Antimicrobial Resistance of Clinical and Commensal Escherichia coli Canine Isolates: Profile Characterization and Comparison of Antimicrobial Susceptibility Results According to Different Guidelines
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Antimicrobial Susceptibility Testing
2.3. Biofilm Formation in the Air–Liquid Interface
2.4. Multiplex Polymerase Chain Reaction Assay of β-lactamases-Encoding Genes blaTEM, blaSHV and blaOXA
2.4.1. DNA Extraction
2.4.2. PCR Amplification
2.5. Statistical Analysis
3. Results
3.1. Antimicrobial Susceptibility Testing
3.1.1. Determination of Multidrug-Resistant Strains
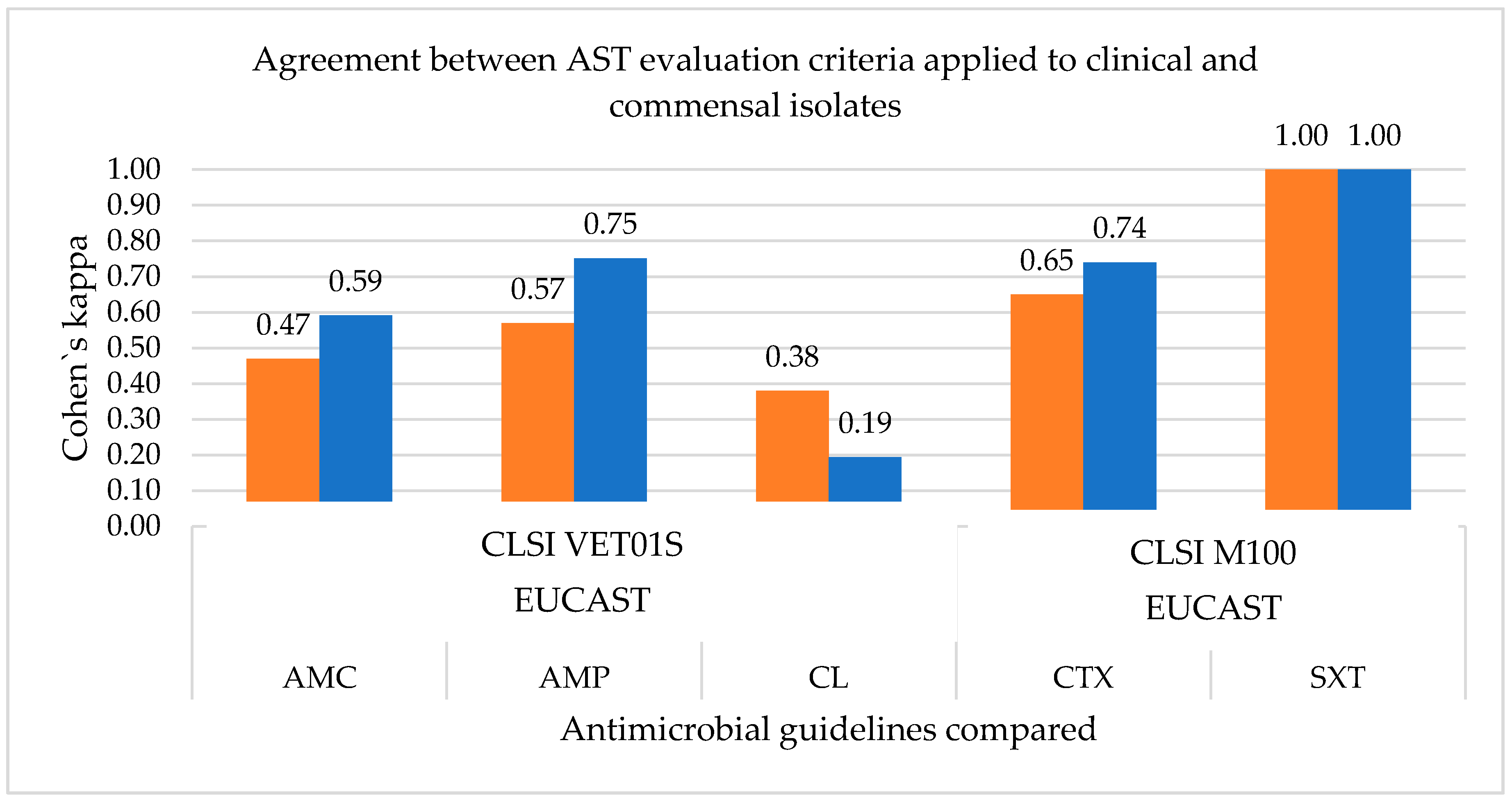
3.1.2. Measurement of the Agreement between the Antimicrobial Susceptibility Testing Guidelines
3.2. Antimicrobial Resistance Profile and Phylogenetic Group
3.3. Biofilm Formation in the Air–Liquid Interface
3.4. Biofilm Formation and Antimicrobial Resistance Profile
3.5. Multiplex Polymerase Chain Reaction Assay of βlactamases-Encoding Genes blaTEM, blaSHV and blaOXA
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- EFSA (European Food Safety Authority); ECDC (European Centre for Disease Prevention and Control). The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2015. EFSA J. 2017, 15, e04694. [Google Scholar]
- Shaikh, S.; Fatima, J.; Shakil, S.; Rizvi, S.; Kamal, M. Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment. Saudi J. Biol. Sci. 2015, 22, 90–101. [Google Scholar] [CrossRef]
- Tenover, F. Mechanisms of antimicrobial resistance in bacteria. Am. J. Infect. Control 2006, 34 (Suppl. 1), S3–S10. [Google Scholar] [CrossRef]
- Adekunle, O. Mechanisms of Antimicrobial Resistance in Bacteria, General Approach. Int. J. Pharm. Med. 2012, 1, 166–187. [Google Scholar]
- World Health Organization. Available online: http://www.who.int/antimicrobial-resistance/global-actionplan/surveillance/glass/en/ (accessed on 12 July 2021).
- Masterton, R. The Importance and Future of Antimicrobial Surveillance Studies. Clin. Infect. Dis. 2008, 47, S21–S31. [Google Scholar] [CrossRef]
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef]
- Heuer, O.; Jensen, V.; Hammerum, A. Antimicrobial Drug Consumption in Companion Animals. Emerg. Infect. Dis. 2005, 11, 344–345. [Google Scholar] [CrossRef]
- Umber, J.; Bender, J. Pets and antimicrobial resistance. Vet. Clin. N. Am. Small Anim. Pract. 2009, 39, 279–292. [Google Scholar] [CrossRef]
- Guardabassi, L.; Jensen, L.B.; Kruse, H. Guide to Antimicrobial Use in Animals; Blackwell Pub.: Oxford, UK, 2008. [Google Scholar]
