An Appropriate Genetic Approach for Improving Reproductive Traits in Crossbred Thai–Holstein Cattle under Heat Stress Conditions
Abstract
1. Introduction
2. Thai–Holstein Crossbred Cattle
3. Reproductive Traits in Dairy Cattle
3.1. Interval Traits
3.1.1. Age at First Calving (AFC)
3.1.2. Calving to First Service Interval (CFI)
3.1.3. Interval from First to Last Insemination (IFL)
3.1.4. Days Open (DO)
3.1.5. Calving Interval (CI)
3.2. Binary Traits
3.2.1. First Service Conception Rate (FSCR)
3.2.2. Conception Rate (CR)
3.2.3. Pregnancy within 90 Days after First Service (P90)
3.3. Count Traits
The Number of Services per Conception (NSPC)
4. Heat Stress and Its Effect on Reproductive Traits
5. Genetic Parameters and a Model for Reproductive Traits in Dairy Cattle Experiencing Heat Stress
5.1. Heritability (h2)
5.2. Genetic Correlation (rg)
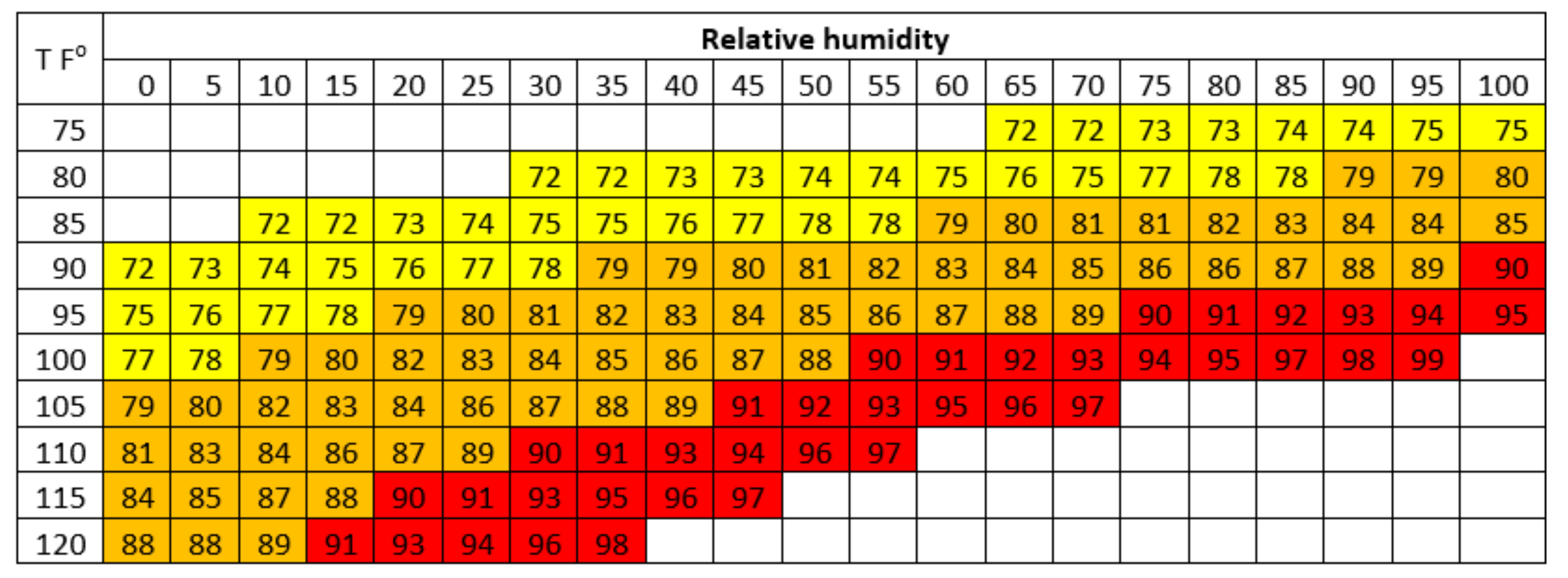
5.3. THI and Genetic Model for Heat Stress
5.3.1. THI Model
5.3.2. Genetic Model
6. The Approach of Genetic Selection for Reproductive Traits in Dairy Cattle
6.1. Traditional Breeding Method
6.2. Marker-Assisted Selection (MAS)
6.3. Genomic Selection (GS) Method
6.3.1. Bayesian Approaches
6.3.2. Multi-Step Genomic Evaluation
6.3.3. Single-Step Genomic Evaluation
- All relatives of genotyped individuals are automatically accounted for, as are their performances.
- Fitting genetic data and estimating additional effects are performed at the same time (e.g., contemporary groups). As a result, there will be no data loss.
- Feedback: all of a genotyped individual’s relatives benefit from the greater accuracy.
- Extensions are simple. Because this is a linear BLUP-like estimator, it can be immediately applied to more complex models (multiple traits, threshold traits, test day records). Any model that can be fitted with relationship matrices may also be fitted with combined relationship matrices.
