Analysis of Uncharacterized mKiaa1211 Expression during Mouse Development and Cardiovascular Morphogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Quantitative PCR Analysis mKiaa1211 Developmental mRNA Expression Levels
2.2. Isolation of Mouse mKiaa1211 cDNA Probe
2.3. In Situ Hybridization and Molecular Marker Immunohistochemistry
2.4. mKiaa1211 Mutant Mouse Analysis
2.5. Statistical Analysis
3. Results
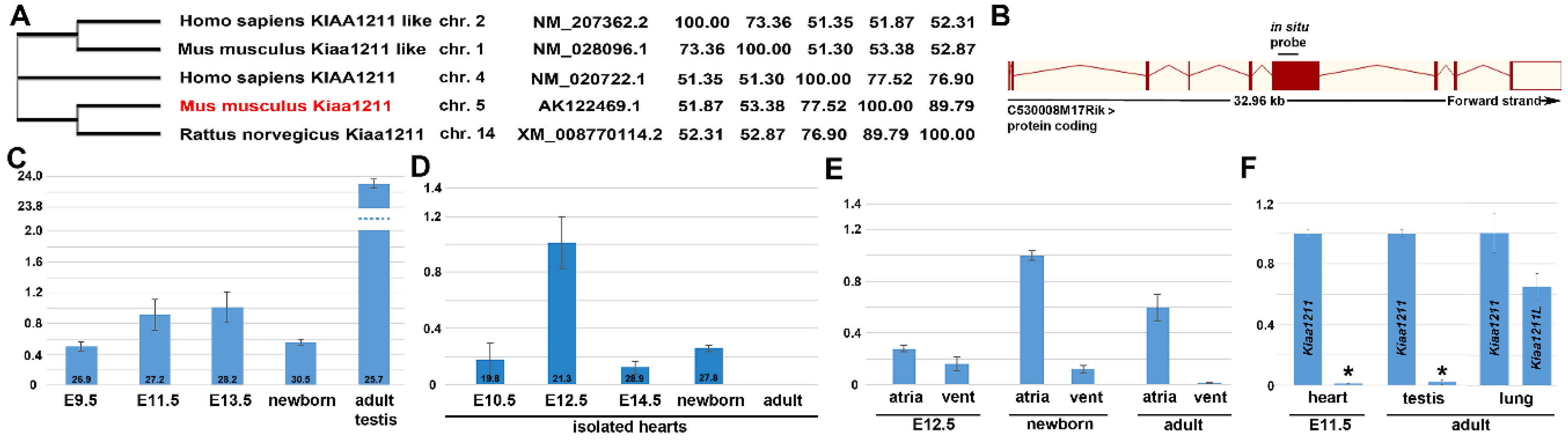
3.1. Phylogenetic and Comparative mRNA Expression Analysis
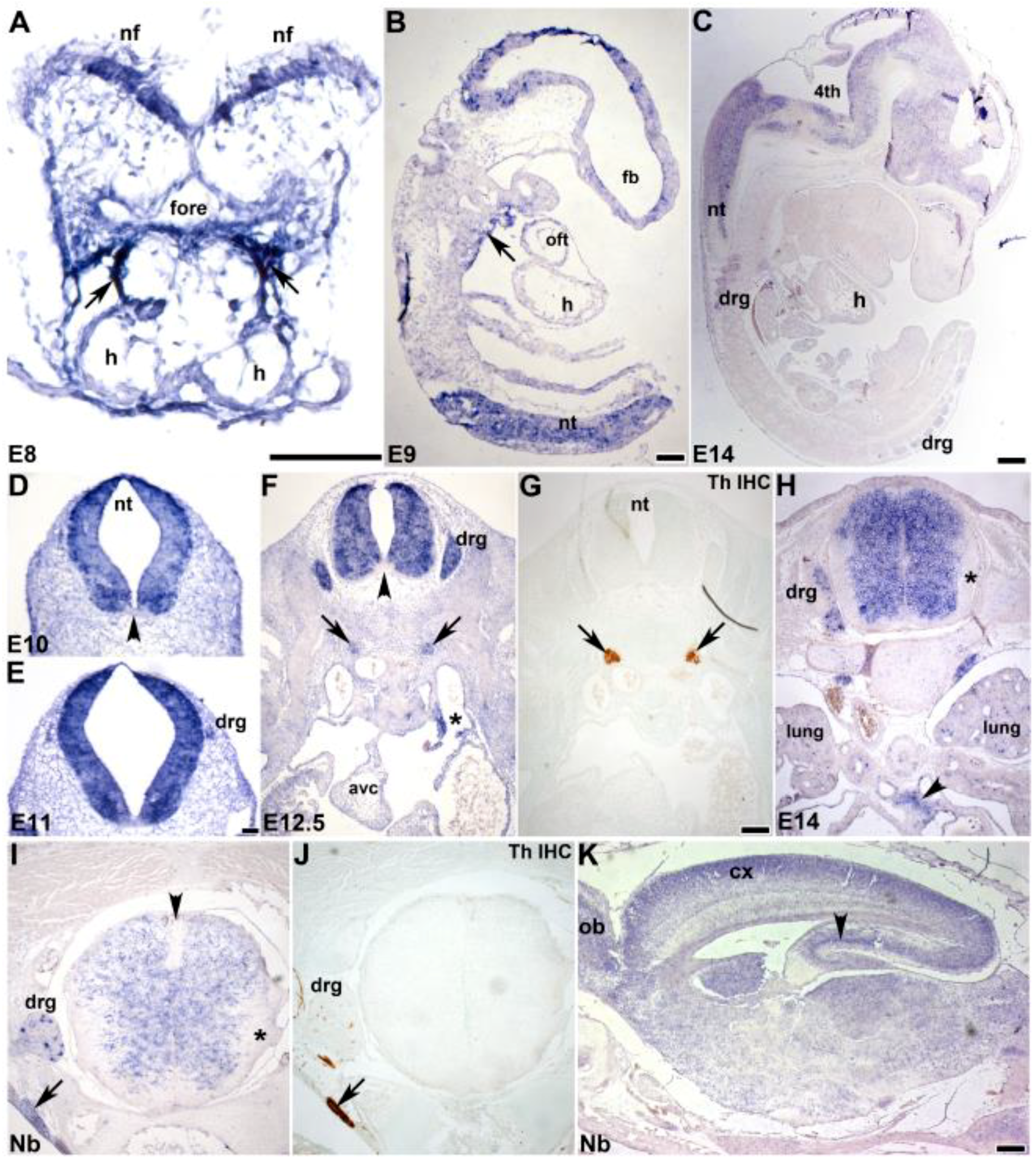
3.2. In Situ Hybridization Analysis of Developmental Spatiotemporal Expression
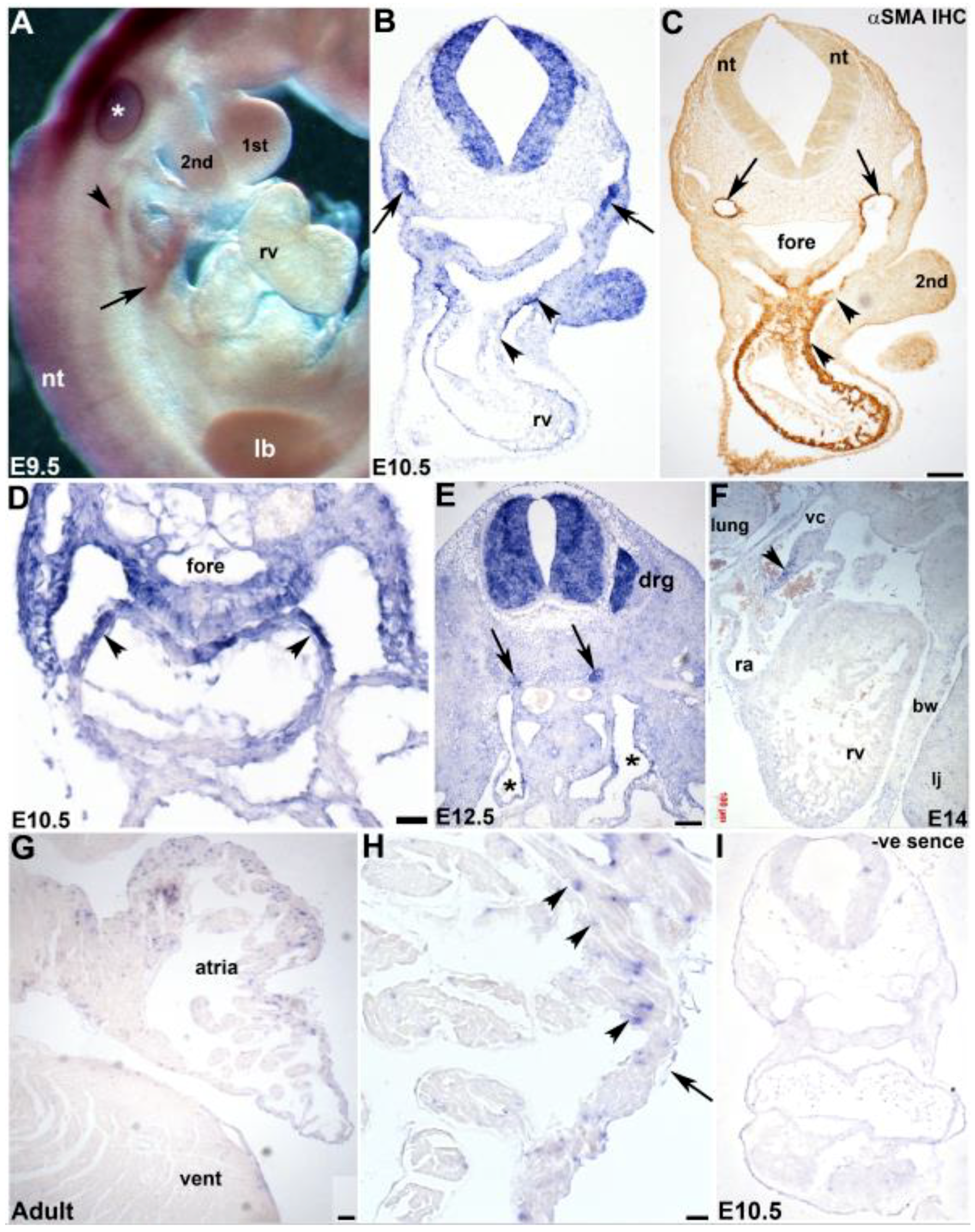
3.3. Cardiovascular Spatiotemporal mKiaa1211 Expression Analysis
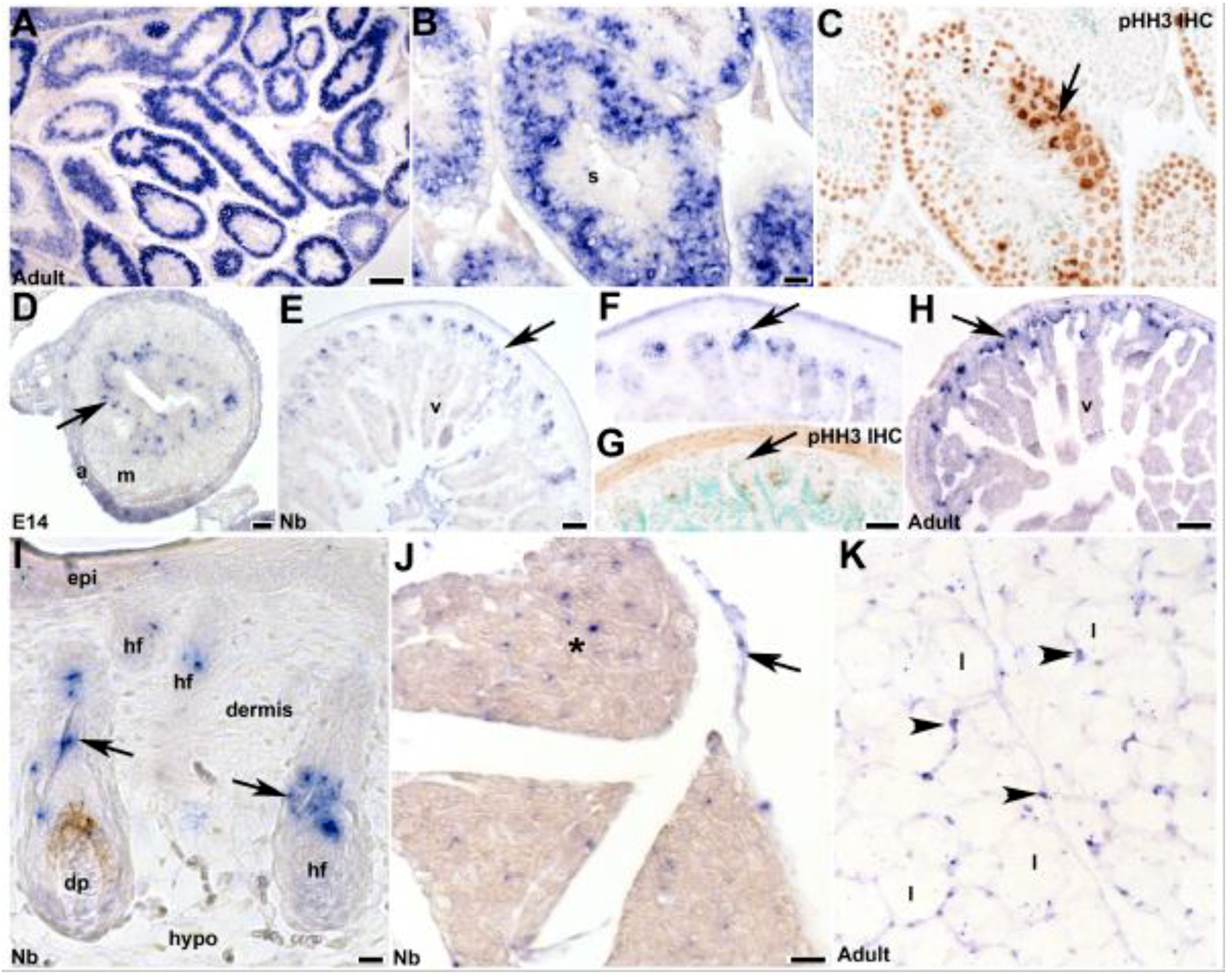
3.4. Fetal and Postnatal Spatiotemporal Analysis of mKiaa1211 mRNA
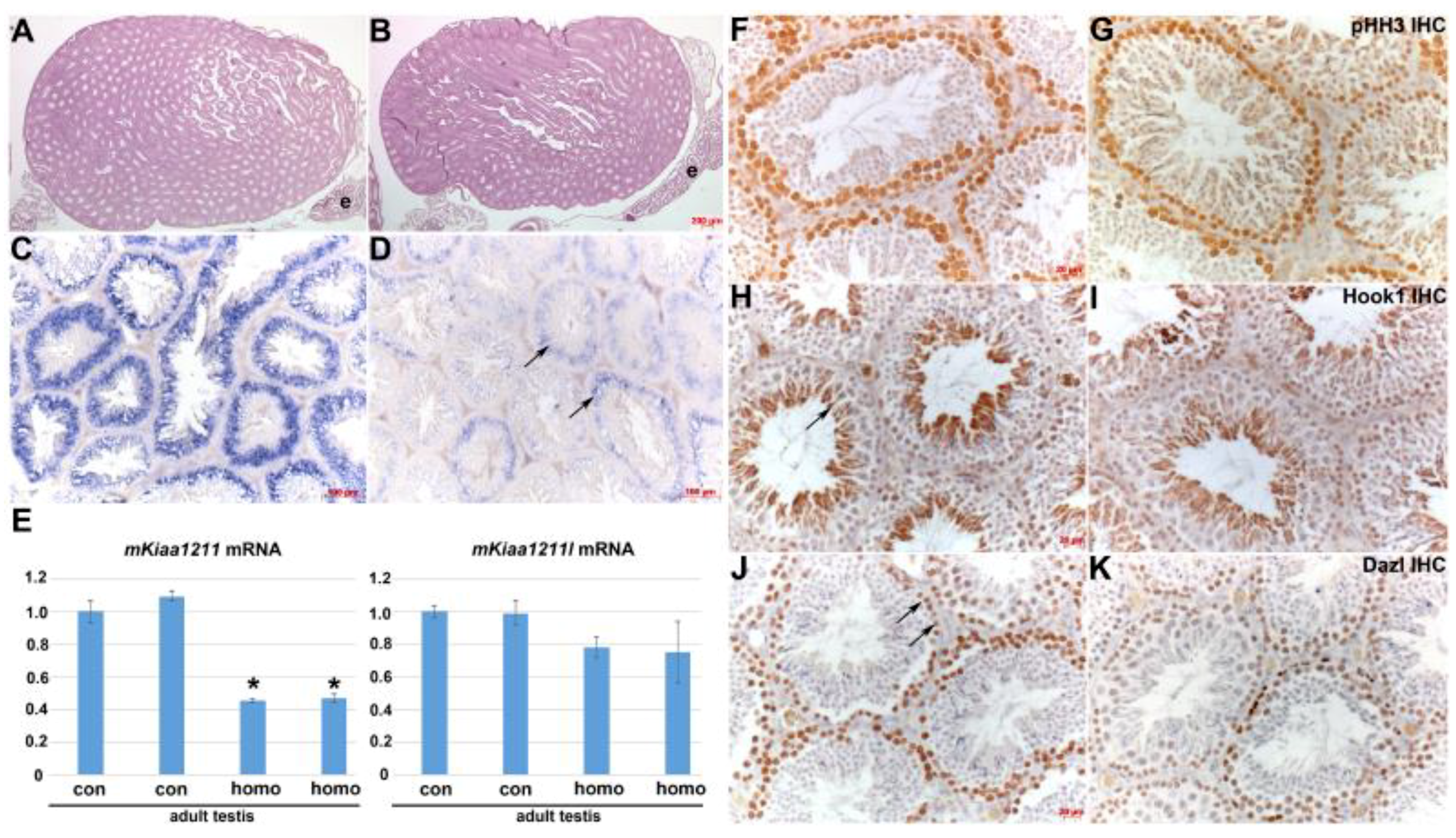
3.5. Analysis of mKiaa1211 Mouse Mutant Phenotype
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bork, P. Powers and pitfalls in sequence analysis: The 70% hurdle. Genome Res. 2000, 10, 398–400. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y.; Koonin, E.V. From complete genome sequence to ‘complete’ understanding? Trends Biotechnol. 2010, 28, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Lee, E.S. Non-coding RNA: What is functional and what is junk? Front. Genet. 2015, 6, 2. [Google Scholar] [CrossRef] [PubMed]
