Sarcomere Disassembly and Transfection Efficiency in Proliferating Human iPSC-Derived Cardiomyocytes
Abstract
:1. Introduction
2. Methods
2.1. Maintenance of hiPSC
2.2. Differentiation of hiPSC into Cardiomyocytes
2.3. Metabolic Maturation of iPSC-CMs
2.4. Immunofluorescence
2.5. Time-Lapse Imaging
2.6. RNA Sequencing for CM Proliferation Genes
2.7. Transfection of hiPSC-CMs with mCherry
2.8. FACS Analysis of Transfection Efficiency
2.9. Statistics
3. Results
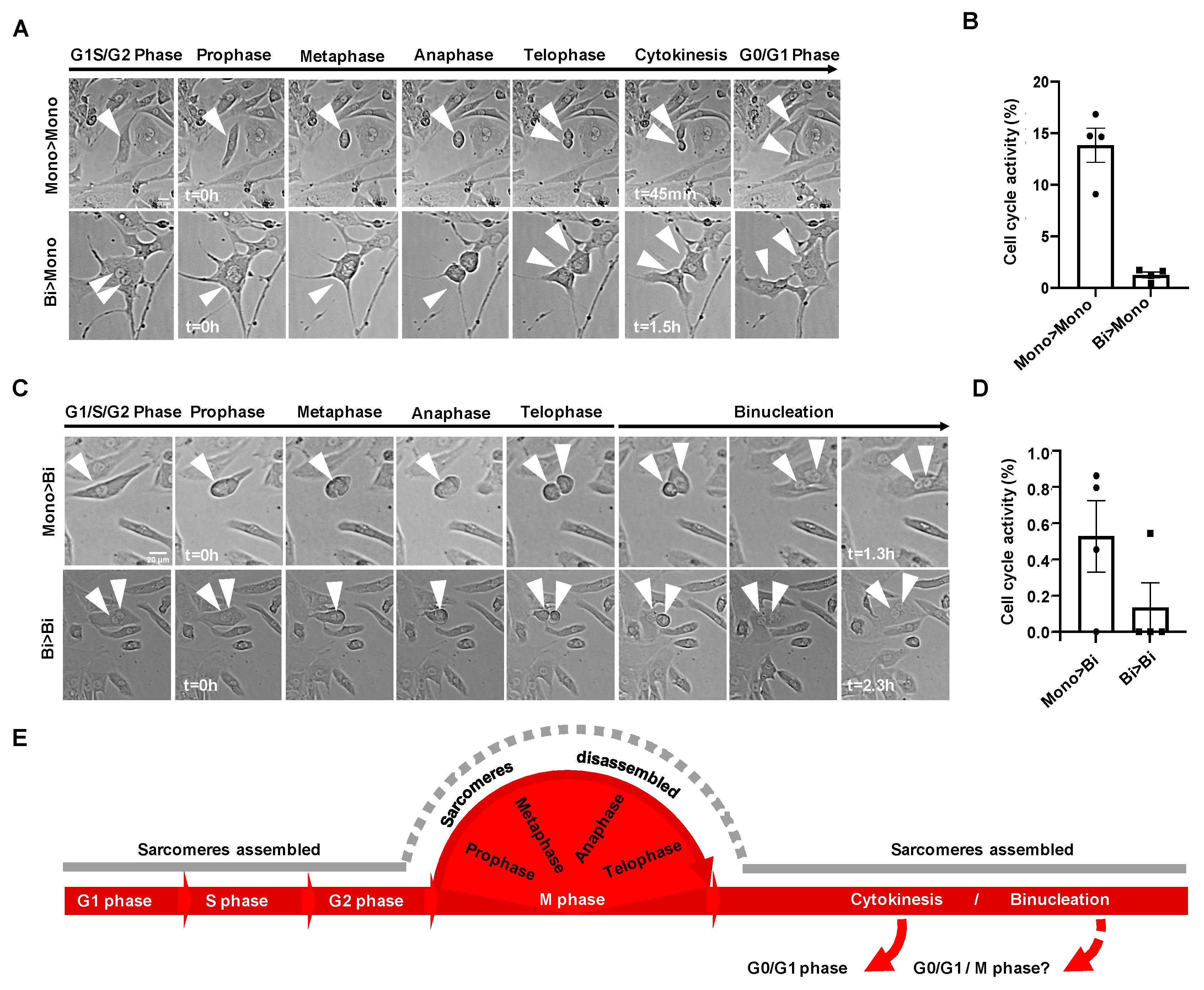
3.1. Sarcomere Organization of hiPSC-CMs during Cell Cycle Phases
3.2. Live Tracing of Mitotic Figures in hiPSC-CMs
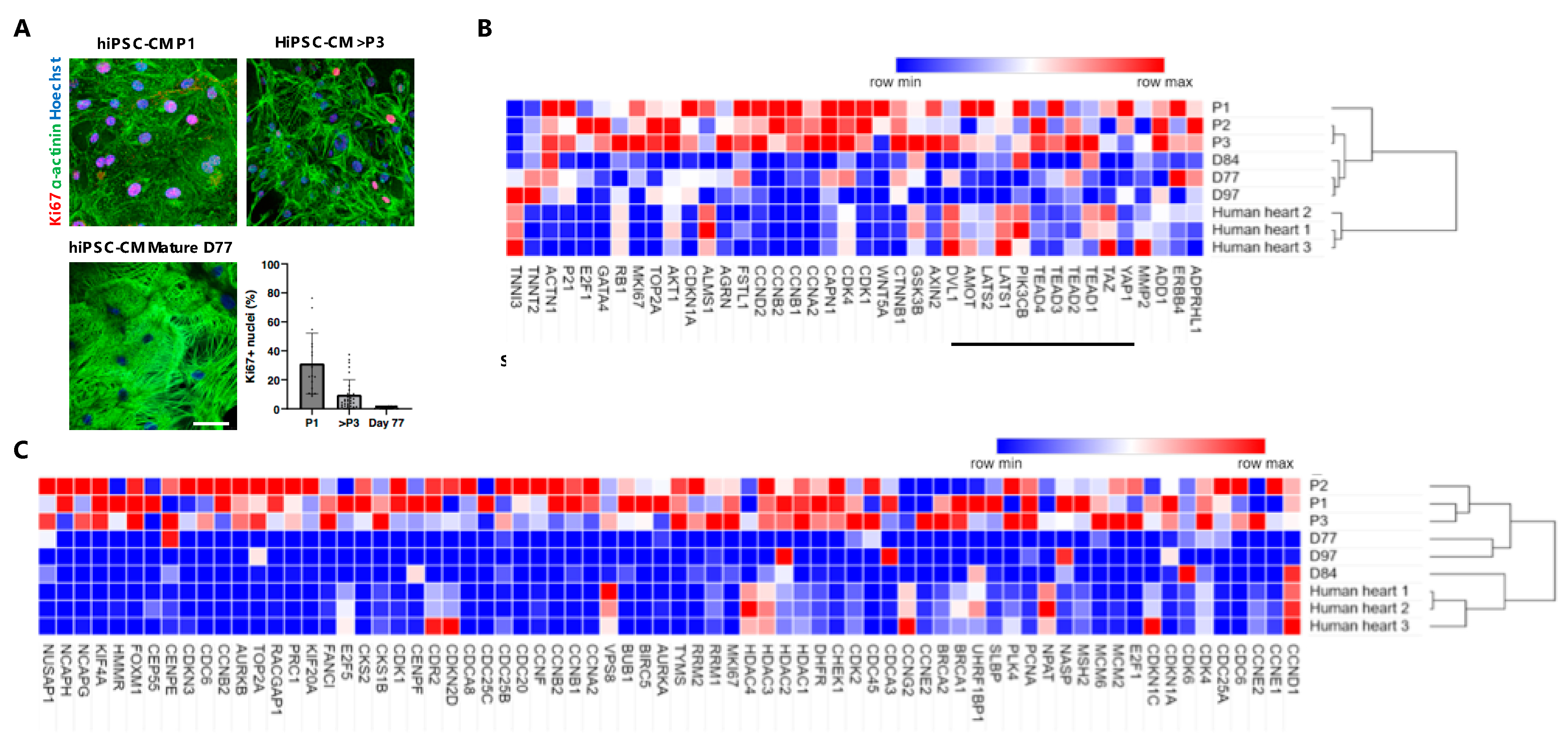
3.3. Gene Expression Analysis of Long-Term Proliferating, Matured Non-Proliferative hiPSC-CMs and Adult Human Heart Tissue
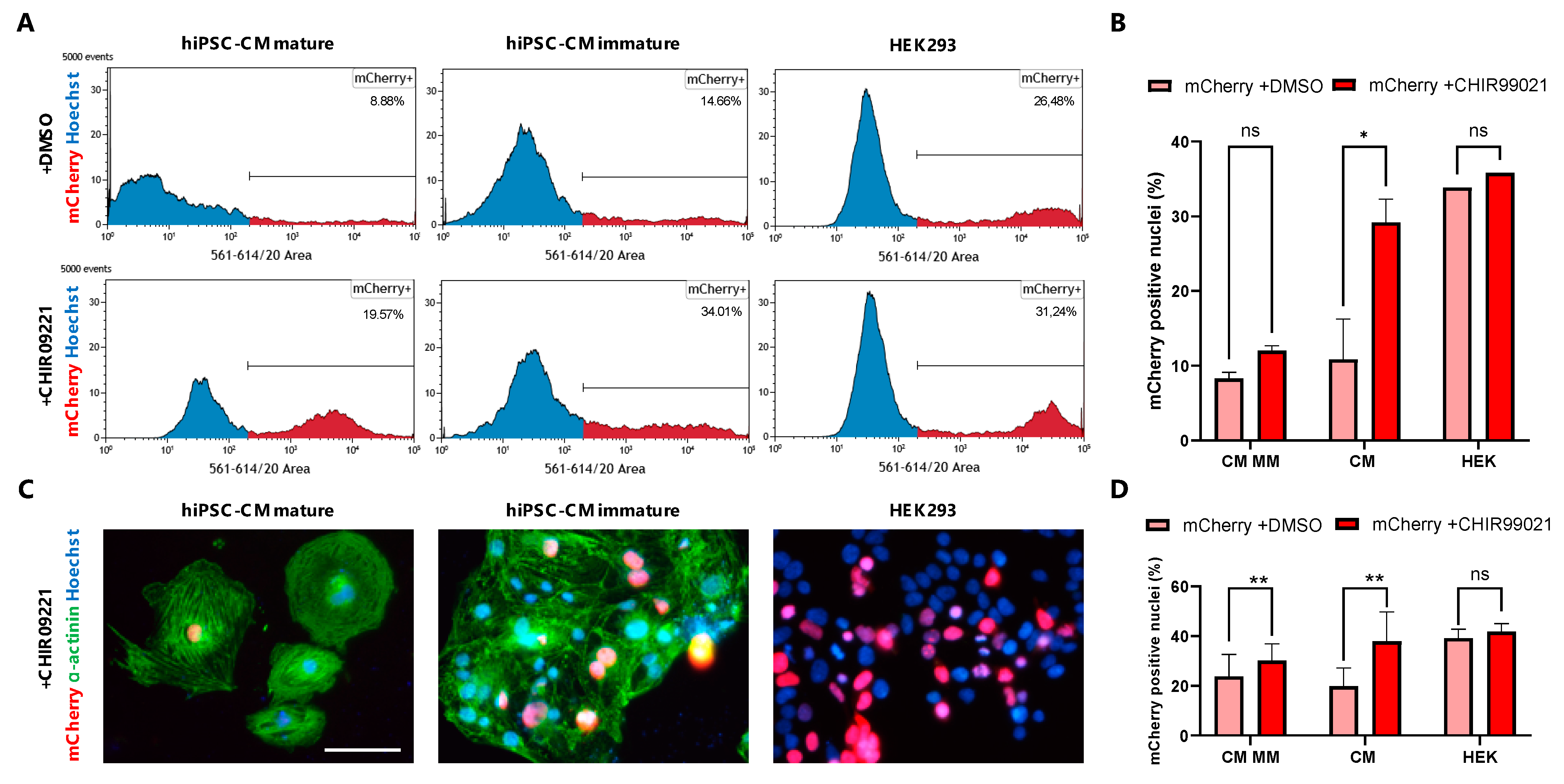
3.4. GSK3β Inhibition Increases Non-Viral Vector Incorporation Efficiency in hiPSC-CMs
3.5. Live Tracing of Non-Viral Vector-Based Fluorescence Expression in hiPSC-CMs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Banerjee, I.; Fuseler, J.W.; Price, R.L.; Borg, T.K.; Baudino, T.A. Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am. J. Physiol. Circ. Physiol. 2007, 293, H1883–H1891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Jardin, B.D.; Zhou, P.; Sethi, I.; Akerberg, B.N.; Toepfer, C.N.; Ai, Y.; Li, Y.; Ma, Q.; Guatimosim, S.; et al. Hierarchical and stage-specific regulation of murine cardiomyocyte maturation by serum response factor. Nat. Commun. 2018, 9, 3837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maas, R.G.; Lee, S.; Harakalova, M.; Blok, C.J.S.; Goodyer, W.R.; Hjortnaes, J.; Doevendans, P.A.; Van Laake, L.W.; van der Velden, J.; Asselbergs, F.W.; et al. Massive expansion and cryopreservation of functional human induced pluripotent stem cell-derived cardiomyocytes. STAR Protoc. 2021, 2, 100334. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Hughes, B.G.; Ali, M.; Cho, W.J.; Lopez, W.; Schulz, R. Dynamic Alterations to α-Actinin Accompanying Sarcomere Disassembly and Reassembly during Cardiomyocyte Mitosis. PLoS ONE 2015, 10, e0129176. [Google Scholar] [CrossRef]
