Abstract
Sparganosis is a neglected zoonotic parasitic disease that poses huge threats to humans worldwide. Snakes play an important role in sparganosis transmission because they are the most common second intermediate hosts for Spirometra parasites. However, the population genetics of Spirometra isolates from snakes is currently not well studied in China. The present study was performed to explore the molecular characteristics and phylogenetic analysis of Spirometra tapeworms from different species of snakes in Hunan Province. This study obtained 49 Spirometra isolates from 15 geographical areas in Hunan Province, Central China. Subsequently, the 18S and 28S ribosomal DNA (rDNA) fragments were amplified from the isolated parasites, and their sequences were analyzed to assess their genetic diversity. Phylogenetic analyses were performed using the maximum likelihood algorithm. The results showed that sequence variations among these isolates were 0–2.3% and 0–0.1% for 18S and 28S rDNA, respectively. The phylogenetic analysis showed that all Spirometra isolates from Hunan Province were clustered into the same branch with Spirometra erinaceieuropaei isolated from other areas (China, Vietnam, Australia). Moreover, the phylogenetic trees revealed that Spirometra is closely related to Adenocephalus, Pyramicocephalus, Ligula, Dibothriocephalus, Schistocephalus, and Diphyllobothrium. The Spirometra isolates of different hosts/regions in Hunan Province are not host segregated or geographically isolated, and support for the taxonomic status of Spirometra tapeworms in China has been added. These results provide reference values for future accurate identification and taxonomic status of Spirometra tapeworms in China.
1. Introduction
Human sparganosis is a worldwide disease caused by the larva (sparganum) of the genus Spirometra [1,2]. Humans can be infected through eating undercooked frog or snake meat and drinking polluted water [3,4]. Although sparganum has been reported to commonly reside in subcutaneous tissues and muscles, they can also migrate to the abdominal cavity, internal organs, eyes, and brain, which can form masses or space-occupying lesions in the body that cause local tissue damage and paralysis [5,6].
More than 10 species of the genus Spirometra have been reported, of which Spirometra erinaceieuropaei mainly infects humans. The first reported human case of sparganosis was discovered in 1882 by Patrick Manson from a man’s autopsy in Xiamen, and was named Ligula mansoni a year later [7]. Sparganosis has been mainly reported in China and can also be found in Europe (Poland, Italy, France, and the Czech Republic), Asia (Korea, Japan, Thailand, and Laos), South America (Ecuador, Paraguay, and Venezuela), and North America [5,8,9]. The reason for the high infection rate in China is mainly related to local customs. Superstitious people stick raw frog or snake flesh on skin wounds and even swallow tadpoles or snake bile in remote regions of China [4,10,11]. Another reason is the high infection rate of frogs and snakes in China. A survey showed that 14.3% (31/217) and 91.7% (344/375) of frogs and snakes, respectively, were infected in Hunan Province [11,12].
Although an important genus in zoonosis, the taxonomy of the Spirometra species has been controversial for a long time. It has also been suggested in some studies that the genus Spirometra belongs to the genus Diphyllobothrium, and should not form a separate genus [13,14]. Meanwhile, the valid species of Spirometra has also been unclear. This is still a mystery whether the pathogen of Chinese sparganosis is S. erinaceieuropaei, Spirometra decipiens, or both [11]. In the recent study of Yamasaki, it was found that two Spirometra species in Asia, neither of which is close to likely S. erinaceieuropaei originating from Poland, and lineage Type I is genetically diverse and widely distributed, however Type II is known so far only from Japan and Korea [15]. The primary and secondary ribosomal DNA (rDNA) structures remain stable during the long evolutionary process, which is one of the tools for studying phylogenetic evolution in parasites [16]. In the last few years of studies, ITS, 16S rDNA, 18S rDNA, and 28S rDNA have been used to establish the phylogenetic relationship of Taenia species [9,17,18,19,20,21]. The 18S and 28S rDNA contain both variable and conserved regions, which make them handy molecular markers to solve phylogenetic relationships at different levels [22]. This study analyzed the genetic diversity of the 18S and 28S rDNA sequences of Spirometra isolates from seven different hosts in 15 geographical regions in Hunan Province, and constructed the Diphyllobothriidae evolutionary tree. The main objectives of this study were as follows: (1) describe sample morphology; (2) perform a genetic diversity analysis of the collected isolates from different geographical locations and hosts in Hunan Province, China; and (3) investigate the taxonomic status of Spirometra isolates using 18S and 28S rDNA sequences from snakes in Hunan Province.
