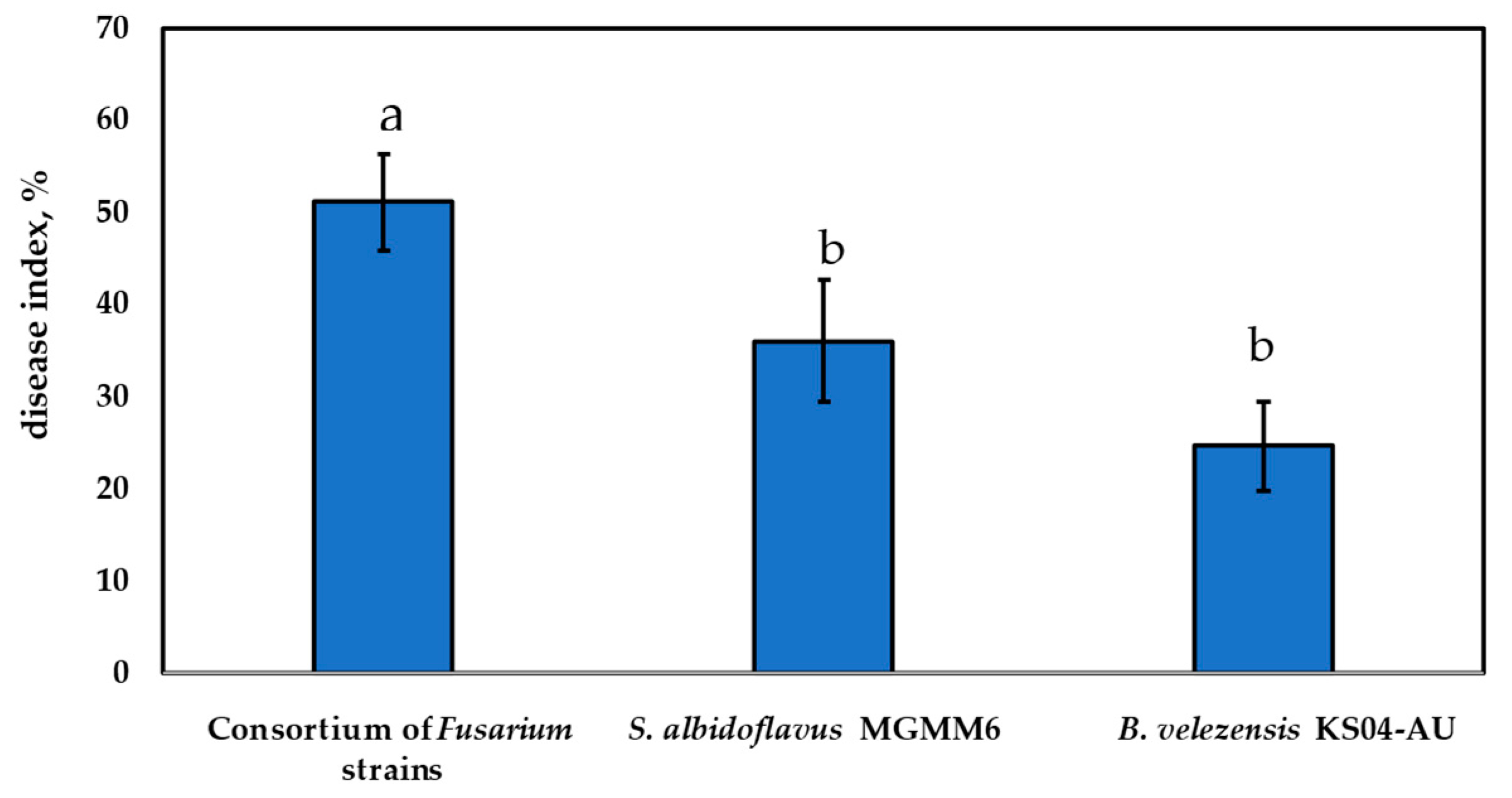
3.6. In Planta Control of Fusarium Isolates through Bacillus velezensis KS04-AU and Streptomyces albidoflavus MGMM6
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tournas, V.H. Spoilage of vegetable crops by bacteria and fungi and related health hazards. Crit. Rev. Microbiol. 2005, 31, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Sharma, S.; Neeraj, B.; Narashans, A.S. Postharvest management of fungal diseases in onion—A review. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 737–752. [Google Scholar]
- Sakhi, S.S.; Sohi, S.S.; Singh, D.P.; Joshi, M.C. Source of resistance to basal rot of onion caused by Fusarium oxysporum. Indian J. Mycol. Plant Pathol. 1973, 4, 214–215. [Google Scholar] [CrossRef]
- Boyhan, G.E.; Torrance, R.L. Vidalia onions—Sweet onion production in southeastern Georgia. HortTechnology 2002, 12, 196–202. [Google Scholar] [CrossRef]
- Anbukkarasi, V.; Paramaguru, P.; Pugalendhi, L.; Ragupathi, N.; Jeyakumar, P. Studies on pre- and postharvest treatments for extending shelf life in onion–A review. Agric. Rev. 2013, 34, 256–268. [Google Scholar] [CrossRef]
- Gupta, R.P.; Verma, L.R. Problem of diseases during storage in onion and garlic and their strategic management. In Implication of Plant Diseases on Produce Quality; Kalyani Publishers: Ludhiana, Indian, 2002. [Google Scholar]
- Oerke, E.C. Crop losses to pests. J. Agric. Sci. 2006, 144, 31–43. [Google Scholar] [CrossRef]
- Saremi, H.; Okhovvat, S.M.; Ashrafi, S.J. Fusarium diseases as the main soil-borne fungal pathogen on plants and their control management with soil solarization in Iran. Afr. J. Biotechnol. 2011, 10, 18391–18398. [Google Scholar] [CrossRef]
- Arie, T. Fusarium diseases of cultivated plants, control, diagnosis, and molecular and genetic studies. J. Pestic. Sci. 2019, 44, 275–281. [Google Scholar] [CrossRef]
- Summerell, B.A. Resolving Fusarium: Current status of the genus. Annu. Rev. Phytopathol. 2019, 57, 323–339. [Google Scholar] [CrossRef]
- Smith, S.N. An overview of ecological and habitat aspects in the genus Fusarium with special emphasis on the soil-borne pathogenic forms. Plant Pathol. Bull. 2007, 16, 97–120. [Google Scholar]
- Caligiore Gei, P.F.; Valdez, J.G.; Piccolo, R.J.; Galmarini, C.R. Influence of Fusarium spp. isolate and inoculum density on resistance screening tests in onion. Trop. Plant Pathol. 2014, 39, 19–27. [Google Scholar] [CrossRef]
- Schwartz, H.F.; Mohan, S.K. Compendium of Onion and Garlic Diseases and Pests, 2nd ed.; American Phytopathological Society: St. Paul, MN, USA, 2008; pp. 11–15. [Google Scholar]
- Kamle, M.; Mahato, D.K.; Devi, S.; Lee, K.E.; Kang, S.G.; Kumar, P. Fumonisins: Impact on agriculture, food, and human health and their management strategies. Toxins 2019, 11, 328. [Google Scholar] [CrossRef] [PubMed]