- Rodloff, A.; Bauer, T.; Ewig, S.; Kujath, P.; Müller, E. Susceptible, Intermediate, and Resistant—The Intensity of Antibiotic Action. Dtsch. Arztebl. Int. 2008, 105, 657–662. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Antimicrobial Resistance Surveillance in Europe 2014. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net); ECDC: Stockholm, Sweden, 2015.
- European Committee on Antimicrobial Susceptibility Testing. Veterinary Committee on AST; EUCAST, Sweden. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_11.0_Breakpoint_Tables.pdf (accessed on 20 December 2020).
- Allocati, N.; Masulli, M.; Alexeyev, M.; Di Illio, C. Escherichia coli in Europe: An Overview. Int. J. Environ. Res. Public Health 2013, 10, 6235–6254. [Google Scholar] [CrossRef]
- Johnston, S.D.; Kustritz, M.V.; Olson, P.S. Canine and Feline Theriogenology; Saunders: Philadelphia, PA, USA, 2001. [Google Scholar]
- Jitpean, S.; Ström-Holst, B.; Emanuelson, U.; Höglund, O.V.; Pettersson, A.; Alneryd-Bull, C.; Hagman, R. Outcome of pyometra in female dogs and predictors of peritonitis and prolonged postoperative hospitalization in surgically treated cases. BMC Vet. Res. 2014, 10, 6. [Google Scholar] [CrossRef]
- Wood, T. Insights on Escherichia coli Biofilm Formation and Inhibition from Whole-Transcriptome Profiling. Environ. Microbiol. 2009, 11, 1–15. [Google Scholar] [CrossRef]
- Schembri, M.; Kjaergaard, K.; Klemm, P. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 2003, 48, 253–267. [Google Scholar] [CrossRef]
- Allewell, N. Introduction to Biofilms Thematic Minireview Series. JBC 2016, 291, 12527–12528. [Google Scholar] [CrossRef]
- Stewart, P.; Costerton, J. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; Sintim, H. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med. Chem. 2015, 7, 493–512. [Google Scholar] [CrossRef]
- Li, X.; Mehrotra, M.; Ghimire, S.; Adewoye, L. β-Lactam resistance and β-lactamases in bacteria of animal origin. Vet. Microbiol. 2007, 121, 197–214. [Google Scholar] [CrossRef]
- Dallence, C.; Da Costa, A.; Decré, D.; Favier, C.; Arlet, G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010, 65, 490–495. [Google Scholar] [CrossRef]
- Mateus, L.; Henriques, S.; Merino, C.; Pomba, C.; Lopes da Costa, L.; Silva, E. Virulence genotypes of Escherichia coli canine isolates from pyometra, cystitis and fecal origin. Vet. Microbiol. 2013, 166, 590–594. [Google Scholar] [CrossRef]