2. Materials and Methods
2.1. Sample Collection
This study collected 49 samples from the field site in 15 geographical locations of Hunan Province in Southern China between April and September 2018 (Table 1). Figure 1 provides a scheme of the geographical locations of the collected Spirometra tapeworms. Spirometra tapeworms were isolated from muscles and subcutaneous tissues of three snake species of the family Colubridae, i.e., Ptyas dhumnades (Cantor, 1842), Elaphe carinata (Günther, 1864), and Elaphe taeniura (Cope, 1861), as well as from the intestines of the family Felidae, i.e., Panthera tigris (Linnaeus, 1758), Prionailurus bengalensis (Kerr, 1792), Felis silvestris (Schreber, 1777), and feral domestic cats. The collected samples were then fixed in 70% ethanol and kept at −20 °C for the molecular analysis.

Table 1.
Geographical origins (different locations in Hunan Province, China) of Spirometra tapeworms isolates used in this study, as well as their GenBank accession numbers for the 18S and 28S sequences.
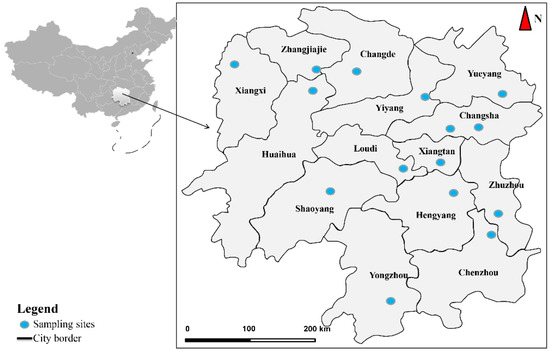
Figure 1.
The sampling sites of Spirometra isolates in Hunan Province.
2.2. Morphological Observations
The live worms were washed by water three times, and then sprayed with heavy metal on the surface. The morphology was made using the SEM-6380LV scanning electron microscope (JEOL, Akishima, Japan). The scolex of the sparganum and the scolex, gravid proglottid, and egg of Spirometra tapeworms were directly glued to the sample table and sprayed with a gold coating, and photographs were taken using a JSM-6380LV scanning electron microscope.
2.3. DNA Extraction and Enzymatic Amplification
The total genomic DNA was extracted from individual samples using the Wizard® SV Genomic DNA Purification System (Promega Corporation, Madison, WI, USA) following the manufacturer’s protocol. Two ribosome markers (18S and 28S rDNA) were amplified by polymerase chain reaction (PCR) using the primer combinations listed in Appendix A. PCR reactions were carried out in a 25 μL reaction mixture containing 8.5 μL distilled water, 12.5 μL Taq PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA), 1 μL of each primer (25 pmol/L), and 2 μL DNA template in a thermal cycler (Biometra, Göttingen, Germany). For the 18S rDNA, the steps were 94 °C for 5 min (first denaturation) and five cycles of 96 °C for 1 min, 44 °C for 1 min, and 72 °C for 2 min, followed by 25 cycles with annealing temperature increased to 48 °C and then by 5 min at 72 °C (final extension). For the 28S rDNA, the steps were 94 °C for 5 min and 35 periods of 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 1 min, followed by 72 °C for 5 min. A negative sample (no DNA) was used in each amplification run. Positive PCR products were purified and then sequenced in both directions by the Tsingke Company (Changsha, China).