- Conway, S.J. In situ hybridization of cells and tissue sections. Methods Mol. Med. 1996, 6, 193–206. [Google Scholar] [CrossRef]
- Geffers, L.; Herrmann, B.; Eichele, G. Web-based digital gene expression atlases for the mouse. Mamm. Genome 2012, 23, 525–538. [Google Scholar] [CrossRef]
- Visel, A.; Thaller, C.; Eichele, G. GenePaint.org: An atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004, 32, D552–D556. [Google Scholar] [CrossRef] [PubMed]
- Krumlauf, R. Hox genes in vertebrate development. Cell 1994, 78, 191–201. [Google Scholar] [CrossRef]
- Parker, H.J.; Pushel, I.; Krumlauf, R. Coupling the roles of Hox genes to regulatory networks patterning cranial neural crest. Dev. Biol. 2018, 444, S67–S78. [Google Scholar] [CrossRef]
- Nikolopoulou, E.; Galea, G.L.; Rolo, A.; Greene, N.D.; Copp, A.J. Neural tube closure: Cellular, molecular and biomechanical mechanisms. Development 2017, 144, 552–566. [Google Scholar] [CrossRef]
- Krumlauf, R. Hox Genes and the Hindbrain: A Study in Segments. Curr. Top. Dev. Biol. 2016, 116, 581–596. [Google Scholar] [CrossRef]