6.3.4. Accuracy of GS
6.3.5. Challenges and Opportunities in Applying GS
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Masson-Delmotte, V.; Zhai, P.; Pirani, A.; Connors, S.L.; Péan, C.; Berger, S.; Caud, N.; Chen, Y.; Goldfarb, L.; Gomis, M.I.; et al. Climate Change 2021: The Physical Science Basis, the Working Group I contribution to the Sixth Assessment Report; Cambridge University Press: Cambridge, UK, 2022; pp. 1–35. [Google Scholar]
- Thammahakin, P.; Yawongsa, A.; Rukkwamsuk, T. Effect of Heat Stress on Reproductive Performance of Dairy Cows under Tropical Climate: A Review. J. Kasetsart Vet. 2020, 30, 122–133. [Google Scholar]
- Kachenchart, B.; Kamlangkla, C.; Puttanapong, N.; Limsakul, A. Urbanization effects on surface air temperature trends in Thailand during 1970–2019. Environ. Eng. Res. 2020, 26, 200378. [Google Scholar] [CrossRef]
- Mader, T.L.; Davis, M.S.; Brown-Brandl, T. Environmental factors influencing heat stress in feedlot cattle. J. Anim. Sci. 2006, 84, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Berman, A.; Horovitz, T.; Kaim, M.; Gacitua, H. A comparison of THI indices leads to a sensible heat-based heat stress index for shaded cattle that aligns temperature and humidity stress. Int. J. Biometeorol. 2016, 60, 1453–1462. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bjerg, B.S.; Choi, C.Y.; Zong, C.; Zhang, G. A review and quantitative assessment of cattle-related thermal indices. J. Therm. Biol. 2018, 77, 24–37. [Google Scholar] [CrossRef]
- Schüller, L.; Michaelis, I.; Heuwieser, W. Impact of heat stress on estrus expression and follicle size in estrus under field conditions in dairy cows. Theriogenology 2017, 102, 48–53. [Google Scholar] [CrossRef]
- Nasr, M.A.; El-Tarabany, M.S. Impact of three THI levels on somatic cell count, milk yield and composition of multiparous Holstein cows in a subtropical region. J. Therm. Biol. 2017, 64, 73–77. [Google Scholar] [CrossRef]
- Boonkum, W.; Misztal, I.; Duangjinda, M.; Pattarajinda, V.; Tumwasorn, S.; Buaban, S. Short communication: Genetic effects of heat stress on days open for Thai Holstein crossbreds. J. Dairy Sci. 2011, 94, 1592–1596. [Google Scholar] [CrossRef]
- Ravagnolo, O.; Misztal, I. Effect of Heat Stress on Nonreturn Rate in Holsteins: Fixed-Model Analyses. J. Dairy Sci. 2002, 85, 3101–3106. [Google Scholar] [CrossRef]
- Thulani, S.; Maliviwe, M.; Ayodeji, I.P. Heat tolerance level in dairy herds: A review on coping strategies to heat stress and ways of measuring heat tolerance. J. Anim. Behav. Biometeorol. 2019, 7, 39–51. [Google Scholar] [CrossRef]
- Boonkum, W.; Misztal, I.; Duangjinda, M.; Pattarajinda, V.; Tumwasorn, S.; Sanpote, J. Genetic effects of heat stress on milk yield of Thai Holstein crossbreds. J. Dairy Sci. 2011, 94, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Bohlouli, M.; Shodja, J.; Alijani, S.; Eghbal, A. The relationship between temperature-humidity index and test-day milk yield of Iranian Holstein dairy cattle using random regression model. Livest. Sci. 2013, 157, 414–420. [Google Scholar] [CrossRef]
- Rahbar, R.; Aminafshar, M.; Abdullahpour, R.; Chamani, M. Genetic analysis of fertility traits of Holstein dairy cattle in warm and temperate climate. Acta Sci. Anim. Sci. 2016, 38, 333–340. [Google Scholar] [CrossRef]
- Sigdel, A.; Liu, L.; Abdollahi-Arpanahi, R.; Aguilar, I.; Peñagaricano, F. Genetic dissection of reproductive performance of dairy cows under heat stress. Anim. Genet. 2020, 51, 511–520. [Google Scholar] [CrossRef]
- Wall, E.; Simm, G.; Moran, D. Developing breeding schemes to assist mitigation of greenhouse gas emissions. Animal 2010, 4, 366–376. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Bowman, P.J.; Haile-Mariam, M.; Pryce, J.; Hayes, B. Genomic selection for tolerance to heat stress in Australian dairy cattle. J. Dairy Sci. 2016, 99, 2849–2862. [Google Scholar] [CrossRef]
- Silva, M.V.; dos Santos, D.J.; Boison, S.A.; Utsunomiya, A.; Carmo, A.S.; Sonstegard, T.S.; Cole, J.B.; Van Tassell, C.P. The development of genomics applied to dairy breeding. Livest. Sci. 2014, 166, 66–75. [Google Scholar] [CrossRef]
- Department of Livestock Development. Summary of Livestock Information by Animal Species, Fiscal Year 2021. Available online: https://ict.dld.go.th/webnew/images/stories/stat_web/monthly/2564/jun64/2---Dairy.pdf (accessed on 30 July 2021).
- Buaban, S.; Puangdee, S.; Duangjinda, M.; Boonkum, W. Estimation of genetic parameters and trends for production traits of dairy cattle in Thailand using a multiple-trait multiple-lactation test day model. Asian-Australas. J. Anim. Sci. 2020, 33, 1387–1399. [Google Scholar] [CrossRef]
- VanRaden, P.; Sanders, A. Economic Merit of Crossbred and Purebred US Dairy Cattle. J. Dairy Sci. 2003, 86, 1036–1044. [Google Scholar] [CrossRef]
- Chanvijit, K.; Duangjinda, M.; Pattarajinda, V.; Reodecha, C. Model comparison for genetic evaluation of milk yield in crossbred Holsteins in the tropics. J. Appl. Genet. 2005, 46, 387–393. [Google Scholar]
- Adisu, A.; Zewdu, W. Reproductive Performance of Indigenous Cow Breeds of Ethiopia: A Review. J. Anim. Health Behav. Sci. 2021, 5, 105. [Google Scholar]
- Tadesse, G. Reproductive Performance and Wastage in Large Ruminant (Cattle) in Ethiopia-Review. J. Dairy Vet. Sci. 2018, 8, 555729. [Google Scholar] [CrossRef]
- Alem, W.T. Review on Reproductive and Productive Performance of Dairy Cow in Ethiopia. Int. J. Ecotoxicol. Ecobiol. 2021, 6, 14. [Google Scholar]