2.4. Sequence Analysis
The obtained sequences in this study and the reference sequences were aligned using Clustal X 1.7 software [23]. The DAMBE v.5.2 program was used to measure the nucleotide substitution saturation [24]. In addition, the obtained sequences in this research were also compared with S. erinaceieuropaei isolates from Australia (Canis familiaris), Vietnam (Xenochrophis flavipunctatus), and China (Amphiesma stolatum and Rana nigromaculata) for 18S rDNA sequences, and Australia (C. familiaris), Vietnam (X. flavipunctatus), and China (A. stolatum) for 28S rDNA sequences, using the Megalign procedure in DNASTAR 5.0 software [25]. Moreover, DnaSP 5.0 was used to analyze the diversity indices (nucleotide diversity (π) and haplotype diversity (Hd)) of these three gene sequences obtained in the current research [26].
2.5. Phylogenetic Analysis
All of the sequences are aligned using Clustal W in MAGE7.0. The best nucleotide substitution models were selected using JModelTest0.1. Phylogeny was estimated using a maximum likelihood algorithm (ML) in MEGA7.0. The stability of the tree was calculated based on 1000 bootstrap replicates. Genetic relationships with other Diphyllobothriidae species as in-group and Bothriocotyle solinosomum as out-group were evaluated (Appendix B).
3. Results
3.1. Morphological Characteristics
In the scanning electron microscope study, the egg of Spirometra tapeworms was olive-shaped with slightly pointed ends and a slightly raised side, filled with many pores on the surface (Figure 2A–C). The scolex of the sparganum was flat, unsegmented, and with a wide front end, horizontal stripes, and apparent depression in the middle of the top end (Figure 2D–F). The adults were flat and segmented. The top of the adult scolex was sunken inward, and without other structure (Figure 2G). Moreover, many eggs existed in utero at the gravid proglottids (Figure 2H).
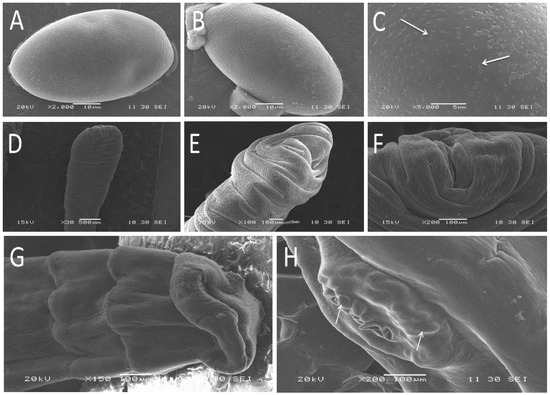
Figure 2.
Scanning electron micrographs of Spirometra tapeworms collected from different hosts in Hunan Province, China. Egg (A,B). Detail of egg surface filled with pores (C). The scolex of larva, front view (D) and lateral view (E). Detail view of scolex (F). The scolex of adult (G). Detail view of egg in utero at the gravid proglottids (H).
3.2. Genetic Characterisations of Spirometra Tapeworms
In this study, 49 and 49 PCR amplicons from 49 isolated samples were successfully amplified for 18S and 28S rDNA, respectively. No size differences were observed for any rDNA region among the amplicons tested (data not shown). The deletions and alignment lengths of the 18S and 28S rDNA were 2006–2010 and 1014 bp, respectively. The 28S rDNA target fragment amplified in this study is the front part of the entire 28S gene (highly protected area).
This study analyzed 49 18S sequences of Spirometra isolates. Intraspecific nucleotide variations within all isolates obtained in the present study were 0–2.3%. However, the 18S sequences obtained in the current study showed lower nucleotide variations of 0–1.6% compared with those of S. erinaceieuropaei from GenBank (China (KX528089 and HQ228991), Vietnam (KY552802), and Australia (KY552801). The pairwise comparison of the 28S rDNA sequences in the present paper showed 0–0.1% nucleotide variations. The sequence variation analysis for the 28S rDNA sequences showed higher nucleotide variations of 0–0.2% compared with those of S. erinaceieuropaei from GenBank (China (HQ228992), Vietnam (KY552835), and Australia (KY552836), and 0.60-0.90% compared with Diphyllobothriidea tapeworms (Schistocephalus solidus, Diphyllobothrium scoticum, Diphyllobothrium sprakeri, Diphyllobothrium tetrapterum, Diphyllobothrium lanceolatum, Diphyllobothrium cordatum, Pyramicocephalus phocarum, Adenocephalus pacificus, and Ligula pavlovskii).