- Ahuja, P.; Perriard, E.; Perriard, J.-C.; Ehler, E. Sequential myofibrillar breakdown accompanies mitotic division of mammalian cardiomyocytes. J. Cell Sci. 2004, 117, 3295–3306. [Google Scholar] [CrossRef] [Green Version]
- Porrello, E.R.; Mahmoud, A.I.; Simpson, E.; Hill, J.A.; Richardson, J.A.; Olson, E.N.; Sadek, H.A. Transient Regenerative Potential of the Neonatal Mouse Heart. Science 2011, 331, 1078–1080. [Google Scholar] [CrossRef] [Green Version]
- Jopling, C.; Sleep, E.; Raya, M.; Martí, M.; Raya, A.; Belmonte, J.C.I. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010, 464, 606–609. [Google Scholar] [CrossRef]
- Bergmann, O.; Zdunek, S.; Felker, A.; Salehpour, M.; Alkass, K.; Bernard, S.; Sjostrom, S.L.; Szewczykowska, M.; Jackowska, T.; Dos Remedios, C.; et al. Dynamics of Cell Generation and Turnover in the Human Heart. Cell 2015, 161, 1566–1575. [Google Scholar] [CrossRef] [Green Version]
- Senyo, S.E.; Steinhauser, M.L.; Pizzimenti, C.L.; Yang, V.K.; Cai, L.; Wang, M.; Wu, T.-D.; Guerquin-Kern, J.-L.; Lechene, C.P.; Lee, R.T. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 2013, 493, 433–436. [Google Scholar] [CrossRef] [Green Version]
- Patterson, M.; Barske, L.; Van Handel, B.; Rau, C.D.; Gan, P.; Sharma, A.; Parikh, S.; Denholtz, M.; Huang, Y.; Yamaguchi, Y.; et al. Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat. Genet. 2017, 49, 1346–1353. [Google Scholar] [CrossRef]
- Derks, W.; Bergmann, O. Polyploidy in Cardiomyocytes. Circ. Res. 2020, 126, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Ponnusamy, M.; Li, P.-F.; Wang, K. Understanding cardiomyocyte proliferation: An insight into cell cycle activity. Cell. Mol. Life Sci. 2017, 74, 1019–1034. [Google Scholar] [CrossRef]