- Rather, I.A.; Koh, W.Y.; Paek, W.K.; Lim, J. The sources of chemical contaminants in food and their health implications. Front. Pharmacol. 2017, 8, 830. [Google Scholar] [CrossRef]
- Walsh, T.J.; Groll, A.; Hiemenz, J.; Fleming, R.; Roilides, E.; Anaissie, E. Infections due to emerging and uncommon medically important fungal pathogens. Clin. Microbiol. Infect. 2004, 10, 48–66. [Google Scholar] [CrossRef] [PubMed]
- Nucci, M.; Anaissie, E. Fusarium infections in immunocompromised patients. Clin. Microbiol. Rev. 2007, 20, 695–704. [Google Scholar] [CrossRef] [PubMed]
- Vahling-Armstrong, C.; Dung, J.K.S.; Humann, J.L.; Schroeder, B.K. Effects of postharvest onion curing parameters on bulb rot caused by Pantoea agglomerans, Pantoea ananatis and Pantoea allii in storage. Plant Pathol. 2016, 65, 536–544. [Google Scholar] [CrossRef]
- Galván, G.A.; Koning-Boucoiran, C.F.; Koopman, W.J.; Burger-Meijer, K.; González, P.H.; Waalwijk, C.; Kik, C.; Scholten, O.E. Genetic variation among Fusarium isolates from onion, and resistance to Fusarium basal rot in related Allium species. Eur. J. Plant Pathol. 2008, 121, 499–512. [Google Scholar] [CrossRef]
- Ji, S.H.; Kim, T.K.; Keum, Y.S.; Chun, S.C. The major postharvest disease of onion and its control with thymol fumigation during low-temperature storage. Mycobiology 2018, 46, 242–253. [Google Scholar] [CrossRef]
- Kadam, D.M.; Nangare, D.D.; Oberoi, H.S. Influence of pre-treatments on microbial load of stored dehydrated onion slices. Int. J. Food Sci.Technol. 2009, 44, 1902–1908. [Google Scholar] [CrossRef]
- Brent, K.J.; Hollomon, D.W. Fungicide Resistance: The Assessment of Risk; Global Crop Protection Federation: Brussels, Belgium, 1998; Volume 2. [Google Scholar]
- Nelson, P.E.; Dignani, M.C.; Anaissie, E.J. Taxonomy, biology, and clinical aspects of Fusarium species. Clin. Microbiol. Rev. 1994, 7, 479–504. [Google Scholar] [CrossRef]
- Missmer, S.A.; Suarez, L.; Felkner, M.; Wang, E.; Merrill, A.H., Jr.; Rothman, K.J.; Hendricks, K.A. Exposure to fumonisins and the occurrence of neural tube defects along the Texas–Mexico border. Environ. Health Perspect. 2006, 114, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Antonissen, G.; Martel, A.; Pasmans, F.; Ducatelle, R.; Verbrugghe, E.; Vandenbroucke, V.; Croubels, S. The impact of Fusarium mycotoxins on human and animal host susceptibility to infectious diseases. Toxins 2014, 6, 430–452. [Google Scholar] [CrossRef] [PubMed]
- Lahlali, R.; Ezrari, S.; Radouane, N.; Kenfaoui, J.; Esmaeel, Q.; El Hamss, H.; Belabess, Z.; Barka, E.A. Biological Control of Plant Pathogens: A Global Perspective. Microorganisms 2022, 10, 596. [Google Scholar] [CrossRef] [PubMed]
- Narayanasamy, P. Biological Management of Diseases of Crops; Springer: Dordrecht, The Netherlands, 2013; p. 673. [Google Scholar] [CrossRef]