The amplified 18S gene fragment sequence was 2006–2010 bp in length with 18 polymorphic sites. Moreover, insertions or deletions were found within the amplified fragments. Table 2 shows that the nucleotide diversity of the 18S sequences was 0.00062, which defined eight haplotypes with a haplotype diversity of 0.392. For 28S rDNA sequences (1014 bp), one polymorphic site was detected among 49 specimens examined in the present study, with no insertion or deletion. The diversity indices are shown in Table 2. The nucleotide diversity for the 28S rDNA sequences was 0.00021, defining two haplotypes with a haplotype diversity of 0.215.

Table 2.
Diversity indices for Spirometra tapeworms using nucleotide data of the ribosomal 18S rRNA (2006–2010 bp) and 28S rRNA (1013 bp) gene sequences obtained in the present paper.
3.3. Phylogenetic Relationship of S. erinaceieuropaei
A phylogenetic tree based on the 18S and 28S sequences was constructed using the ML method under the general time-reversible (GTR) model by MEGA7.0 (Figure 3). Data showed that all the isolated samples recorded in this study were grouped into one group, and clustered into the same branch with the S. erinaceieuropaei in Genbank from other countries (China, Vietnam, and Australia). In addition, a relatively complete phylogenetic Diphyllobothriidae tree was constructed based on the 18S and 28S sequences. In the current study, Spirometra spp. formed a separate group and were closely related to Schistocephalus spp. Moreover, the genus Diphyllobothrium occupied most of the phylogenetic tree, which was made up of Adenocephalus spp., Pyramicocephalus spp., Ligula spp., Dibothriocephalus spp., and Schistocephalus spp. However, the relationships among the species of Diphyllobothrium by 18S and 28S sequence were not established. Duthiersia fimbriata and Duthiersia expansa formed the Duthiersia spp. branch and then formed a sister group, the Bothridium pithonis branch.
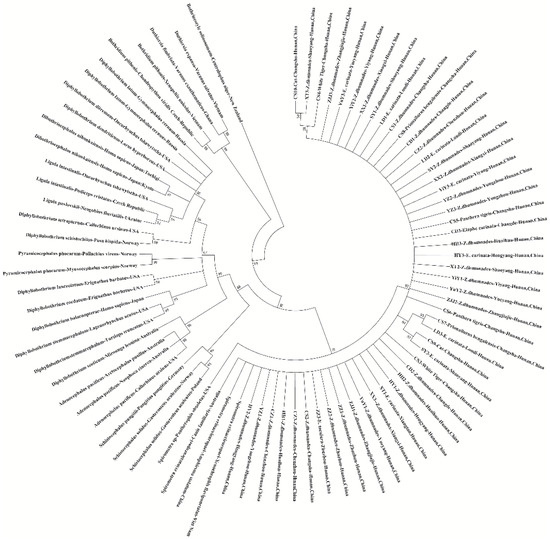
Figure 3.
Maximum likelihood estimates of the phylogenetic relationships of Spirometra tapeworms based on 18S and 28S sequences computed in MEGA version 7.0.26 under the GTR model. The confidence levels in each node were assessed with the boot-strap method (1000 pseudo replicates) and bootstrap values >50.
4. Discussion
The species classification of Spirometra has been controversial. For many years, many researchers considered S. erinaceieuropaei as a global species [5,15]. As more and more mitochondrial gene sequences of S. erinaceieuropaei have been reported globally in recent years, studies have found that S. erinaceieuropaei in China and Southeast Asia and S. erinaceieuropaei in Europe do not belong to the same branch, which also means that the Chinese and Southeast Asia region may not be the previously thought S. erinaceieuropaei [7]. The present study aimed to analyze the genetic diversity of Spirometra tapeworms from snakes and to explore the taxonomic status of Spirometra isolates from Hunan Province on a molecular level. At the same time, this study provides the description of the morphology of Spirometra isolates from snakes in Hunan Province based on scanning electron microscopy, which will lay the foundation for future Spirometra tapeworm species classification in China.