- Neininger, A.; Long, J.H.; Baillargeon, S.M.; Burnette, D.T. A simple and flexible high-throughput method for the study of cardiomyocyte proliferation. Sci. Rep. 2019, 9, 15917. [Google Scholar] [CrossRef]
- Paige, S.L.; Osugi, T.; Afanasiev, O.K.; Pabon, L.; Reinecke, H.; Murry, C.E. Endogenous Wnt/β-Catenin Signaling Is Required for Cardiac Differentiation in Human Embryonic Stem Cells. PLoS ONE 2010, 5, e11134. [Google Scholar] [CrossRef] [PubMed]
- Lian, X.; Hsiao, C.; Wilson, G.; Zhu, K.; Hazeltine, L.B.; Azarin, S.M.; Raval, K.K.; Zhang, J.; Kamp, T.J.; Palecek, S.P. Cozzarelli Prize Winner: Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA 2012, 109, E1848–E1857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lian, X.; Zhang, J.; Azarin, S.M.; Zhu, K.; Hazeltine, L.B.; Bao, X.; Hsiao, C.; Kamp, T.; Palecek, S.P. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat. Protoc. 2013, 8, 162–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buikema, J.W.; Zwetsloot, P.-P.M.; Doevendans, P.A.; Domian, I.J.; Sluijter, J.P.G. Wnt/β-Catenin Signaling during Cardiac Development and Repair. J. Cardiovasc. Dev. Dis. 2014, 1, 98–110. [Google Scholar] [CrossRef] [Green Version]
- Buikema, J.W.; Wu, S.M. Untangling the Biology of Genetic Cardiomyopathies with Pluripotent Stem Cell Disease Models. Curr. Cardiol. Rep. 2017, 19, 30. [Google Scholar] [CrossRef]
- Titmarsh, D.M.; Glass, N.R.; Mills, R.J.; Hidalgo, A.; Wolvetang, E.J.; Porrello, E.R.; Hudson, J.E.; Cooper-White, J.J. Induction of Human iPSC-Derived Cardiomyocyte Proliferation Revealed by Combinatorial Screening in High Density Microbioreactor Arrays. Sci. Rep. 2016, 6, 24637. [Google Scholar] [CrossRef]
- Sharma, A.; Zhang, Y.; Buikema, J.W.; Serpooshan, V.; Chirikian, O.; Kosaric, N.; Churko, J.M.; Dzilic, E.; Shieh, A.; Burridge, P.W.; et al. Stage-specific Effects of Bioactive Lipids on Human iPSC Cardiac Differentiation and Cardiomyocyte Proliferation. Sci. Rep. 2018, 8, 6618. [Google Scholar] [CrossRef] [Green Version]