- Franco, D.; Kelly, R.; Lamers, W.H.; Buckingham, M.; Moorman, A.F. Regionalized transcriptional domains of myosin light chain 3f transgenes in the embryonic mouse heart: Morphogenetic implications. Dev. Biol. 1997, 188, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Franco, D.; Markman, M.M.; Wagenaar, G.T.; Ya, J.; Lamers, W.H.; Moorman, A.F. Myosin light chain 2a and 2v identifies the embryonic outflow tract myocardium in the developing rodent heart. Anat. Rec. 1999, 254, 135–146. [Google Scholar] [CrossRef]
- Kelly, R.G.; Brown, N.A.; Buckingham, M.E. The arterial pole of the mouse heart forms from Fgf10-expressing cells in pharyngeal mesoderm. Dev. Cell 2001, 1, 435–440. [Google Scholar] [CrossRef]
- Cai, C.L.; Liang, X.; Shi, Y.; Chu, P.H.; Pfaff, S.L.; Chen, J.; Evans, S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 2003, 5, 877–889. [Google Scholar] [CrossRef]
- Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature 2002, 420, 520–562. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Wheeler, J.; Grimwood, J.; Dickson, M.; Yang, J.; Caoile, C.; Bajorek, E.; Black, S.; Chan, Y.M.; Denys, M.; et al. Quality assessment of the human genome sequence. Nature 2004, 429, 365–368. [Google Scholar] [CrossRef]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 2004, 431, 931–945. [Google Scholar] [CrossRef]
- Salzberg, S.L. Open questions: How many genes do we have? BMC Biol. 2018, 16, 94. [Google Scholar] [CrossRef]