- Fenta, L.; Mekonnen, H.; Kabtimer, N. The Exploitation of Microbial Antagonists against Postharvest Plant Pathogens. Microorganisms 2023, 11, 1044. [Google Scholar] [CrossRef]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Ravensberg. Mode of action of microbial biological control agents against plant diseases: Relevance beyond efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef]
- Compant, S.; Duffy, B.; Nowak, J.; Clément, C.; Barka, E.A. Use of plant growth-promoting bacteria for biocontrol of plant diseases: Principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 2005, 71, 4951–4959. [Google Scholar] [CrossRef]
- Leslie, J.F.; Summerell, B.A. The Fusarium Laboratory Manual; John Wiley & Sons: Hoboken, NJ, USA, 2008. [Google Scholar]
- Green, M.R.; Sambrook, J. Isolation of High-Molecular-Weight DNA Using Organic Solvents. Cold Spring Harb Protoc. 2017, 2017, pdb.prot093450. [Google Scholar] [CrossRef]
- Desjardins, P.; Conklin, D. NanoDrop microvolume quantitation of nucleic acids. JoVE 2010, 22, e2565. [Google Scholar] [CrossRef]
- Altinok, H.H. Fusarium species isolated from common weeds in eggplant fields and symptomless hosts of Fusarium oxysporum f. sp. melongenae in Turkey. J. Phytopathol. 2013, 161, 335–340. [Google Scholar] [CrossRef]
- Zarrin, M.; Ganj, F.; Faramarzi, S. Analysis of the rDNA internal transcribed spacer region of the Fusarium species by polymerase chain reaction-restriction fragment length polymorphism. Biomed. Rep. 2016, 4, 471–474. [Google Scholar] [CrossRef]
- Rzhetsky, A.; Nei, M. A simple method for estimating and testing minimum evolutionary trees. Mol. Biol. Evol. 1992, 9, 945–967. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.; van der Bij, A.J.; Brand, J.; de Weger, L.A.; Wijffelman, C.A.; Lugtenberg, B.J.J. Gnotobiotic system for studying rhizosphere colonization by plant growth-promoting Pseudomonas bacteria. Mol. Plant Microbe Interact 1996, 9, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Afordoanyi, D.M.; Diabankana, R.G.C.; Akosah, Y.A.; Validov, S.Z. Are formae speciales pathogens truly host specific? A broadened host specificity in Fusarium oxysporum f.sp. radicis-cucumerinum. Braz. J. Microbiol. 2022, 53, 1745–1759. [Google Scholar] [CrossRef]
- Diabankana, R.G.C.; Afordoanyi, D.M.; Safin, R.I.; Nizamov, R.M.; Karimova, L.Z.; Validov, S.Z. Antifungal properties, abiotic stress resistance, and biocontrol ability of Bacillus mojavensis PS17. Curr. Microbiol. 2021, 78, 3124–3132. [Google Scholar] [CrossRef]
- Koyshibayev, M.; Muminjanov, H. Guidelines for Monitoring Diseases, Pest and Weeds in Cereal Crops; Food and Agriculture Organization of the United Nations: Ankara, Turkey, 2016; p. 8. Available online: http://www.fao.org/3/a-i5550e (accessed on 1 February 2016).
- Tirado-Ramirez, A.; López-Urquídez, G.A.; Amarillas-Bueno, L.A.; Retes-Manjarrez, J.E.; Vega-Gutiérrez, T.A.; López Avendaño, J.E.; López-Orona, C.A. Identification and virulence of Fusarium falciforme and Fusarium brachygibbosum as causal agents of basal rot on onion in Mexico. Can. J. Plant Pathol. 2021, 43, 722–733. [Google Scholar] [CrossRef]