- Buaban, S.; Sanpote, J.; Duangjinda, M. Estimates of genetic parameters for dairy crossbred female fertility traits. Khon Kaen Agric. J. 2018, 46, 767–778. [Google Scholar]
- Yodklaew, P.; Koonawootrittriron, S.; Elzo, M.A.; Suwanasopee, T.; Laodim, T. Genome-wide association study for lactation characteristics, milk yield and age at first calving in a Thai multibreed dairy cattle population. Agric. Nat. Resour. 2017, 51, 223–230. [Google Scholar] [CrossRef]
- Hutchison, J.; VanRaden, P.; Null, D.; Cole, J.; Bickhart, D. Genomic evaluation of age at first calving. J. Dairy Sci. 2017, 100, 6853–6861. [Google Scholar] [CrossRef]
- Ettema, J.; Santos, J. Impact of Age at Calving on Lactation, Reproduction, Health, and Income in First-Parity Holsteins on Commercial Farms. J. Dairy Sci. 2004, 87, 2730–2742. [Google Scholar] [CrossRef]
- Evans, R.; Wallace, M.; Garrick, D.; Dillon, P.; Berry, D.; Olori, V. Effects of calving age, breed fraction and month of calving on calving interval and survival across parities in Irish spring-calving dairy cows. Livest. Sci. 2006, 100, 216–230. [Google Scholar] [CrossRef]
- Pirlo, G.; Miglior, F.; Speroni, M. Effect of Age at First Calving on Production Traits and on Difference Between Milk Yield Returns and Rearing Costs in Italian Holsteins. J. Dairy Sci. 2000, 83, 603–608. [Google Scholar] [CrossRef]
- Cooke, J.S.; Cheng, Z.; Bourne, N.E.; Wathes, D.C. Association between growth rates, age at first calving and subsequent fertility, milk production and survival in Holstein-Friesian heifers. Open J. Anim. Sci. 2013, 3, 1–12. [Google Scholar] [CrossRef]
- Softic, A.; Asmare, K.; Granquist, E.G.; Godfroid, J.; Fejzić, N.; Skjerve, E. The serostatus of Brucella spp., Chlamydia abortus, Coxiella burnetii and Neospora caninum in cattle in three cantons in Bosnia and Herzegovina. BMC Vet. Res. 2018, 14, 40. [Google Scholar] [CrossRef] [PubMed]
- Yusuf, M.; Nakao, T.; Yoshida, C.; Long, S.T.; Gautam, G.; Ranasinghe, R.B.K.; Koike, K.; Hayashi, A. Days in Milk at First AI in Dairy Cows; Its Effect on Subsequent Reproductive Performance and Some Factors Influencing It. J. Reprod. Dev. 2011, 57, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Haile, B. Reproductive Performance of Holstein Friesian Dairy Cows at Alage Dairy Farm, Ethiopia. J. Dairy Vet. Sci. 2018, 7, 555713. [Google Scholar] [CrossRef]
- Semara, L.; Mouffok, C.; Madani, T.; Radi, F.; Rezig, N. Environmental Factors Affecting Reproductive Traits in Cows on Algerian Smallholder Farms. Int. J. Agric. Sci. Vet. Med. 2014, 2, 85–95. [Google Scholar]
- Softic, A.; Martin, A.; Skjerve, E.; Fejzic, N.; Goletic, T.; Kustura, A.; Granquist, E.G. Reproductive Performance in a Selected Sample of Dairy Farms in Una-Sana Canton, Bosnia and Herzegovina. Vet. Med. Int. 2020, 2020, 1–10. [Google Scholar] [CrossRef]
- Elkjær, K.; Ancker, M.-L.; Gustafsson, H.; Friggens, N.; Waldmann, A.; Mølbak, L.; Callesen, H. Uterine bacterial flora in postpartum Danish Holstein dairy cows determined using DNA-based fingerprinting: Correlation to uterine condition and calving management. Anim. Reprod. Sci. 2013, 138, 39–48. [Google Scholar] [CrossRef][Green Version]
- Setiaji, A.; Oikawa, T. Genetics of heifer reproductive traits in Japanese Black cattle. Asian-Australas. J. Anim. Sci. 2020, 33, 197–202. [Google Scholar] [CrossRef]
- Zavadilová, L.; Zink, V. Genetic relationship of functional longevity with female fertility and milk production traits in Czech Holsteins. Czech J. Anim. Sci. 2013, 58, 554–565. [Google Scholar] [CrossRef]
- Kaewlamun, W.; Chayaratanasin, R.; Virakul, P.; Ponter, A.A.; Humblot, P.; Suadsong, S.; Tummaruk, P.; Techakumphu, M. Differences of Periods of Calving on Days Open of Dairy Cows in Different Regions and Months of Thailand. Thai J. Vet. Med. 2011, 41, 315–320. [Google Scholar]
- Negussie, E.; Brännäng, E.; Banjaw, K.; Rottmann, O.J. Reproductive performance of dairy cattle at Asella livestock farm, Arsi, Ethiopia. I: Indigenous cows versus their F1 crosses. J. Anim. Breed. Genet. 1998, 115, 267–280. [Google Scholar] [CrossRef]
- Ibrahim, N.; Seid, A. Review on Reproductive Performance of Crossbred Dairy Cattle in Ethiopia. J. Reprod. Infertil. 2017, 8, 88–94. [Google Scholar]
- Kalantari, A.; Armentano, L.; Shaver, R.; Cabrera, V. Economic impact of nutritional grouping in dairy herds. J. Dairy Sci. 2016, 99, 1672–1692. [Google Scholar] [CrossRef] [PubMed]
- De Vries, A. Economic Value of Pregnancy in Dairy Cattle. J. Dairy Sci. 2006, 89, 3876–3885. [Google Scholar] [CrossRef]
- Hall, D.; Ehui, S.; Shapiro, B. Economic analysis of the impact of adopting herd health control programs on smallholder dairy farms in Central Thailand. Agric. Econ. 2004, 31, 335–342. [Google Scholar] [CrossRef]
- Fodor, I.; Abonyi-Tóth, Z.; Ózsvári, L. Management practices associated with reproductive performance in Holstein cows on large commercial dairy farms. Animal 2018, 12, 2401–2406. [Google Scholar] [CrossRef]
- Mostert, B.; Van Der Westhuizen, R.; Theron, H. Calving interval genetic parameters and trends for dairy breeds in South Africa. S. Afr. J. Anim. Sci. 2010, 40, 57288. [Google Scholar] [CrossRef]
- Duguma, B.; Kechero, Y.; Janssens, G.P.J. Productive and Reproductive Performance of Zebu X Holstein-Friesian Crossbred Dairy Cows in Jimma Town, Oromia, Ethiopia. Glob. Vet. 2012, 8, 67–72. [Google Scholar]