- Machado, I. Piometra na Cadela e na Gata: Diferenças e Semelhanças. Dissertação de Mestrado Integrado em Medicina Veterinária, Faculdade de Medicina Veterinária, Universidade de Lisboa, Lisboa, Portugal, 2017. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals, 5th ed.; VET01S; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2021. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Tests, 31st ed.; M100; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2021. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals, 4th ed.; VET01-A4; Clinical and Laboratory Standards Institute (CLSI): Wayne, PA, USA, 2013. [Google Scholar]
- Magiorakos, A.; Srinivasan, A.; Carey, R.; Carmeli, Y.; Falagas, M.; Giske, C.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrugresistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Seixas, R.; Machado, J.; Bernardo, F.; Vilela, C.; Oliveira, M. Biofilm Formation by Salmonella Enterica Serovar 1,4,[5],12:i:—Portuguese Isolates: A Phenotypic, Genotypic, and Sociogeographic Analysis. Curr. Microbiol. 2014, 68, 670–677. [Google Scholar] [CrossRef]
- Landis, J.; Koch, G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef]
- Jones-Dias, D.; Manageiro, V.; Ferreira, E.; Louro, D.; Antibiotic Resistance Surveillance Program in Portugal (ARSIP) Participants; Caniça, M. Diversity of Extended-Pectrum and Plasmid-Mediated AmpC β-Lactamases in Enterobacteriaceae Isolates from Portuguese Health Care Facilities. J. Microbiol. 2014, 52, 496–503. [Google Scholar] [CrossRef]
- Picard, B.; Garcia, J.S.; Gouriou, S.; Duriez, P.; Brahimi, N.; Bingen, E.; Elion, J.; Denamur, E. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 1999, 67, 546–553. [Google Scholar] [CrossRef]
- Stewart, P. Antimicrobial Tolerance in Biofilms. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef]
- Weese, J. Antimicrobial resistance in companion animals. Anim. Health Res. Rev. 2008, 9, 169–176. [Google Scholar] [CrossRef]
- Cummings, K.J.; Aprea, V.A.; Altier, C. Antimicrobial resistance trends among canine Escherichia coli isolates obtained from clinical samples in the northeastern USA, 2004–2011. Can. Vet. J. 2015, 56, 393–398. [Google Scholar]
- Guardabassi, L.; Schwarz, S.; Lloyd, D. Pet animals as reservoirs of antimicrobial-resistant bacteria. J. Antimicrob. Chemother. 2004, 54, 321–332. [Google Scholar] [CrossRef]
- Thungrat, K.; Price, S.; Carpenter, M.; Boothe, D. Antimicrobial susceptibility patterns of clinical Escherichia coli isolates from dogs and cats in the United States: January 2008 through January 2013. Vet. Microbiol. 2015, 179, 287–295. [Google Scholar] [CrossRef]