- Diabankana, R.G.C.; Shulga, E.U.; Validov, S.Z.; Afordoanyi, D.M. Genetic Characteristics and Enzymatic Activities of Bacillus velezensis KS04AU as a Stable Biocontrol Agent against Phytopathogens. Int. J. Plant Biol. 2022, 13, 201–222. [Google Scholar] [CrossRef]
- Diabankana, R.G.C.; Frolov, M.; Keremli, S.; Validov, S.Z.; Afordoanyi, D.M. Genomic Insights into the Microbial Agent Streptomyces albidoflavus MGMM6 for Various Biotechnology Applications. Microorganisms 2023, 11, 2872. [Google Scholar] [CrossRef] [PubMed]
- Heras, J.; Domínguez, C.; Mata, E.; Pascual, V.; Lozano, C.; Torres, C.; Zarazaga, M. GelJ–a tool for analyzing DNA fingerprint gel images. BMC Bioinform. 2015, 16, 270. [Google Scholar] [CrossRef] [PubMed]
- Waller, J.M.; Brayford, D. Fusarium diseases in the tropics. Int. J. Pest Manag. 1990, 36, 181–194. [Google Scholar] [CrossRef]
- Anand, G.; Rajeshkumar, K.C. Challenges and Threats Posed by Plant Pathogenic Fungi on Agricultural Productivity and Economy. In Fungal Diversity, Ecology and Control Management; Fungal Biology; Rajpal, V.R., Singh, I., Navi, S.S., Eds.; Springer: Singapore, 2022. [Google Scholar] [CrossRef]
- Oldenburg, E.; Höppner, F.; Ellner, F.; Weinert, J. Fusarium diseases of maize associated with mycotoxin contamination of agricultural products intended to be used for food and feed. Mycotoxin Res. 2017, 33, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Rojo, F.G.; Reynoso, M.M.; Ferez, M.; Chulze, S.N.; Torres, A.M. Biological control by Trichoderma species of Fusarium solani causing peanut brown root rot under field conditions. Crop Prot. 2007, 26, 549–555. [Google Scholar] [CrossRef]
- Kintega, K.R.; Zida, P.E.; Tarpaga, V.W.; Sankara, P.; Sereme, P. Identification of Fusarium species associated with onion (Allium cepa L.) plants in field in Burkina Faso. Adv. Biosci. Biotechnol. 2020, 11, 94–110. [Google Scholar] [CrossRef][Green Version]
- Zhan, J.; Mundt, C.C.; Hoffer, M.E.; McDonald, B.A. Local adaptation and effect of host genotype on the rate of pathogen evolution: An experimental test in a plant pathosystem. J. Evol. Biol. 2002, 15, 634–647. [Google Scholar] [CrossRef]
- Zhan, J.; McDonald, B.A. Thermal adaptation in the fungal pathogen Mycosphaerella graminicola. Mol. Ecol. 2011, 20, 1689–1701. [Google Scholar] [CrossRef] [PubMed]
- Wu, E.J.; Wang, Y.P.; Yahuza, L.; He, M.H.; Sun, D.L.; Huang, Y.M.; Liu, Y.C.; Yang, L.N.; Zhu, W.; Zhan, J. Rapid adaptation of the Irish potato famine pathogen Phytophthora infestans to changing temperature. Evol. Appl. 2020, 13, 769–781. [Google Scholar] [CrossRef]
- Yang, L.N.; Nkurikiyimfura, O.; Pan, Z.C.; Wang, Y.P.; Waheed, A.; Chen, R.S.; Burdon, J.J.; Sui, Q.J.; Zhan, J. Plantdiversity ameliorates the evolutionary development of fungicide resistance in an agricultural ecosystem. J. Appl. Ecol. 2021, 58, 2566–2578. [Google Scholar] [CrossRef]