The study used 18S and 2S rDNA genes to explore the intraspecific nucleotide variations of the Spirometra isolates in Hunan Province, China. The results show that the maximum variation values for the 18S and 28S rDNA sequences were 0–2.3% and 0–0.1%, respectively, among the Spirometra isolates from different hosts examined (Zaocys dhumnades, Elaphe carinata, Elaphe taeniura, Panthera tigris, Prionailurus bengalensis, Felis silvestris, and cat). The sequence variation analysis for the 18S gene showed 0–2.3% nucleotide divergence compared with those of S. erinaceieuropaei in China (R. nigromaculata KX528089 and A. stolatum HQ228991), Vietnam (X. flavipunctatus KY552802), and Australia (C. familiaris KY552801). This suggests that both host specificity and geographical effects are not the main factors contributing to the genetic variation of S. erinaceieuropaei, which can also be based on the results of the sequence variation analysis of 28S rDNA. This conclusion is in accordance with recently conducted research [9,21,27].
Haplotype and nucleotide diversities are two important indicators to measure the genetic variation of a gene. A base change can form a haploid type, and haploid type diversity can rapidly rise in a concise time. However, nucleotide base changes have little effect on nucleotide diversity. The rise of nucleotide diversity needs a long accumulation time. Thus, nucleotide diversity is more applicable for measuring the genetic diversity of a species [28]. For most organisms, a nucleotide diversity of >0.01 is considered a large variation [29]. In the current study, the nucleotide diversity of 18S and 28S rDNA genes of the Spirometra isolates was 0.00062 and 0.00028, respectively, which was lower than 0.01. The results showed that the genetic variation of Spirometra isolates from different hosts in Hunan Province was low.
In recent years, it has been shown by the molecular genetic evolution analysis that China and Poland are in different branches. Some scholars have proposed that Spirometra tapeworms should be restored to the title of Spirometra mansoni in China and Southeast Asia [7,15,30]. The phylogenetic tree based on 18S and 28S sequences showed that all the Spirometra isolates from different regions in Hunan Province formed a branch with S. erinaceieuropaei from Genbank from other countries (China, Vietnam, and Australia), except for the S. erinaceieuropaei reported in the United States. This result is consistent with Kuchta et al.’s proposal that China and Southeast Asia should be classified as S. mansoni, North America should be classified as S. decipiens, and Europe should be classified as S. erinaceieuropai. In the current study, phylogenetic trees revealed that Spirometra is closely related to Adenocephalus, Pyramicocephalus, Ligula, Dibothriocephalus, Schistocephalus, and Diphyllobothrium and forms a branch, which is similar to the study of Waeschenbach and Hernandez [18,21].
5. Conclusions
In our study, the genetic variability among different distinct developmental stages (larvae and adults) of Spirometra tapeworms isolated from 15 geographical areas in Hunan Province was analyzed for the 18S and 28S rDNA genes. The results revealed genetic variability in 18S and 28S rDNA. The phylogenetic tree based on 18S and 28S sequences revealed that the Spirometra isolates of different hosts/regions in Hunan Province are not host segregated or geographically isolated, and support for the taxonomic status of Spirometra tapeworms in China was thus added. These results provide reference values for future accurate identification and taxonomic status of Spirometra tapeworms in China.
Author Contributions
Conceptualization, Y.-F.Z. and W.L.; methodology, S.-Y.C., T.-F.G., J.-L.H., F.L., Y.-S.L. and W.L.; software, S.-Y.C., T.-F.G., J.-L.H., L.-X.X., W.-C.L. and Y.-S.L.; validation, F.L.,Y.-S.L., Y.-F.Z. and W.L.; formal analysis, S.-Y.C., T.-F.G., J.-L.H., L.-X.X. and X.-R.X.; investigation, S.-Y.C., T.-F.G., L.-X.X., W.-C.L. and X.-R.X.; data curation, S.-Y.C., T.-F.G. and W.L.; writing—original draft preparation, S.-Y.C. and T.-F.G.; writing—review and editing, F.L., Y.-S.L., Y.-F.Z. and W.L.; visualization, S.-Y.C. and W.L.; supervision, W.L.; project administration, Y.-F.Z. and W.L.; funding acquisition, W.L. All authors have read and agreed to the published version of the manuscript.