- Nagase, T.; Koga, H.; Ohara, O. Kazusa mammalian cDNA resources: Towards functional characterization of KIAA gene products. Brief. Funct. Genomic. Proteom. 2006, 5, 4–7. [Google Scholar] [CrossRef][Green Version]
- Nagase, T.; Ishikawa, K.; Kikuno, R.; Hirosawa, M.; Nomura, N.; Ohara, O. Prediction of the coding sequences of unidentified human genes. XV. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1999, 6, 337–345. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Q.; Xu, S.; Jin, P.; Luan, P.; Li, Y.; Cao, Z.; Leng, L.; Wang, Y.; Wang, S. Differential expression of six chicken genes associated with fatness traits in a divergently selected broiler population. Mol. Cell. Probes 2016, 30, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.S.; Wang, W.; Jun, S.; Zhang, J.; Srivastava, M.; Kim, M.J.; Lien, E.M.; Shang, J.; Chen, J.; McCrea, P.D.; et al. Deregulation of CRAD-controlled cytoskeleton initiates mucinous colorectal cancer via β-catenin. Nat. Cell Biol. 2018, 20, 1303–1314. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, K.; Kakiuchi, C.; Bundo, M.; Ikeda, K.; Kato, T. Molecular characterization of bipolar disorder by comparing gene expression profiles of postmortem brains of major mental disorders. Mol. Psychiatry 2004, 9, 406–416. [Google Scholar] [CrossRef] [PubMed]
- Snider, P.; Simmons, O.; Wang, J.; Hoang, C.; Conway, S.J. Ectopic Noggin in a Population of Nfatc1 Lineage Endocardial Progenitors Induces Embryonic Lethality. J. Cardiovasc. Dev. Dis. 2014, 1, 214–236. [Google Scholar] [CrossRef] [PubMed]