- Marques, A.S.; Marchaison, A.; Gardan, L.; Samson, R. BOX-PCR-based identification of bacterial species belonging to Pseudomonas syringae: P. viridiflava group. Genet. Mol. Biol. 2008, 31, 106–115. [Google Scholar] [CrossRef][Green Version]
- Tacão, M.; Alves, A.; Saavedra, M.J.; Correia, A. BOX-PCR is an adequate tool for typing Aeromonas spp. Antonie Leeuwenhoek 2005, 88, 173–179. [Google Scholar] [CrossRef]
- Proctor, R.H.; Desjardins, A.E.; Moretti, A. Biological and chemical complexity of Fusarium proliferatum. In The Role of Plant Pathology in Food Safety; Strange, R.N., Gullino, M.L., Eds.; Springer: Berlin, Germany, 2009; pp. 97–111. [Google Scholar] [CrossRef]
- Niehaus, E.M.; Münsterkötter, M.; Proctor, R.H.; Brown, D.W.; Sharon, A.; Idan, Y.; Tudzynski, B. Comparative “omics” of the Fusarium fujikuroi species complex highlights differences in genetic potential and metabolite synthesis. Genome Biol. Evol. 2016, 8, 3574–3599. [Google Scholar] [CrossRef]
- Dong, T.; Qiao, S.; Xu, J.; Shi, J.; Qiu, J.; Ma, G. Effect of Abiotic Conditions on Growth, Mycotoxin Production, and Gene Expression by Fusarium fujikuroi Species Complex Strains from Maize. Toxins 2023, 15, 260. [Google Scholar] [CrossRef]
- O’Donnell, K.; Cigelnik, E.; Nirenberg, H.I. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 1998, 90, 465–493. [Google Scholar] [CrossRef]
- Huss, M.J.; Campbell, C.L.; Jennings, D.B.; Leslie, J.F. Isozyme variation among biological species in the Gibberella fujikuroi species complex (Fusarium section Liseola). Appl. Environ. Microbiol. 1996, 62, 3750–3756. [Google Scholar] [CrossRef]
- Stankovic, S.; Levic, J.; Petrovic, T.; Logrieco, A.; Moretti, A. Pathogenicity and mycotoxin production by Fusarium proliferatum isolated from onion and garlic in Serbia. Eur. J. Plant Pathol. 2007, 118, 165–172. [Google Scholar] [CrossRef]
- Heidarian, Z.; Javan-Nikkhah, M.; Fotouhifar, K.B.; Ahmadpour, A. Morphological and phylogenetic investigation on selected Fusarium species belong to Gibberella fujikuroi species complex in Iran. Rostaniha 2013, 14, 163–174. [Google Scholar] [CrossRef]
- Dissanayake, M.L.M.C.; Kashima, R.; Tanaka, S.; and Ito, S.I. Pathogenic variation and molecular characterization of Fusarium species isolated from wilted Welsh onion in Japan. Plant Pathol. J. 2009, 75, 37–45. [Google Scholar] [CrossRef]
- Haapalainen, M.; Latvala, S.; Kuivainen, E.; Qiu, Y.; Segerstedt, M.; Hannukkala, A.O. Fusarium oxysporum, F. proliferatum and F. redolens associated with basal rot of onion in Finland. Plant Pathol. 2016, 65, 1310–1320. [Google Scholar] [CrossRef]
- Taylor, A.; Vagany, V.; Barbara, D.J.; Thomas, B.; Pink, D.A.C.; Jones, J.E.; Clarkson, J.P. Identification of differential resistance to six Fusarium oxysporum f. sp. cepae isolates in commercial onion cultivars through the development of a rapid seedling assay. Plant Pathol. 2013, 62, 103–111. [Google Scholar] [CrossRef]