Funding
Project supported by the Natural Science Foundation of Hunan Province, China (2021JJ30335), Scientific Research Fund of Hunan Provincial Education Department, China (21A0141) and the Research and Innovation Project of Hunan Agricultural University (202147).
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and all procedures involving animals were approved by the Animal Ethics Committee of the Hunan Agricultural University, Changsha, China (43321503).
Informed Consent Statement
Not applicable.
Data Availability Statement
Please refer to suggested Data Availability Statements at https://www.ncbi.nlm.nih.gov/nuccore/?term=18S+and+Spirometra+erinaceieuropaei and https://www.ncbi.nlm.nih.gov/nuccore/?term=28S+and+Spirometra+erinaceieuropaei.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Appendix A

Table A1.
Primers used to amplify the sequences studied.
Table A1.
Primers used to amplify the sequences studied.
| Gene | Name | Sequence (5′–3′) | References |
|---|---|---|---|
| 18S | PL3F | ACCTGGTTGATCCTGCCAG | Barta et al., 1997 |
| PL3R | CTTCCGCTGGTTCACCTACGG | ||
| 28S | 28S-F | TGATAGGTTATTTAAACTGGC | This study |
| 28S-R | ACCCGACCCGTCTTGAAACA |
Appendix B

Table A2.
Spirometra isolates included in the molecular analysis, and accession numbers of the corresponding individual sequence.
Table A2.
Spirometra isolates included in the molecular analysis, and accession numbers of the corresponding individual sequence.
| Species | Country of Origin | Host | Sample Codes | Accession Number | References | |
|---|---|---|---|---|---|---|
| 18S | 28S | |||||
| Spirometra erinaceieuropaei | Yiyang City, Hunan Province, China | Zaocys dhumnades | HuN-YiY1 | MZ267595 | MZ293029 | This study |
| Z. dhumnades | HuN-YiY2 | MZ267596 | MZ293030 | This study | ||
| Elaphe carinata | HuN-YiY3 | MZ267597 | MZ293031 | This study | ||
| Changde City, Hunan Province, China | Z. dhumnades | HuN-CD1 | MZ267569 | MZ293003 | This study | |
| Z. dhumnades | HuN-CD2 | MZ267570 | MZ293004 | This study | ||
| E. carinata | HuN-CD3 | MZ267571 | MZ293005 | This study | ||
| Yongzhou City, Hunan Province, China | Z. dhumnades | HuN-YZ1 | MZ267598 | MZ293035 | This study | |
| Z. dhumnades | HuN-YZ2 | MZ267599 | MZ293036 | This study | ||
| Z. dhumnades | HuN-YZ3 | MZ267600 | MZ293037 | This study | ||
| Hengyang City, Hunan Province, China | Z. dhumnades | HuN-HY1 | MZ267583 | MZ293017 | This study | |
| Z. dhumnades | HuN-HY2 | MZ267584 | MZ293018 | This study | ||
| E. carinata | HuN-HY3 | MZ267585 | MZ293019 | This study | ||
| Xiangtan City, Hunan Province, China | Z. dhumnades | HuN-XT1 | MZ267589 | MZ293023 | This study | |
| Z. dhumnades | HuN-XT2 | MZ267590 | MZ293024 | This study | ||
| E. carinata | HuN-XT3 | MZ267591 | MZ293025 | This study | ||
| Shaoyang City, Hunan Province, China | Z. dhumnades | HuN-SY1 | MZ267586 | MZ293020 | This study | |
| Z. dhumnades | HuN-SY2 | MZ267587 | MZ293021 | This study | ||