- Kruzynska-Frejtag, A.; Machnicki, M.; Rogers, R.; Markwald, R.; Conway, S.J. Periostin (an osteoblast-specific factor) is expressed within the embryonic mouse heart during valve formation. Mech. Dev. 2001, 103, 183–188. [Google Scholar] [CrossRef]
- Simmons, O.; Bolanis, E.; Wang, J.; Conway, S.J. In situ hybridization (both radioactive and nonradioactive) and spatiotemporal gene expression analysis. Methods Mol. Biol. 2014, 1194, 225–244. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef]
- Lallemend, F.; Ernfors, P. Molecular interactions underlying the specification of sensory neurons. Trends Neurosci. 2012, 35, 373–381. [Google Scholar] [CrossRef]
- Cochard, P.; Goldstein, M.; Black, I.B. Ontogenetic appearance and disappearance of tyrosine hydroxylase and catecholamines in the rat embryo. Proc. Natl. Acad. Sci. USA 1978, 75, 2986–2990. [Google Scholar] [CrossRef]
- Zammit, P.S.; Kelly, R.G.; Franco, D.; Brown, N.; Moorman, A.F.; Buckingham, M.E. Suppression of atrial myosin gene expression occurs independently in the left and right ventricles of the developing mouse heart. Dev. Dyn. 2000, 217, 75–85. [Google Scholar] [CrossRef]