- Özer, N.; Köycü, N.D.; Chilosi, G.; Magro, P. Resistance to Fusarium basal rot of onion in greenhouse and field and associated expression of antifungal compounds. Phytoparasitica 2004, 32, 388–394. [Google Scholar] [CrossRef]
- Singer, M. Pathogen-pathogen interaction: A syndemic model of complex biosocial processes in disease. Virulence 2010, 1, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Braga, R.M.; Dourado, M.N.; Araújo, W.L. Microbial interactions: Ecology in a molecular perspective. Braz. J. Microbiol. 2016, 47, 86–98. [Google Scholar] [CrossRef] [PubMed]
- Bektas, I.; Kusek, M. Phylogenetic and morphological characterization of Fusarium oxysporum f. sp. cepae the causal agent of basal rot on onion isolated from Turkey. Fresenius Environ. Bull. 2019, 28, 1733–1742. [Google Scholar]
- Beck, K.D.; Reyes-Corral, C.; Rodriguez-Rodriguez, M.; May, C.; Barnett, R.; Thornton, M.K.; Schroeder, B.K. First report of Fusarium proliferatum causing necrotic leaf lesions and bulb rot on storage onion (Allium cepa) in southwestern Idaho. Plant Dis. 2021, 105, 494. [Google Scholar] [CrossRef]
- Abdullah, A.S.; Moffat, C.S.; Lopez-Ruiz, F.J.; Gibberd, M.R.; Hamblin, J.; Zerihun, A. Host–multipathogen warfare: Pathogen interactions in coinfected plants. Front. Plant Sci. 2017, 8, 1806. [Google Scholar] [CrossRef]
- Dissanayake, M.; Kashima, R.; Tanaka, S.; Ito, S.I. Genetic diversity and pathogenicity of Fusarium oxysporum isolated from wilted Welsh onion in Japan. J. Gen. Plant Pathol. 2009, 75, 125–130. [Google Scholar] [CrossRef]
- Sasaki, K.; Nakahara, K.; Tanaka, S.; Shigyo, M.; Ito, S.I. Genetic and pathogenic variability of Fusarium oxysporum f. sp. cepae isolated from onion and Welsh onion in Japan. Phytopathology 2015, 105, 525–532. [Google Scholar] [CrossRef]
- Alabouvette, C.; Olivain, C.; Migheli, Q.; Steinberg, C. Microbiological control of soil-borne phytopathogenic fungi with special emphasis on wilt-inducing Fusarium oxysporum. New Phytol. 2009, 184, 529–544. [Google Scholar] [CrossRef]
- De Lamo, F.J.; Takken, F.L. Biocontrol by Fusarium oxysporum using endophyte-mediated resistance. Front. Plant Sci. 2020, 11, 37. [Google Scholar] [CrossRef]
- Gärber, U.; Grosch, R.; Goßmann, M.; Büttner, C. Auftreten und Nachweis von Fusarium oxysporum und F. proliferatum an Steck-und Saatzwiebeln. Gesunde Pflanzen 2012, 64, 1–10. [Google Scholar] [CrossRef]
- Labanska, M.; Van Amsterdam, S.; Jenkins, S.; Clarkson, J.P.; Covington, J.A. Preliminary studies on detection of Fusarium basal rot infection in onions and shallots using electronic nose. Sensors 2022, 22, 5453. [Google Scholar] [CrossRef] [PubMed]
- Lager, S. Survey of Fusarium Species on Yellow Onion (Allium cepa) on Öland; Uppsala Swedish University of Agricultural Science: Uppsala, Sweden, 2011. [Google Scholar]