- Buikema, J.W.; Mady, A.S.; Mittal, N.; Atmanli, A.; Caron, L.; Doevendans, P.A.; Sluijter, J.; Domian, I.J. Wnt/β-catenin signaling directs the regional expansion of first and second heart field-derived ventricular cardiomyocytes. Development 2013, 140, 4165–4176. [Google Scholar] [CrossRef] [Green Version]
- Mills, R.J.; Titmarsh, D.M.; Koenig, X.; Parker, B.L.; Ryall, J.G.; Quaife-Ryan, G.A.; Voges, H.K.; Hodson, M.P.; Ferguson, C.; Drowley, L.; et al. Functional screening in human cardiac organoids reveals a metabolic mechanism for cardiomyocyte cell cycle arrest. Proc. Natl. Acad. Sci. USA 2017, 114, E8372–E8381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunner, S.; Fürtbauer, E.; Sauer, T.; Kursa, M.; Wagner, E. Overcoming the Nuclear Barrier: Cell Cycle Independent Nonviral Gene Transfer with Linear Polyethylenimine or Electroporation. Mol. Ther. 2002, 5, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Feyen, D.A.M.; McKeithan, W.L.; Bruyneel, A.A.N.; Spiering, S.; Hörmann, L.; Ulmer, B.; Zhang, H.; Briganti, F.; Schweizer, M.; Hegyi, B.; et al. Metabolic Maturation Media Improve Physiological Function of Human iPSC-Derived Cardiomyocytes. Cell Rep. 2020, 32, 107925. [Google Scholar] [CrossRef] [PubMed]
- Buikema, J.W.; Lee, S.; Goodyer, W.R.; Maas, R.G.; Chirikian, O.; Li, G.; Miao, Y.; Paige, S.L.; Lee, D.; Wu, H.; et al. Wnt Activation and Reduced Cell-Cell Contact Synergistically Induce Massive Expansion of Functional Human iPSC-Derived Cardiomyocytes. Cell Stem Cell 2020, 27, 50–63.e5. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.H.; Tao, Z.; Loo, S.; Su, L.; Chen, X.; Ye, L. Non-viral vector based gene transfection with human induced pluripotent stem cells derived cardiomyocytes. Sci. Rep. 2019, 9, 14404. [Google Scholar] [CrossRef]