- Rochais, F.; Mesbah, K.; Kelly, R.G. Signaling pathways controlling second heart field development. Circ. Res. 2009, 104, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Cohen, E.D.; Wang, Z.; Lepore, J.J.; Lu, M.M.; Taketo, M.M.; Epstein, D.J.; Morrisey, E.E. Wnt/beta-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J. Clin. Investig. 2007, 117, 1794–1804. [Google Scholar] [CrossRef] [PubMed]
- Klaus, A.; Müller, M.; Schulz, H.; Saga, Y.; Martin, J.F.; Birchmeier, W. Wnt/β-catenin and Bmp signals control distinct sets of transcription factors in cardiac progenitor cells. Proc. Natl. Acad. Sci. USA 2012, 109, 10921–10926. [Google Scholar] [CrossRef] [PubMed]
- Brafman, D.; Willert, K. Wnt/β-catenin signaling during early vertebrate neural development. Dev. Neurobiol. 2017, 77, 1239–1259. [Google Scholar] [CrossRef]
- Snider, P.; Olaopa, M.; Firulli, A.B.; Conway, S.J. Cardiovascular development and the colonizing cardiac neural crest lineage. Sci. World J. 2007, 7, 1090–1113. [Google Scholar] [CrossRef] [PubMed]
- Mayor, R.; Theveneau, E. The role of the non-canonical Wnt-planar cell polarity pathway in neural crest migration. Biochem. J. 2014, 457, 19–26. [Google Scholar] [CrossRef]
- Lajiness, J.D.; Snider, P.; Wang, J.; Feng, G.S.; Krenz, M.; Conway, S.J. SHP-2 deletion in postmigratory neural crest cells results in impaired cardiac sympathetic innervation. Proc. Natl. Acad. Sci. USA 2014, 111, E1374–E1382. [Google Scholar] [CrossRef]