- Tsitsigiannis, D.I.; Dimakopoulou, M.; Antoniou, P.P.; Tjamos, E.C. Biological control strategies of mycotoxigenic fungi and associated mycotoxins in Mediterranean basin crops. Phytopathol. Mediterr. 2012, 51, 158–174. [Google Scholar]
- Carrieri, R.; Raimo, F.; Pentangelo, A.; Lahoz, E. Fusarium proliferatum and Fusarium tricinctum as causal agents of pink rot of onion bulbs and the effect of soil solarization combined with compost amendment in controlling their infections in field. Crop Prot. 2013, 43, 31–37. [Google Scholar] [CrossRef]
- Chernin, L.; Chet, I. Microbial enzymes in biocontrol of plant pathogens and pests. In Enzymes in the Environment: Activity, Ecology, and Applications; Marcel Dekker, Inc.: New Yoer, NY, USA, 2002; pp. 171–225. [Google Scholar]
- Du, Y.; Wang, T.; Jiang, J.; Wang, Y.; Lv, C.; Sun, K.; Huang, L. Biological control and plant growth promotion properties of Streptomyces albidoflavus St-220 isolated from Salvia miltiorrhiza rhizosphere. Front. Plant Sci. 2022, 13, 976813. [Google Scholar] [CrossRef]
- Carlucci, A.; Raimondo, M.L.; Colucci, D.; Lops, F. Streptomyces albidoflavus strain CARA17 as a biocontrol agent against fungal soil-borne pathogens of fennel plants. Plants 2022, 11, 1420. [Google Scholar] [CrossRef]
- Rabbee, M.F.; Hwang, B.S.; Baek, K.-H. Bacillus velezensis: A Beneficial Biocontrol Agent or Facultative Phytopathogen for Sustainable Agriculture. Agronomy 2023, 13, 840. [Google Scholar] [CrossRef]
- Djebaili, R.; Pellegrini, M.; Ercole, C.; Farda, B.; Kitouni, M.; Del Gallo, M. Biocontrol of soil-borne pathogens of Solanum lycopersicum L. and Daucus carota L. by plant growth-promoting actinomycetes: In vitro and in planta antagonistic activity. Pathogens 2021, 10, 1305. [Google Scholar] [CrossRef]
- Sidharthan, V.K.; Pothi-raj, G.; Suryaprakash, V.; Singh, A.K.; Aggarwal, R.; Shanmugam, V. A synergic and compatible microbial-based consortium for biocontrol of Fusarium wilt of tomato. Phytopathol. Mediterr. 2023, 62, 183–197. [Google Scholar] [CrossRef]


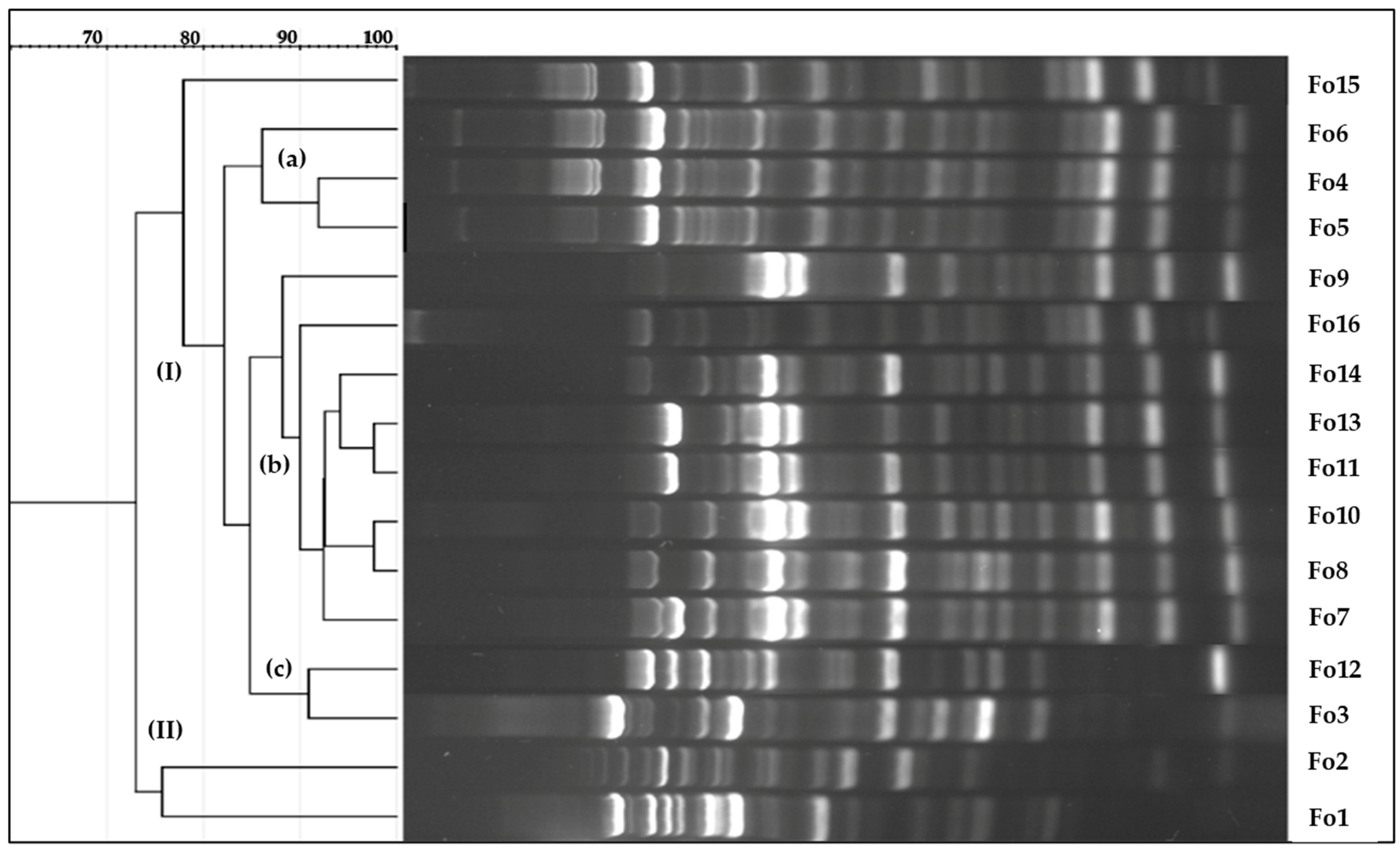
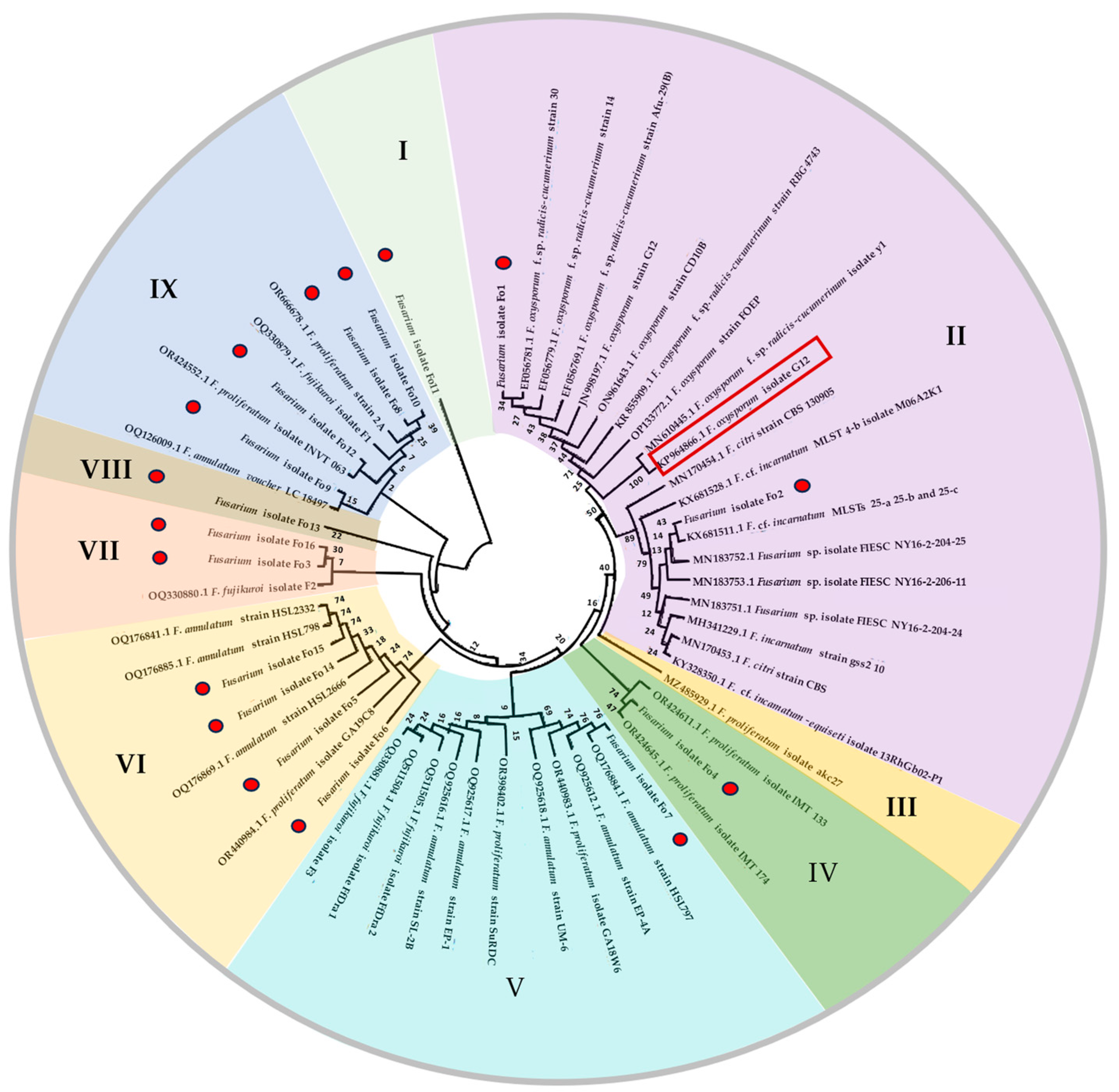

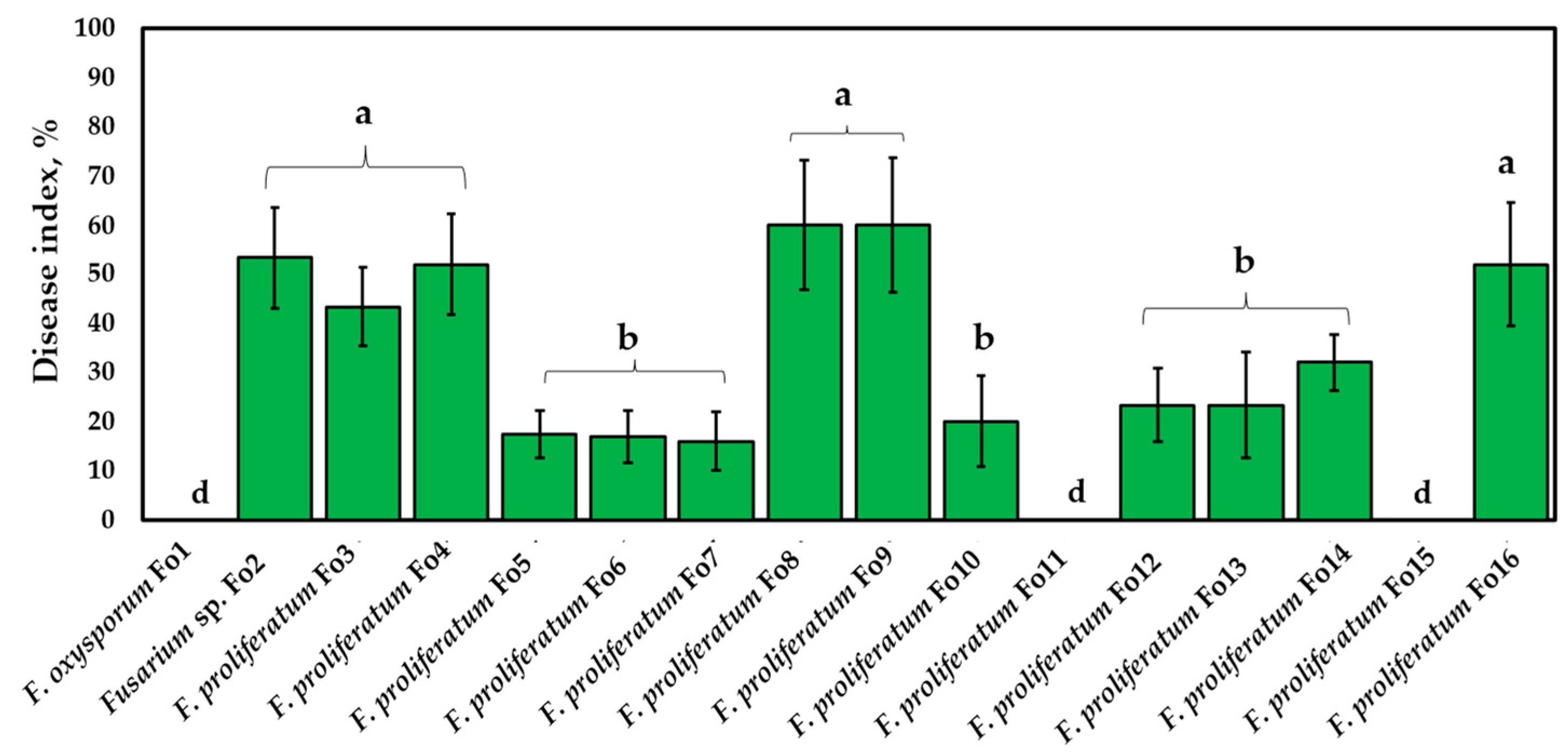
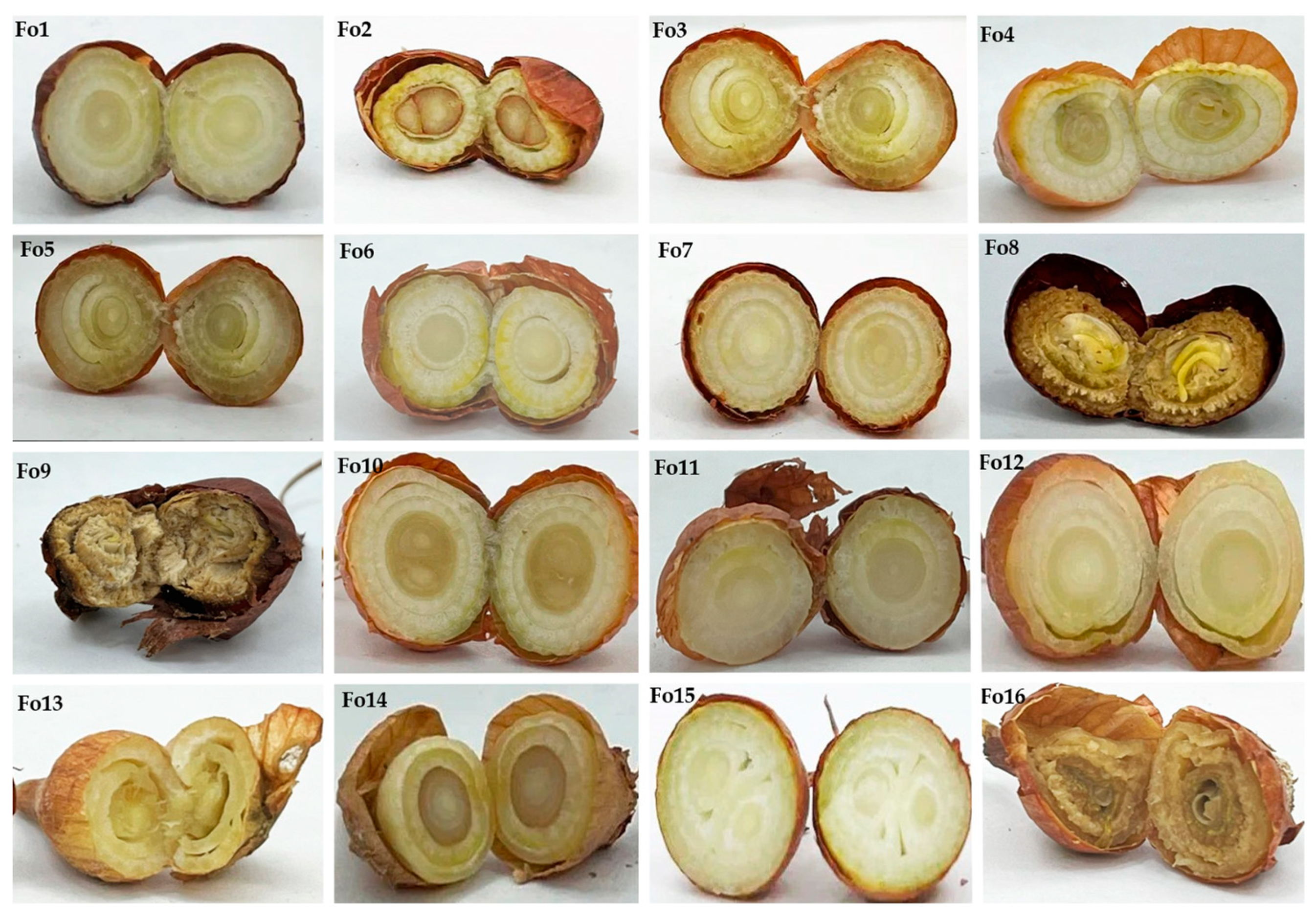
| Isolates | Marker | Species | Accession Number Deposited in NCBI Genbank | Similarity | Species Complex | Reference Accession Number |
|---|---|---|---|---|---|---|
| Fo1 | tef-1α | F. oxysporum | PP140391 | 96.95% | F. oxysporum | FJ985420 |
| ITS | F. oxysporum | 98.51% | F. oxysporum | DQ790536 | ||
| Fo2 | tef-1α | Fusarium sp. | PP140392 | 96.60% | F. incarnatum-equiseti | JF740715 |
| ITS | 98.50% | F. oxysporum | DQ790536 | |||
| Fo3 | tef-1α | F. proliferatum | PP140393 | 97.67% | F. fujikuroi | MH582346 |
| ITS | 98.00% | F. oxysporum | DQ790536 | |||
| Fo4 | tef-1α | F. proliferatum | PP140394 | 97.66% | F. fujikuroi | MH582346 |
| ITS | 99.04% | Gibberella fujikuroi | U34569 | |||