- Buaban, S.; Duangjinda, M.; Suzuki, M.; Masuda, Y.; Sanpote, J.; Kuchida, K. Genetic relationships of fertility traits with test-day milk yield and fat-to-protein ratio in tropical smallholder dairy farms. Anim. Sci. J. 2015, 87, 627–637. [Google Scholar] [CrossRef]
- Mureda, E.; Mekuriaw, Z.Z. Reproductive Performance of Crossbred Dairy Cows in Eastern Lowlands of Ethiopia. Livest. Res. Rural Dev. 2007, 19, 11. [Google Scholar]
- Hammoud, M.H.; El-Zarkouny, S.Z.; Oudah, E.Z.M. Effect of Sire, Age at First Calving, Season and Year of Calving and Parity on Reproductive Performance of Friesian Cows under Semiarid Conditions in Egypt. Arch. Zootech. 2010, 13, 60–82. [Google Scholar]
- Lemma, H.; Belihu, K.; Sheferaw, D. Study on the Reproductive Performance of Jersey Cows at Wolaita Sodo Dairy Farm, Southern Ethiopia. Ethiop. Vet. J. 2010, 14, 53–70. [Google Scholar]
- Galliou, J.M.; Kiser, J.N.; Oliver, K.F.; Seabury, C.M.; Moraes, J.G.N.; Burns, G.W.; Spencer, T.E.; Dalton, J.; Neibergs, H.L. Identification of Loci and Pathways Associated with Heifer Conception Rate in U.S. Holsteins. Genes 2020, 11, 767. [Google Scholar] [CrossRef] [PubMed]
- McWhorter, T.; Hutchison, J.; Norman, H.; Cole, J.; Fok, G.; Lourenco, D.; VanRaden, P. Investigating conception rate for beef service sires bred to dairy cows and heifers. J. Dairy Sci. 2020, 103, 10374–10382. [Google Scholar] [CrossRef] [PubMed]
- Suadsong, S. Alleviating Heat Stress Leads to Improved Cow Reproductive Performance; IntechOpen: London, UK, 2012. [Google Scholar] [CrossRef]
- Shiferaw, Y.; Tenhagen, B.-A.; Bekana, M.; Kassa, T. Reproductive performance of crossbred dairy cows in different production systems in the Central Highlands of Ethiopia. Trop. Anim. Health Prod. 2003, 35, 551–561. [Google Scholar] [CrossRef]
- González-Recio, O.; Alenda, R. Genetic Parameters for Female Fertility Traits and a Fertility Index in Spanish Dairy Cattle. J. Dairy Sci. 2005, 88, 3282–3289. [Google Scholar] [CrossRef]
- Kumar, N.; Eshetie, A.; Tesfaye, A.; Yizengaw, H.A. Productive performance of indigenous and HF crossbred dairy cows in Gondar, Ethiopia. Vet. World 2014, 7, 177–181. [Google Scholar] [CrossRef]
- Ayeneshet, B.; Abera, M.; Wondifraw, Z. Reproductive and Productive Performance of Indigenous Dairy Cows under Smallholder Farmers Management System in North. J. Fish. Livest. Prod. 2018, 6, 261. [Google Scholar] [CrossRef]
- Ill-Hwa, K.; Jeong, J.K. Risk factors limiting first service conception rate in dairy cows, and their economic impact. Asian-Australas. J. Anim. Sci. 2019, 32, 519–526. [Google Scholar] [CrossRef]
- De Rensis, F.; Scaramuzzi, R. Heat stress and seasonal effects on reproduction in the dairy cow—A review. Theriogenology 2003, 60, 1139–1151. [Google Scholar] [CrossRef]
- Bernabucci, U.; Lacetera, N.; Baumgard, L.H.; Rhoads, R.P.; Ronchi, B.; Nardone, A. Metabolic and hormonal acclimation to heat stress in domesticated ruminants. Animal 2010, 4, 1167–1183. [Google Scholar] [CrossRef]
- Kadzere, C.; Murphy, M.; Silanikove, N.; Maltz, E. Heat stress in lactating dairy cows: A review. Livest. Prod. Sci. 2002, 77, 59–91. [Google Scholar] [CrossRef]
- Jian, W.; Duangjinda, M.; Vajrabukka, C.; Katawatin, S. Differences of skin morphology in Bos indicus, Bos taurus, and their crossbreds. Int. J. Biometeorol. 2013, 58, 1087–1094. [Google Scholar] [CrossRef] [PubMed]
- Habeeb, A.A.; Gad, A.E.; Atta, M.A. Temperature-Humidity Indices as Indicators to Heat Stress of Climatic Conditions with Relation to Production and Reproduction of Farm Animals. Int. J. Biotechnol. Recent Adv. 2018, 1, 35–50. [Google Scholar] [CrossRef]
- Ozawa, M.; Tabayashi, D.; Latief, T.; Shimizu, T.; Oshima, I.; Kanai, Y. Alterations in follicular dynamics and steroidogenic abilities induced by heat stress during follicular recruitment in goats. Reproduction 2005, 129, 621–630. [Google Scholar] [CrossRef]
- Kornmatitsuk, B.; Chantaraprateep, P.; Kindahl, H. Different Types of Postpartum Luteal Activity Affected by the Exposure of Heat Stress and Subsequent Reproductive Performance in Holstein Lactating Cows. Reprod. Domest. Anim. 2008, 43, 515–519. [Google Scholar] [CrossRef]
- Orihuela, A. Some factors affecting the behavioural manifestation of oestrus in cattle: A review. Appl. Anim. Behav. Sci. 2000, 70, 1–16. [Google Scholar] [CrossRef]
- Dash, S.; Chakravarty, A.K.; Singh, A.; Upadhyay, A.; Singh, M.; Yousuf, S. Effect of heat stress on reproductive performances of dairy cattle and buffaloes: A review. Vet. World 2016, 9, 235–244. [Google Scholar] [CrossRef]
- Kananub, S.; VanLeeuwen, J.A.; Arunvipas, P. Association between milk urea nitrogen and first service conception in smallholder dairy farms under heat and humidity stress. Vet. World 2018, 11, 1604–1608. [Google Scholar] [CrossRef]
- Recce, S.; Huber, E.; Notaro, U.S.; Rodríguez, F.M.; Ortega, H.H.; Rey, F.; Signorini, M.L.; Salvetti, N.R. Association between heat stress during intrauterine development and the calving-to-conception and calving-to-first-service intervals in Holstein cows. Theriogenology 2021, 162, 95–104. [Google Scholar] [CrossRef]
- Djelailia, H.; Bouraoui, R.; Jemmali, B.; Najar, T. Effects of heat stress on reproductive efficiency in Holstein dairy cattle in the North African arid region. Reprod. Domest. Anim. 2020, 55, 1250–1257. [Google Scholar] [CrossRef]