- Boothe, D.; Smaha, T.; Carpenter, D.; Shaheen, B.; Hatchcock, T. Antimicrobial Resistance and Pharmacodynamics of Canine and Feline Pathogenic E. coli in the United States. J. Am. Anim. Hosp. Assoc. 2012, 48, 379–389. [Google Scholar] [CrossRef]
- Tadesse, D.; Zhao, S.; Tong, E.; Ayers, S.; Singh, A.; Bartholomew, M.J.; McDermott, P.F. Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950–2002. Emerg. Infect. Dis. 2012, 18, 741–749. [Google Scholar] [CrossRef]
- Ettiger, S.J.; Feldman, E.C. Textbook of Veterinary Internal Medicine; Saunders: Philadelphia, PA, USA, 2010; Volume 2. [Google Scholar]
- Tenaillon, O.; Skurnik, D.; Picard, B.; Denamur, E. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 2010, 8, 207–217. [Google Scholar] [CrossRef]
- Lees, P.; Shojaee, F. Rational dosing of antimicrobial drugs: Animals versus humans. Int. J. Antimicrob. Agents 2002, 19, 269–284. [Google Scholar] [CrossRef]
- Labreche, M.; Graber, C.; Nguyen, H. Recent Upgrades on the Role of Pharmacokinetic-pharmacodynamics in Antimicrobial Susceptibility Testing as Applied to Clinical Practice. Clin. Infect. Dis. 2015, 61, 1446–1452. [Google Scholar]
- Jenkins, S.; Jerris, R. Critical Assessment of Issues Applicable to Development of Antimicrobial Susceptibility Testing Breakpoints. J. Clin. Microbiol. 2011, 49, S5–S10. [Google Scholar] [CrossRef][Green Version]
- Silley, P. Susceptibility testing methods, resistance and breakpoints: What do these terms really mean? Rev. Sci. Tech. Off. Int. Epiz. 2012, 31, 33–41. [Google Scholar] [CrossRef]
- Valsesia, G.; Roos, M.; Böttger, E.; Hombach, M. A Statistical Approach for Determination of Disk Diffusion-Based Cutoff Values for Systematic Characterization of Wild-Type and Non-Wild-Type Bacterial Populations in Antimicrobial Susceptibility Testing. J. Clin. Microbiol. 2015, 53, 1812–1822. [Google Scholar] [CrossRef]
- Matusckek, E.; Brown, D.; Kahlmeter, G. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin. Microbiol. Infect. 2014, 20, O255–O266. [Google Scholar] [CrossRef]
- Toutain, P.L.; Bousquet-Mélou, A.; Damborg, P.; Ferran, A.A.; Mevius, D.; Pelligand, L.; Veldman, K.T.; Lees, P. En Route towards European Clinical Breakpoints for Veterinary Antimicrobial Susceptibility Testing: A Position Paper Explaining the VetCAST Approach. Front. Microbiol. 2017, 8, 2344. [Google Scholar] [CrossRef]