- Nam, H.-S.; Benezra, R. High Levels of Id1 Expression Define B1 Type Adult Neural Stem Cells. Cell Stem Cell 2009, 5, 515–526. [Google Scholar] [CrossRef] [Green Version]
- Bergmann, O.; Bhardwaj, R.D.; Bernard, S.; Zdunek, S.; Barnabé-Heider, F.; Walsh, S.; Zupicich, J.; Alkass, K.; Buchholz, B.A.; Druid, H.; et al. Evidence for Cardiomyocyte Renewal in Humans. Science 2009, 324, 98–102. [Google Scholar] [CrossRef] [Green Version]
- Bersell, K.; Arab, S.; Haring, B.; Kühn, B. Neuregulin1/ErbB4 Signaling Induces Cardiomyocyte Proliferation and Repair of Heart Injury. Cell 2009, 138, 257–270. [Google Scholar] [CrossRef] [Green Version]
- Kühn, B.; Del Monte, F.; Hajjar, R.J.; Chang, Y.-S.; Lebeche, D.; Arab, S.; Keating, M.T. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat. Med. 2007, 13, 962–969. [Google Scholar] [CrossRef]
- Wang, W.E.; Li, L.; Xia, X.; Fu, W.; Liao, Q.; Lan, C.; Yang, D.; Chen, H.; Yue, R.; Zeng, C.S.; et al. Dedifferentiation, Proliferation, and Redifferentiation of Adult Mammalian Cardiomyocytes After Ischemic Injury. Circulation 2017, 136, 834–848. [Google Scholar] [CrossRef] [PubMed]
- D’Uva, G.; Aharonov, A.; Lauriola, M.; Kain, D.; Yahalom-Ronen, Y.; Carvalho, S.; Weisinger, K.; Bassat, E.; Rajchman, D.; Yifa, O.; et al. ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat. Cell Biol. 2015, 17, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Van der Wal, E.; Herrero-Hernandez, P.; Wan, R.; Broeders, M.; In’t Groen, S.L.; van Gestel, T.J.; van IJcken, W.F.J.; Cheung, T.H.; van der Ploeg, A.T.; Schaaf, G.J.; et al. Large-Scale Expansion of Human iPSC-Derived Skeletal Muscle Cells for Disease Modeling and Cell-Based Therapeutic Strategies. Stem Cell Rep. 2018, 10, 1975–1990. [Google Scholar] [CrossRef] [PubMed]