- Toghiani, S. Quantitative Genetic Application in the Selection Process for Livestock Production; IntechOpen: London, UK, 2012. [Google Scholar] [CrossRef]
- Bennett, G.; Pollak, E.; Kuehn, L.; Snelling, W. Breeding: Animals. In Encyclopedia of Agriculture and Food Systems; Elsevier: Amsterdam, The Netherlands, 2014; pp. 173–186. [Google Scholar] [CrossRef]
- Almeida, T.P.; Kern, E.L.; Daltro, D.D.S.; Neto, J.B.; McManus, C.; Neto, A.T.; Cobuci, J.A. Genetic associations between reproductive and linear-type traits of Holstein cows in Brazil. Rev. Bras. Zootec. 2017, 46, 91–98. [Google Scholar] [CrossRef]
- Eriksson, S.; Johansson, K.; Axelsson, H.H.; Fikse, W. Genetic trends for fertility, udder health and protein yield in Swedish red cattle estimated with different models. J. Anim. Breed. Genet. 2017, 134, 308–321. [Google Scholar] [CrossRef]
- Þórarinsdóttir, Þ.; Eriksson, S.; Albertsdóttir, E. Genetic parameters and genetic trends of female fertility in Icelandic dairy cattle. Livest. Sci. 2021, 251, 104628. [Google Scholar] [CrossRef]
- Häggman, J.; Christensen, J.M.; Mäntysaari, E.A.; Juga, J. Genetic parameters for endocrine and traditional fertility traits, hyperketonemia and milk yield in dairy cattle. Animal 2019, 13, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Muuttoranta, K.; Tyrisevä, A.-M.; Mäntysaari, E.A.; Pösö, J.; Aamand, G.P.; Lidauer, M.H. Genetic parameters for female fertility in Nordic Holstein and Red Cattle dairy breeds. J. Dairy Sci. 2019, 102, 8184–8196. [Google Scholar] [CrossRef] [PubMed]
- Berry, D.P.; Wall, E.; Pryce, J. Genetics and genomics of reproductive performance in dairy and beef cattle. Animal 2014, 8, 105–121. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Zhao, F.; Wang, Y.; Zhang, Y.; Du, L.; Su, G. Comparison of single-trait and multiple-trait genomic prediction models. BMC Genet. 2014, 15, 30. [Google Scholar] [CrossRef]
- Karaman, E.; Lund, M.S.; Su, G. Multi-trait single-step genomic prediction accounting for heterogeneous (co)variances over the genome. Heredity 2019, 124, 274–287. [Google Scholar] [CrossRef]
- Budhlakoti, N.; Mishra, D.C.; Rai, A.; Lal, S.; Chaturvedi, K.K.; Kumar, R.R. A Comparative Study of Single-Trait and Multi-Trait Genomic Selection. J. Comput. Biol. 2019, 26, 1100–1112. [Google Scholar] [CrossRef]
- Sodini, S.M.; Kemper, K.; Wray, N.R.; Trzaskowski, M. Comparison of Genotypic and Phenotypic Correlations: Cheverud’s Conjecture in Humans. Genetics 2018, 209, 941–948. [Google Scholar] [CrossRef]
- Ghiasi, H.; Pakdel, A.; Nejati-Javaremi, A.; Mehrabani-Yeganeh, H.; Honarvar, M.; Gonzalez-Recio, O.; Carabaño, M.J.; Alenda, R. Genetic variance components for female fertility in Iranian Holstein cows. Livest. Sci. 2011, 139, 277–280. [Google Scholar] [CrossRef]
- Ayalew, W.; Aliy, M.; Negussie, E. Estimation of genetic parameters of the productive and reproductive traits in Ethiopian Holstein using multi-trait models. Asian-Australas. J. Anim. Sci. 2017, 30, 1550–1556. [Google Scholar] [CrossRef] [PubMed]
- Nascimento, F.G.D.O.; Aguiar, H.C.P.; Rodrigues, G.M.; Guimarães, E.C.; Nascimento, M.R.B.D.M. What is the best temperature-humidity index equation to indicate heat stress in crossbred dairy calves in a tropical environment? Cienc. Rural 2019, 49, 132. [Google Scholar] [CrossRef]
- Aguilar, I.; Misztal, I.; Tsuruta, S. Genetic components of heat stress for dairy cattle with multiple lactations. J. Dairy Sci. 2009, 92, 5702–5711. [Google Scholar] [CrossRef] [PubMed]
- Boonkum, W.; Duangjinda, M. Estimation of genetic parameters for heat stress, including dominance gene effects, on milk yield in Thai Holstein dairy cattle. Anim. Sci. J. 2014, 86, 245–250. [Google Scholar] [CrossRef]
- Oseni, S.; Misztal, I.; Tsuruta, S.; Rekaya, R. Genetic Components of Days Open Under Heat Stress. J. Dairy Sci. 2004, 87, 3022–3028. [Google Scholar] [CrossRef]
- Misztal, I.; Tsuruta, S.; Strabel, T.; Auvray, B.; Druet, T.; Lee, D.H. Blupf90 and Related Programs (Bgf90). In Proceedings of the 7th World Congress on Genetics Applied to Livestock Production, Montpellier, France, 19–23 August 2002. [Google Scholar]
- Madsen, P.; Jensen, J. DMU-A Package for Analyzing Multivariate Mixed Models in quantitative Genetics and Genomics. In Proceedings of the 10th World Congress of Genetics Applied to Livestock Production (WCGALP), Vancouver, BC, Canada, 17–22 August 2013. [Google Scholar]
- Ibtisham, F.; Zhang, L.; Xiao, M.; An, L.; Ramzan, M.B.; Nawab, A.; Zhao, Y.; Li, G.; Xu, Y. Genomic Selection and Its Application in Animal Breeding. Thai J. Vet. Med. 2017, 47, 301–310. [Google Scholar]
- Calus, M.; de Haas, Y.; Pszczola, M.; Veerkamp, R. Predicted accuracy of and response to genomic selection for new traits in dairy cattle. Animal 2013, 7, 183–191. [Google Scholar] [CrossRef]
- Dekkers, J.C. Application of Genomics Tools to Animal Breeding. Curr. Genom. 2012, 13, 207–212. [Google Scholar] [CrossRef]
- Cowling, W.A.; Stefanova, K.T.; Beeck, C.P.; Nelson, M.N.; Hargreaves, B.L.W.; Sass, O.; Gilmour, A.R.; Siddique, K. Using the Animal Model to Accelerate Response to Selection in a Self-Pollinating Crop. G3 Genes Genomes Genet. 2015, 5, 1419–1428. [Google Scholar] [CrossRef]
- Sun, C.; Madsen, P.; Nielsen, U.; Zhang, Y.; Lund, M.; Su, G. Comparison between a sire model and an animal model for genetic evaluation of fertility traits in Danish Holstein population. J. Dairy Sci. 2009, 92, 4063–4071. [Google Scholar] [CrossRef] [PubMed]