- Nave, P.; Prado, G.; Huelves, L.; Gracia, M.; Ruiz, V.; Blanco, J.; Dahbi, G.; Blanco, M.; Mdel, C.P.; Soriano, F. Correlation between virulence factors and in vitro biofilm formation by Escherichia coli strains. Microb. Pathog. 2008, 45, 86–91. [Google Scholar] [CrossRef]
- Lopes, C.E.; De Carli, S.; Riboldi, C.I.; De Lorenzo, C.; Panziera, W.; Driemeier, D.; Siqueira, F.M. Pet Pyometra: Correlating Bacteria Pathogenicity to Endometrial Histological Changes. Pathogens 2021, 10, 833. [Google Scholar] [CrossRef]
- Mackenzie, K.; Palmer, M.; Köster, W.; White, A. Examining the Link between Biofilm Formation and the Ability of Pathogenic Salmonella Strains to Colonize Multiple Host Species. Front. Vet. Sci. 2017, 4, 138. [Google Scholar] [CrossRef]
- Clemente, L.; Manageiro, V.; Jones-Dias, D.; Correia, I.; Themudo, P.; Albuquerque, T.; Geraldes, M.; Matos, F.; Almendra, C.; Ferreira, E.; et al. Antimicrobial susceptibility and oxymino-β-lactam resistance mechanisms in Salmonella enterica and Escherichia coli isolates from different animal sources. Res. Microbiol. 2015, 166, 574–583. [Google Scholar] [CrossRef]
- Féria, C.; Ferreira, E.; Correia, J.; Gonçalves, J.; Caniça, M. Patterns and mechanisms of resistance to β-lactamase inhibitors in uropathogenic Escherichia coli isolated from dogs in Portugal. J. Antimicrob. Chemother. 2002, 49, 77–85. [Google Scholar] [CrossRef]
- Ghafourian, S.; Sadegjifard, N.; Soheili, S.; Sekawi, Z. Extended Spectrum Beta-lactamases: Definition, Classification and Epidemiology. Curr. Issues Mol. Biol. 2015, 17, 11–22. [Google Scholar]

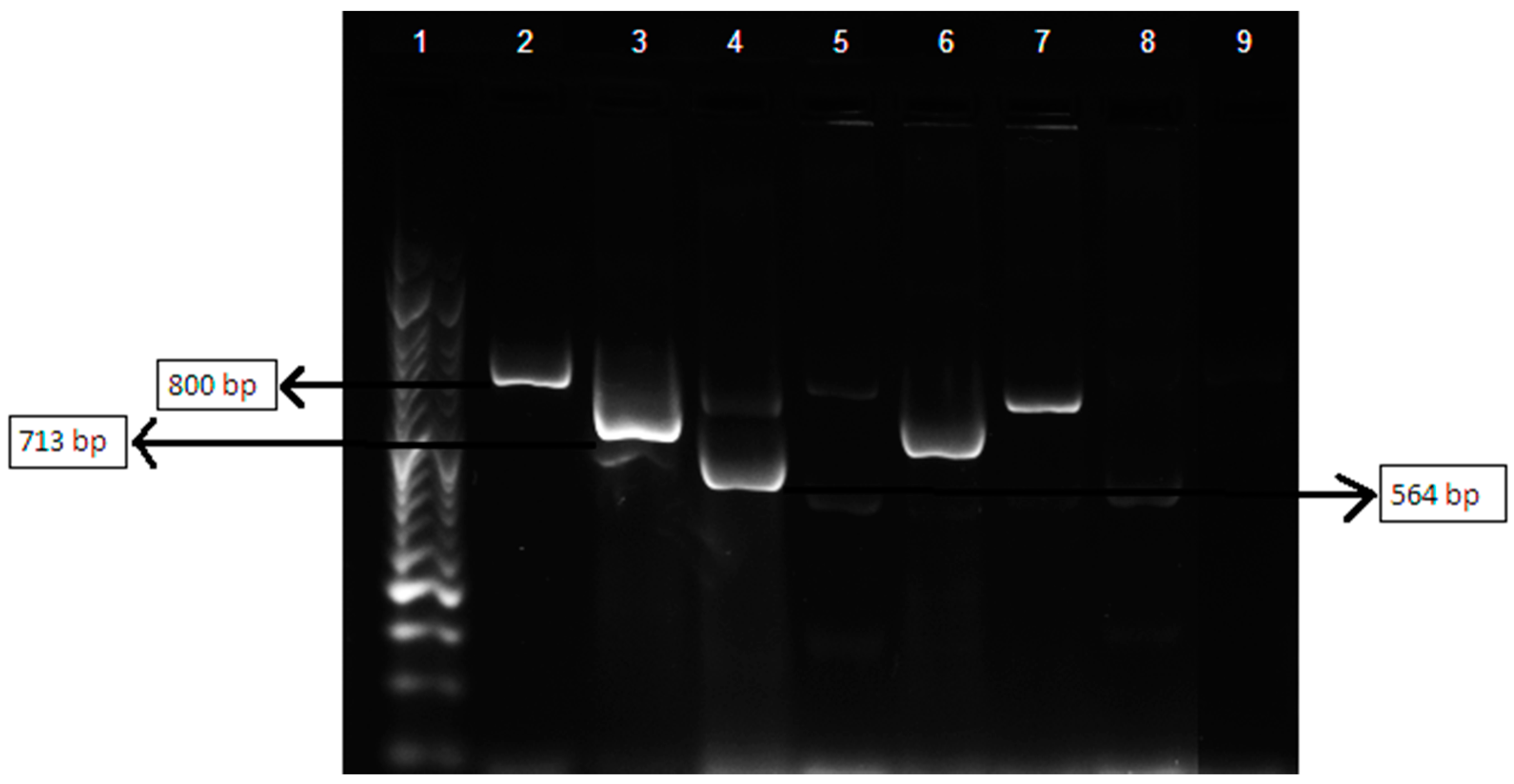
| β-lactamases Targeted | Sequence (5′–3′) | Product Size (bp) |
|---|---|---|
| TEM variants | F_CATTTCCGTGTCGCCCTTATTC | 800 |
| R_CGTTCATCCATAGTTGCCTGAC | ||
| SHV variants | F_AGCCGCTTGAGCAAATTAAAC | 713 |
| R_ATCCCGCAGATAAATCACCAC | ||
| OXA-1, OXA-4 and OXA-30 | F_GGCACCAGATTCAACTTTCAAG | 564 |
| R_GACCCCAAGTTTCCTGTAAGTG |
| E. coli Total Strains (n = 74) | E. coli Strains from Pyometra-Presenting Dogs (n = 33) | E. coli Commensal Strains (n = 41) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Drug/Interpretation (%) | M | V | EUC | M | V | EUC | M | V | EUC | |
| R | 59.5 | 59.5 | 59.5 | 51.5 | 51.5 | 51.5 | 65.9 | 65.9 | 65.9 | |
| Ampicillin | I | 18.9 | 18.9 | 0.0 | 27.3 | 27.3 | 0.0 | 12.2 | 12.2 | 0.0 |
| S | 21.6 | 21.6 | 40.5 | 21.2 | 21.2 | 48.5 | 22.0 | 22.0 | 34.1 | |
| Amoxicillin | R | 29.7 | 29.7 | 54.1 | 21.2 | 21.2 | 51.5 | 36.6 | 36.6 | 56.1 |