| Fo5 | tef-1α | F. proliferatum | PP140395 | 97.30% | F. fujikuroi | MH582346 |
| ITS | 98.57% | Gibberella fujikuroi | U34569 | |||
| Fo6 | tef-1α | F. proliferatum | PP140396 | 98.07% | F. fujikuroi | MH582347 |
| ITS | 99.04% | Gibberella fujikuroi | U34569 | |||
| Fo7 | tef-1α | F. proliferatum | PP140397 | 95.80% | F. fujikuroi | MH582347 |
| ITS | 98.56% | Gibberella fujikuroi | U34569 | |||
| Fo8 | tef-1α | F. proliferatum | PP140398 | 96.91% | F. fujikuroi | MH582346 |
| ITS | 99.04% | Gibberella fujikuroi | U34569 | |||
| Fo9 | tef-1α | F. proliferatum | PP140399 | 94.02% | F. fujikuroi | MH582346 |
| ITS | 97.00% | F. incarnatum-equiseti | DQ790541 | |||
| Fo10 | tef-1α | F. proliferatum | PP140400 | 95.74% | F. fujikuroi | MH582346 |
| ITS | 99.04% | Gibberella fujikuroi | U34569 | |||
| Fo11 | tef-1α | F. proliferatum | PP140401 | 97.67% | F. fujikuroi | MH582346 |
| ITS | 99.04% | Gibberella fujikuroi | U34569 | |||
| Fo12 | tef-1α | F. proliferatum | PP140402 | 95.38% | F. fujikuroi | MH582346 |
| ITS | 98.56% | Gibberella fujikuroi | U34569 | |||
| Fo13 | tef-1α | F. proliferatum | PP140403 | 95.38% | F. fujikuroi | MH582346 |
| ITS | 97.52% | Gibberella fujikuroi | U34569 | |||
| Fo14 | tef-1α | F. proliferatum | PP140404 | 95.38% | F. fujikuroi | MH582346 |
| ITS | 98.61% | Gibberella fujikuroi | U34569 | |||
| Fo15 | tef-1α | F. proliferatum | PP140405 | 98.05% | F. fujikuroi | MH582346 |
| ITS | 97.02% | Gibberella fujikuroi | U34569 | |||
| Fo16 | tef-1α | F. proliferatum | PP140406 | 98.07% | F. fujikuroi | MH582347 |
| ITS | 98.54% | Gibberella fujikuroi | U34569 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Diabankana, R.G.C.; Frolov, M.; Islamov, B.; Shulga, E.; Filimonova, M.N.; Afordoanyi, D.M.; Validov, S. Identification and Aggressiveness of Fusarium Species Associated with Onion Bulb (Allium cepa L.) during Storage. J. Fungi 2024, 10, 161. https://doi.org/10.3390/jof10020161
Diabankana RGC, Frolov M, Islamov B, Shulga E, Filimonova MN, Afordoanyi DM, Validov S. Identification and Aggressiveness of Fusarium Species Associated with Onion Bulb (Allium cepa L.) during Storage. Journal of Fungi. 2024; 10(2):161. https://doi.org/10.3390/jof10020161
Chicago/Turabian StyleDiabankana, Roderic Gilles Claret, Mikhail Frolov, Bakhtiyar Islamov, Elena Shulga, Maria Nikolaevna Filimonova, Daniel Mawuena Afordoanyi, and Shamil Validov. 2024. "Identification and Aggressiveness of Fusarium Species Associated with Onion Bulb (Allium cepa L.) during Storage" Journal of Fungi 10, no. 2: 161. https://doi.org/10.3390/jof10020161
APA StyleDiabankana, R. G. C., Frolov, M., Islamov, B., Shulga, E., Filimonova, M. N., Afordoanyi, D. M., & Validov, S. (2024). Identification and Aggressiveness of Fusarium Species Associated with Onion Bulb (Allium cepa L.) during Storage. Journal of Fungi, 10(2), 161. https://doi.org/10.3390/jof10020161