- Misztal, I.; Gianola, D.; Foulley, J. Computing Aspects of a Nonlinear Method of Sire Evaluation for Categorical Data. J. Dairy Sci. 1989, 72, 1557–1568. [Google Scholar] [CrossRef]
- Weller, J.I.; Ron, M. Invited review: Quantitative trait nucleotide determination in the era of genomic selection. J. Dairy Sci. 2011, 94, 1082–1090. [Google Scholar] [CrossRef] [PubMed]
- Vanraden, P.M.; Van Tassell, C.P.; Wiggans, G.R.; Sonstegard, T.S.; Schnabel, R.D.; Taylor, J.F.; Schenkel, F.S. Invited Review: Reliability of genomic predictions for North American Holstein bulls. J. Dairy Sci. 2009, 92, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Dekkers, J.C.M. Commercial application of marker- and gene-assisted selection in livestock: Strategies and lessons. J. Anim. Sci. 2004, 82, E313–E328. [Google Scholar]
- Goddard, M.E.; Hayes, B.J. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat. Rev. Genet. 2009, 10, 381–391. [Google Scholar] [CrossRef]
- Ortega, M.S. Identification of genes associated with reproductive function in dairy cattle. Anim. Reprod. 2018, 15, 923–932. [Google Scholar] [CrossRef]
- Ortega, M.S.; Denicol, A.C.; Cole, J.B.; Null, D.J.; Taylor, J.F.; Schnabel, R.D.; Hansen, P.J. Association of single nucleotide polymorphisms in candidate genes previously related to genetic variation in fertility with phenotypic measurements of reproductive function in Holstein cows. J. Dairy Sci. 2017, 100, 3725–3734. [Google Scholar] [CrossRef]
- Jecminkova, K.; Müller, U.; Kyselova, J.; Sztankoova, Z.; Zavadilova, L.; Stipkova, M.; Majzlik, I. Association of leptin, toll-like receptor 4, and chemokine receptor of interleukin 8 C-X-C motif single nucleotide polymorphisms with fertility traits in Czech Fleckvieh cattle. Asian-Australas. J. Anim. Sci. 2018, 31, 1721–1728. [Google Scholar] [CrossRef]
- Cañizares-Martínez, M.A.; Parra-Bracamonte, G.M.; Segura-Correa, J.C.; Magaña-Monforte, J.G. Effect of Leptin, Pituitary Transcription Factor and Luteinizing Hormone Receptor Genes Polymorphisms on Reproductive Traits and Milk Yield in Holstein Cattle. Braz. Arch. Biol. Technol. 2021, 64, 643. [Google Scholar] [CrossRef]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [CrossRef] [PubMed]
- Guarini, A.; Lourenco, D.; Brito, L.; Sargolzaei, M.; Baes, C.; Miglior, F.; Misztal, I.; Schenkel, F. Comparison of genomic predictions for lowly heritable traits using multi-step and single-step genomic best linear unbiased predictor in Holstein cattle. J. Dairy Sci. 2018, 101, 8076–8086. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lourenco, D.; Aguilar, I.; Legarra, A.; Misztal, I. Weighting Strategies for Single-Step Genomic BLUP: An Iterative Approach for Accurate Calculation of GEBV and GWAS. Front. Genet. 2016, 7, 151. [Google Scholar] [CrossRef] [PubMed]
- Alvarenga, A.; Veroneze, R.; De Oliveira, H.R.; Marques, D.B.D.; Lopes, P.S.; Silva, F.F.; Brito, L.F. Comparing Alternative Single-Step GBLUP Approaches and Training Population Designs for Genomic Evaluation of Crossbred Animals. Front. Genet. 2020, 11, 263. [Google Scholar] [CrossRef]
- Pocrnic, I.; Lourenco, D.A.L.; Chen, C.-Y.; Herring, W.; Misztal, I. Crossbred evaluations using single-step genomic BLUP and algorithm for proven and young with different sources of data1. J. Anim. Sci. 2019, 97, 1513–1522. [Google Scholar] [CrossRef] [PubMed]
- Guarini, A.; Lourenco, D.; Brito, L.; Sargolzaei, M.; Baes, C.; Miglior, F.; Misztal, I.; Schenkel, F. Genetics and genomics of reproductive disorders in Canadian Holstein cattle. J. Dairy Sci. 2019, 102, 1341–1353. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, Q.; Ding, X. Advances in genomic selection in domestic animals. Chin. Sci. Bull. 2011, 56, 2655–2663. [Google Scholar] [CrossRef][Green Version]
- Gutierrez-Reinoso, M.; Aponte, P.; Garcia-Herreros, M. Genomic Analysis, Progress and Future Perspectives in Dairy Cattle Selection: A Review. Animals 2021, 11, 599. [Google Scholar] [CrossRef]
- García-Ruiz, A.; Cole, J.B.; VanRaden, P.M.; Wiggans, G.R.; Ruiz-López, F.J.; Van Tassell, C.P. Changes in genetic selection differentials and generation intervals in US Holstein dairy cattle as a result of genomic selection. Proc. Natl. Acad. Sci. USA 2016, 113, E3995–E4004. [Google Scholar] [CrossRef]
- König, S.; Simianer, H.; Willam, A. Economic evaluation of genomic breeding programs. J. Dairy Sci. 2009, 92, 382–391. [Google Scholar] [CrossRef]
- Shumbusho, F.; Raoul, J.; Astruc, J.M.; Palhiere, I.; Elsen, J.M. Potential benefits of genomic selection on genetic gain of small ruminant breeding programs1. J. Anim. Sci. 2013, 91, 3644–3657. [Google Scholar] [CrossRef] [PubMed]
- Jonas, E.; De Koning, D.-J. Genomic selection needs to be carefully assessed to meet specific requirements in livestock breeding programs. Front. Genet. 2015, 6, 49. [Google Scholar] [CrossRef] [PubMed]
- Lund, M.S.; de Roos, A.P.; de Vries, A.G.; Druet, T.; Ducrocq, V.; Fritz, S.; Guillaume, F.; Guldbrandtsen, B.; Liu, Z.; Reents, R.; et al. A common reference population from four European Holstein populations increases reliability of genomic predictions. Genet. Sel. Evol. 2011, 43, 43. [Google Scholar] [CrossRef] [PubMed]
- Wiggans, G.; VanRaden, P.; Cooper, T. The genomic evaluation system in the United States: Past, present, future. J. Dairy Sci. 2011, 94, 3202–3211. [Google Scholar] [CrossRef] [PubMed]