- Barbaric, I.; Miller, G.; Dear, T.N. Appearances can be deceiving: Phenotypes of knockout mice. Brief Funct. Genom. Proteom. 2007, 6, 91–103. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
L. Snider, P.; Snider, E.; Simmons, O.; Lilly, B.; J. Conway, S. Analysis of Uncharacterized mKiaa1211 Expression during Mouse Development and Cardiovascular Morphogenesis. J. Cardiovasc. Dev. Dis. 2019, 6, 24. https://doi.org/10.3390/jcdd6020024
L. Snider P, Snider E, Simmons O, Lilly B, J. Conway S. Analysis of Uncharacterized mKiaa1211 Expression during Mouse Development and Cardiovascular Morphogenesis. Journal of Cardiovascular Development and Disease. 2019; 6(2):24. https://doi.org/10.3390/jcdd6020024
Chicago/Turabian StyleL. Snider, Paige, Elizabeth Snider, Olga Simmons, Brenda Lilly, and Simon J. Conway. 2019. "Analysis of Uncharacterized mKiaa1211 Expression during Mouse Development and Cardiovascular Morphogenesis" Journal of Cardiovascular Development and Disease 6, no. 2: 24. https://doi.org/10.3390/jcdd6020024
APA StyleL. Snider, P., Snider, E., Simmons, O., Lilly, B., & J. Conway, S. (2019). Analysis of Uncharacterized mKiaa1211 Expression during Mouse Development and Cardiovascular Morphogenesis. Journal of Cardiovascular Development and Disease, 6(2), 24. https://doi.org/10.3390/jcdd6020024