- Chal, J.; Oginuma, M.; Al Tanoury, Z.; Gobert, B.; Sumara, O.; Hick, A.; Bousson, F.; Zidouni, Y.; Mursch, C.; Moncuquet, P.; et al. Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat. Biotechnol. 2015, 33, 962–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariizumi, T.; Ogose, A.; Kawashima, H.; Hotta, T.; Umezu, H.; Endo, N. Multinucleation followed by an acytokinetic cell division in myxofibrosarcoma with giant cell proliferation. J. Exp. Clin. Cancer Res. 2009, 28, 44. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, Q.; Maas, R.G.C.; Brouwer, E.C.J.; Pei, J.; Blok, C.S.; Popovic, M.A.; Paauw, N.J.; Bovenschen, N.; Hjortnaes, J.; Harakalova, M.; et al. Sarcomere Disassembly and Transfection Efficiency in Proliferating Human iPSC-Derived Cardiomyocytes. J. Cardiovasc. Dev. Dis. 2022, 9, 43. https://doi.org/10.3390/jcdd9020043
Yuan Q, Maas RGC, Brouwer ECJ, Pei J, Blok CS, Popovic MA, Paauw NJ, Bovenschen N, Hjortnaes J, Harakalova M, et al. Sarcomere Disassembly and Transfection Efficiency in Proliferating Human iPSC-Derived Cardiomyocytes. Journal of Cardiovascular Development and Disease. 2022; 9(2):43. https://doi.org/10.3390/jcdd9020043
Chicago/Turabian StyleYuan, Qianliang, Renee G. C. Maas, Ellen C. J. Brouwer, Jiayi Pei, Christian Snijders Blok, Marko A. Popovic, Nanne J. Paauw, Niels Bovenschen, Jesper Hjortnaes, Magdalena Harakalova, and et al. 2022. "Sarcomere Disassembly and Transfection Efficiency in Proliferating Human iPSC-Derived Cardiomyocytes" Journal of Cardiovascular Development and Disease 9, no. 2: 43. https://doi.org/10.3390/jcdd9020043
APA StyleYuan, Q., Maas, R. G. C., Brouwer, E. C. J., Pei, J., Blok, C. S., Popovic, M. A., Paauw, N. J., Bovenschen, N., Hjortnaes, J., Harakalova, M., Doevendans, P. A., Sluijter, J. P. G., van der Velden, J., & Buikema, J. W. (2022). Sarcomere Disassembly and Transfection Efficiency in Proliferating Human iPSC-Derived Cardiomyocytes. Journal of Cardiovascular Development and Disease, 9(2), 43. https://doi.org/10.3390/jcdd9020043