- Ben Zaabza, H.; Ben Gara, A.; Rekik, B. Bayesian Modeling in Genetics and Genomicsvvv; IntechOpen: London, UK, 2017. [Google Scholar] [CrossRef]
- Habier, D.; Fernando, R.L.; Kizilkaya, K.; Garrick, D.J. Extension of the bayesian alphabet for genomic selection. BMC Bioinform. 2011, 12, 186. [Google Scholar] [CrossRef]
- Campos, G.D.L.; Naya, H.; Gianola, D.; Crossa, J.; Legarra, A.; Manfredi, E.; Weigel, K.; Cotes, J.M. Predicting Quantitative Traits with Regression Models for Dense Molecular Markers and Pedigree. Genetics 2009, 182, 375–385. [Google Scholar] [CrossRef]
- Erbe, M.; Hayes, B.J.; Matukumalli, L.K.; Goswami, S.; Bowman, P.J.; Reich, C.M.; Mason, B.A.; Goddard, M.E. Improving accuracy of genomic predictions within and between dairy cattle breeds with imputed high-density single nucleotide polymorphism panels. J. Dairy Sci. 2012, 95, 4114–4129. [Google Scholar] [CrossRef]
- Genomic selection of dairy cattle:A review of methods, strategies, and impact. J. Anim. Breed. Genom. 2017, 2017, 1. [CrossRef]
- Legarra, A.; Christensen, O.F.; Aguilar, I.; Misztal, I. Single Step, a general approach for genomic selection. Livest. Sci. 2014, 166, 54–65. [Google Scholar] [CrossRef]
- Misztal, I.; Lourenco, D.; Legarra, A. Current status of genomic evaluation. J. Anim. Sci. 2020, 98, 101. [Google Scholar] [CrossRef]
- Legarra, A.; Aguilar, I.; Misztal, I. A relationship matrix including full pedigree and genomic information. J. Dairy Sci. 2009, 92, 4656–4663. [Google Scholar] [CrossRef] [PubMed]
- Guillaume, F.; Fritz, S.; Boichard, D.; Druet, T. Short Communication: Correlations of Marker-Assisted Breeding Values with Progeny-Test Breeding Values for Eight Hundred Ninety-Nine French Holstein Bulls. J. Dairy Sci. 2008, 91, 2520–2522. [Google Scholar] [CrossRef] [PubMed]
- Neuner, S.; Edel, C.; Emmerling, R.; Thaller, G.; Götz, K.-U. Precision of genetic parameters and breeding values estimated in marker assisted BLUP genetic evaluation. Genet. Sel. Evol. 2009, 41, 26. [Google Scholar] [CrossRef] [PubMed]
- Patry, C.; Ducrocq, V. Evidence of biases in genetic evaluations due to genomic preselection in dairy cattle. J. Dairy Sci. 2011, 94, 1011–1020. [Google Scholar] [CrossRef]
- Lourenco, D.A.L.; Misztal, I.; Tsuruta, S.; Aguilar, I.; Ezra, E.; Ron, M.; Shirak, A.; Weller, J.I. Methods for Genomic Evaluation of a Relatively Small Genotyped Dairy Population and Effect of Genotyped Cow Information in Multiparity Analyses. J. Dairy Sci. 2014, 97, 1742–1752. [Google Scholar] [CrossRef]
- Sungkhapreecha, P.; Chankitisakul, V.; Duangjinda, M.; Buaban, S.; Boonkum, W. Determining Heat Stress Effects of Multiple Genetic Traits in Tropical Dairy Cattle Using Single-Step Genomic BLUP. Vet. Sci. 2022, 9, 66. [Google Scholar] [CrossRef]
- Sungkhapreecha, P.; Misztal, I.; Hidalgo, J.; Steyn, Y.; Buaban, S.; Duangjinda, M.; Boonkum, W. Changes in genetic parameters for milk yield and heat tolerance in the Thai Holstein crossbred dairy population under different heat stress levels and over time. J. Dairy Sci. 2021, 104, 12703–12712. [Google Scholar] [CrossRef]
- Buaban, S.; Prempree, S.; Sumreddee, P.; Duangjinda, M.; Masuda, Y. Genomic prediction of milk-production traits and somatic cell score using single-step genomic best linear unbiased predictor with random regression test-day model in Thai dairy cattle. J. Dairy Sci. 2021, 104, 12713–12723. [Google Scholar] [CrossRef]
- Dekkers, J.C.M.; Su, H.; Cheng, J. Predicting the accuracy of genomic predictions. Genet. Sel. Evol. 2021, 53, 55. [Google Scholar] [CrossRef]
- Dekkers, J.C.M. Prediction of response to marker-assisted and genomic selection using selection index theory. J. Anim. Breed. Genet. 2007, 124, 331–341. [Google Scholar] [CrossRef]
- Daetwyler, H.D.; Villanueva, B.; Woolliams, J.A. Accuracy of Predicting the Genetic Risk of Disease Using a Genome-Wide Approach. PLoS ONE 2008, 3, e3395. [Google Scholar] [CrossRef] [PubMed]
- Moser, G.; Khatkar, M.S.; Hayes, B.J.; Raadsma, H.W. Accuracy of direct genomic values in Holstein bulls and cows using subsets of SNP markers. Genet. Sel. Evol. 2010, 42, 37. [Google Scholar] [CrossRef] [PubMed]
- Wiggans, G.R.; Cole, J.B.; Hubbard, S.M.; Sonstegard, T.S. Genomic Selection in Dairy Cattle: The USDA Experience. Annu. Rev. Anim. Biosci. 2017, 5, 309–327. [Google Scholar] [CrossRef] [PubMed]
- Hayes, B.J.; Bowman, P.; Chamberlain, A.; Goddard, M. Invited review: Genomic selection in dairy cattle: Progress and challenges. J. Dairy Sci. 2009, 92, 433–443. [Google Scholar] [CrossRef]
- Shao, B.; Sun, H.; Ahmad, M.J.; Ghanem, N.; Abdel-Shafy, H.; Du, C.; Deng, T.; Mansoor, S.; Zhou, Y.; Yang, Y.; et al. Genetic Features of Reproductive Traits in Bovine and Buffalo: Lessons from Bovine to Buffalo. Front. Genet. 2021, 12, 617128. [Google Scholar] [CrossRef]
- Laodim, T.; Elzo, M.A.; Koonawootrittriron, S.; Suwanasopee, T.; Jattawa, D. Genomic-polygenic and polygenic predictions for milk yield, fat yield, and age at first calving in Thai multibreed dairy population using genic and functional sets of genotypes. Livest. Sci. 2018, 219, 17–24. [Google Scholar] [CrossRef]
- Boison, S.A.; Utsunomiya, A.T.H.; Santos, D.J.A.; Neves, H.H.R.; Carvalheiro, R.; Mészáros, G.; Utsunomiya, Y.T.; do Carmo, A.S.; Verneque, R.S.; Machado, M.A.; et al. Accuracy of genomic predictions in Gyr (Bos indicus) dairy cattle. J. Dairy Sci. 2017, 100, 5479–5490. [Google Scholar] [CrossRef]