| clavulanic acid | I | 13.5 | 13.5 | 0.0 | 24.2 | 24.2 | 0.0 | 4.9 | 4.9 | 0.0 |
| S | 56.8 | 56.8 | 45.9 | 54.5 | 54.5 | 48.5 | 58.5 | 58.5 | 43.9 | |
| R | - | 5.4 | - | - | 0.0 | - | - | 9.8 | - | |
| Enrofloxacin | I | - | 6.8 | - | - | 3.0 | - | - | 9.8 | - |
| S | - | 87.8 | - | - | 97.0 | - | - | 80.5 | - | |
| R | 16.2 | 16.2 | 10.8 | 18.2 | 18.2 | 15.2 | 14.6 | 14.6 | 7.3 | |
| Cephalexin | I | 32.4 | 32.4 | 0.0 | 30.3 | 30.3 | 0.0 | 34.1 | 34.1 | 0.0 |
| S | 51.4 | 51.4 | 89.2 | 51.5 | 51.5 | 84.8 | 51.2 | 51.2 | 92.7 | |
| R | 4.1 | - | 1.4 | 3.0 | - | 0.0 | 4.9 | - | 2.4 | |
| Cefotaxime | I | 0.0 | - | 1.4 | 0.0 | - | 0.0 | 0.0 | - | 2.4 |
| S | 95.9 | - | 97.3 | 97.0 | - | 100 | 95.1 | - | 95.1 | |
| Sulfamethoxazole | R | 10.8 | 10.8 | 10.8 | 0.0 | 0.0 | 0.0 | 19.5 | 19.5 | 19.5 |
| Trimethoprim | I | 1.4 | 1.4 | 1.4 | 3.0 | 3.0 | 3.0 | 0.0 | 0.0 | 0.0 |
| S | 87.8 | 87.8 | 87.8 | 97.0 | 97.0 | 97.0 | 80.5 | 80.5 | 80.5 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes, V.; Cunha, E.; Nunes, T.; Silva, E.; Tavares, L.; Mateus, L.; Oliveira, M. Antimicrobial Resistance of Clinical and Commensal Escherichia coli Canine Isolates: Profile Characterization and Comparison of Antimicrobial Susceptibility Results According to Different Guidelines. Vet. Sci. 2022, 9, 284. https://doi.org/10.3390/vetsci9060284
Fernandes V, Cunha E, Nunes T, Silva E, Tavares L, Mateus L, Oliveira M. Antimicrobial Resistance of Clinical and Commensal Escherichia coli Canine Isolates: Profile Characterization and Comparison of Antimicrobial Susceptibility Results According to Different Guidelines. Veterinary Sciences. 2022; 9(6):284. https://doi.org/10.3390/vetsci9060284
Chicago/Turabian StyleFernandes, Vera, Eva Cunha, Telmo Nunes, Elisabete Silva, Luís Tavares, Luísa Mateus, and Manuela Oliveira. 2022. "Antimicrobial Resistance of Clinical and Commensal Escherichia coli Canine Isolates: Profile Characterization and Comparison of Antimicrobial Susceptibility Results According to Different Guidelines" Veterinary Sciences 9, no. 6: 284. https://doi.org/10.3390/vetsci9060284
APA StyleFernandes, V., Cunha, E., Nunes, T., Silva, E., Tavares, L., Mateus, L., & Oliveira, M. (2022). Antimicrobial Resistance of Clinical and Commensal Escherichia coli Canine Isolates: Profile Characterization and Comparison of Antimicrobial Susceptibility Results According to Different Guidelines. Veterinary Sciences, 9(6), 284. https://doi.org/10.3390/vetsci9060284