- Buaban, S.; Duangjinda, M.; Suzuki, M.; Masuda, Y.; Sanpote, J.; Kuchida, K. Short communication: Genetic analysis for fertility traits of heifers and cows from smallholder dairy farms in a tropical environment. J. Dairy Sci. 2015, 98, 4990–4998. [Google Scholar] [CrossRef]
- Burrow, H.M.; Mrode, R.; Mwai, A.O.; Coffey, M.P.; Hayes, B.J. Challenges and Opportunities in Applying Genomic Selection to Ruminants Owned by Smallholder Farmers. Agriculture 2021, 11, 1172. [Google Scholar] [CrossRef]
- Koonawootrittriron, S.; Elzo, M.; Thongprapi, T. Genetic trends in a Holstein × other breeds multibreed dairy population in Central Thailand. Livest. Sci. 2009, 122, 186–192. [Google Scholar] [CrossRef]
- Sarakul, M.; Koonawootrittriron, S.; Suwanasopee, T.; Hirunwong, A.; Tongprapi, T. Situation and attitude for production and sire selection of dairy farmers in Thailand. In Proceeding of the 47th Kasetsart University Conference, Bangkok, Thailand, 17–21 March 2009. [Google Scholar]
| Traits 1 | Breeds | h2 | Model 2 | References |
|---|---|---|---|---|
| AFC | Holstein (Brazil) | 0.02 | MTM | [76] |
| NSPC | Thai–Holstein crossbred | 0.04 | UM | [26] |
| Spanish dairy cattle | 0.02 | STM | [58] | |
| Holstein (Iran) | 0.04 | ALM | [14] | |
| Swedish red | 0.02–0.03 | MTM | [77] | |
| Icelandic dairy cattle | 0.02 | MTM | [78] | |
| CFI | Thai–Holstein crossbred | 0.07 | UM | [26] |
| Spanish dairy cattle | 0.05 | STM | [58] | |
| Finland dairy cattle | 0.07 | UM | [79] | |
| Swedish red | 0.02–0.03 | MTM | [77] | |
| Nordic Holstein | 0.05–0.06 | REP | [80] | |
| IFL | Thai–Holstein crossbred | 0.02 | UM | [26] |
| Spanish dairy cattle | 0.03 | STM | [58] | |
| Finland dairy cattle | 0.03 | UM | [79] | |
| Icelandic dairy cattle | 0.02 | MTM | [78] | |
| Nordic Holstein | 0.01–0.04 | REP | [80] | |
| FSCR | Thai–Holstein crossbred | 0.01 | UM | [26] |
| Spanish dairy cattle | 0.04 | STM | [58] | |
| Holstein (Iran) | 0.01 | ALM | [14] | |
| CR | Spanish dairy cattle | 0.04 | STM | [58] |
| Icelandic dairy cattle | 0.02 | MTM | [78] | |
| Nordic Holstein | 0.01–0.03 | REP | [80] | |
| DO | Thai–Holstein crossbred | 0.07–0.08 | MTM | [9] |
| Spanish dairy cattle | 0.04 | STM | [58] | |
| Holstein (Iran) | 0.02 | ALM | [14] | |
| Holstein (Brazil) | 0.03 | MTM | [76] | |
| P90 | Thai–Holstein crossbred | 0.02 | TAM | [50] |
| Spanish dairy cattle | 0.06 | STM | [58] | |
| CI | Thai–Holstein crossbred | 0.07 | UM | [26] |
| Spanish dairy cattle | 0.04 | STM | [58] | |
| Holstein (Iran) | 0.03 | ALM | [14] | |
| Holstein (Brazil) | 0.03 | MTM | [76] | |
| Icelandic dairy cattle | 0.03 | MTM | [78] |
| Traits 1 | NSPC | CFI | IFL | FSCR | DO | P90 | CI |
|---|---|---|---|---|---|---|---|
| NSPC | 0.36 | 0.95 | −0.80 | 0.64 | −0.76 | 0.63 | |
| CFI | 0.63 | −0.58 | 0.90 | −0.64 | 0.91 | ||
| IFL | −0.93 | 0.90 | −0.99 | 0.89 | |||
| FSCR | −0.85 | 0.51 | −0.85 | ||||
| DO | −0.53 | 1.00 | |||||
| P90 | −0.96 | ||||||
| CI |
| No. | Traits 1 | SNP ID | Chromosomes | Genes 2 | Type of Mutation | References |
|---|---|---|---|---|---|---|
| 1 | FSCR | rs109137982 | 3 | FCER1G | missense | [105] |
| rs43745234 | 11 | FSHR | missense | |||
| rs110789098 | 6 | IBSP | missense | |||
| rs109629628 | 25 | PMM2 | missense | |||
| rs110660625 | 25 | TBC1D24 | missense | |||
| rs109956567 | 4 | LEP | missense | [106] | ||
| rs29004508 | 4 | LEP | missense | |||
| 2 | NSPC | rs137601357 | 7 | CAST | missense | [105] |
| rs109621328 | 7 | CD14 | missense | |||
| rs109137982 | 3 | FCER1G | missense | |||
| rs41893756 | 18 | FUT1 | missense | |||
| rs109262355 | 20 | FYB | missense | |||
| 3 | DO | rs109669573 | 13 | BCAS1 | missense | |
| rs110217852 | 6 | BSP3 | missense | |||
| rs137601357 | 7 | CAST | missense | |||
| rs109621328 | 7 | CD14 | missense | |||
| rs109137982 | 3 | FCER1G | missense | |||
| rs109956567 | 4 | LEP | missense | [106] | ||
| rs29004508 | 4 | LEP | missense | |||
| 4 | AFC | rs41256848 | 11 | LHR | missense | [107] |
| rs109956567 | 4 | LEP | missense | [106] | ||
| rs29004508 | 4 | LEP | missense | |||
| 5 | CFI | rs29004508 | 4 | LEP | missense | [107] |
| 6 | CI | rs109956567 | 4 | LEP | missense | [106] |
| rs29004508 | 4 | LEP | missense | |||
| rs29017188 | 8 | TLR4 | missense |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fathoni, A.; Boonkum, W.; Chankitisakul, V.; Duangjinda, M. An Appropriate Genetic Approach for Improving Reproductive Traits in Crossbred Thai–Holstein Cattle under Heat Stress Conditions. Vet. Sci. 2022, 9, 163. https://doi.org/10.3390/vetsci9040163
Fathoni A, Boonkum W, Chankitisakul V, Duangjinda M. An Appropriate Genetic Approach for Improving Reproductive Traits in Crossbred Thai–Holstein Cattle under Heat Stress Conditions. Veterinary Sciences. 2022; 9(4):163. https://doi.org/10.3390/vetsci9040163
Chicago/Turabian StyleFathoni, Akhmad, Wuttigrai Boonkum, Vibuntita Chankitisakul, and Monchai Duangjinda. 2022. "An Appropriate Genetic Approach for Improving Reproductive Traits in Crossbred Thai–Holstein Cattle under Heat Stress Conditions" Veterinary Sciences 9, no. 4: 163. https://doi.org/10.3390/vetsci9040163
APA StyleFathoni, A., Boonkum, W., Chankitisakul, V., & Duangjinda, M. (2022). An Appropriate Genetic Approach for Improving Reproductive Traits in Crossbred Thai–Holstein Cattle under Heat Stress Conditions. Veterinary Sciences, 9(4), 163. https://doi.org/10.3390/vetsci9040163