| E. carinata | HuN-SY3 | MZ267588 | MZ293022 | This study | ||
| Zhuzhou City, Hunan Province, China | Z. dhumnades | HuN-ZZ1 | MZ267604 | MZ293041 | This study | |
| Z. dhumnades | HuN-ZZ2 | MZ267605 | MZ293042 | This study | ||
| E. taeniura | HuN-ZZ3 | MZ267606 | MZ293043 | This study | ||
| Changsha City, Hunan Province, China | Z. dhumnades | HuN-CS1 | MZ267572 | MZ293006 | This study | |
| Z. dhumnades | HuN-CS2 | MZ267573 | MZ293007 | This study | ||
| White Tiger | HuN- CS3 | MZ267607 | MZ292995 | This study | ||
| W. Tiger | HuN- CS4 | MZ267608 | MZ292996 | This study | ||
| Panthera tigris | HuN- CS5 | MZ267609 | MZ292997 | This study | ||
| P. tigris | HuN- CS6 | MZ267610 | MZ292998 | This study | ||
| Prionailurus bengalensis | HuN- CS7 | MZ267611 | MZ292999 | This study | ||
| P. bengalensis | HuN- CS8 | MZ267612 | MZ293000 | This study | ||
| Cat | HuN- CS9 | MZ267613 | MZ293001 | This study | ||
| Cat | HuN- CS10 | MZ267614 | MZ293000 | This study | ||
| Loudi City, Hunan Province, China | E. carinata | HuN-LD1 | MZ267580 | MZ293014 | This study | |
| E. carinata | HuN-LD2 | MZ267581 | MZ293015 | This study | ||
| E. carinata | HuN-LD3 | MZ267582 | MZ293016 | This study | ||
| Chenzhou City, Hunan Province, China | Z. dhumnades | HuN-CZ1 | MZ267574 | MZ293008 | This study | |
| Z. dhumnades | HuN-CZ2 | MZ267575 | MZ293009 | This study | ||
| Z. dhumnades | HuN-CZ3 | MZ267576 | MZ293010 | This study | ||
| Huaihua City, Hunan Province, China | Z. dhumnades | HuN-HH1 | MZ267577 | MZ293011 | This study | |
| Z. dhumnades | HuN-HH2 | MZ267578 | MZ293012 | This study | ||
| Z. dhumnades | HuN-HH3 | MZ267579 | MZ293013 | This study | ||
| Zhangjiajie City, Hunan Province, China | Z. dhumnades | HuN-ZZJ1 | MZ267601 | MZ293038 | This study | |
| Z. dhumnades | HuN-ZZJ2 | MZ267602 | MZ293039 | This study | ||
| Z. dhumnades | HuN-ZZJ3 | MZ267603 | MZ293040 | This study | ||
| Yueyang City, Hunan Province, China | Z. dhumnades | HuN-YuY1 | MZ267566 | MZ293032 | This study | |
| Z. dhumnades | HuN-YuY2 | MZ267567 | MZ293033 | This study | ||
| E. taeniura | HuN-YuY3 | MZ267568 | MZ293034 | This study | ||
| Xiangxi City, Hunan Province, China | Z. dhumnades | HuN-XX1 | MZ267592 | MZ293026 | This study | |
| Z. dhumnades | HuN-XX2 | MZ267593 | MZ293027 | This study | ||
| Z. dhumnades | HuN-XX3 | MZ267594 | MZ293028 | This study | ||
| Guilin City, Guangxi Province, China | Amphiesma stolatum | HQ228991 | HQ288992 | Lee et al., 2010 | ||
| Xiangtan City, Hunan Province, China | Rana nigromaculata | KX528089 | Zhang et al., 2017 | |||
| Australia | Canis familiaris | KY552801 | KY552835 | Kuchta et al., 2017 | ||
| Vietnam | Xenochrophis flavipunctatus | KY552802 | KY552836 | Kuchta et al., 2017 | ||
| Adenocephalus pacificus | Australia | Arctocephalus pusillus | KY552774 | KY552808 | Kuchta et al., 2017 | |
| USA | Callorhinus ursinus | KY552775 | KY552810 | Kuchta et al., 2017 | ||
| Australia | Neophoca cinerea | KY552776 | KY552809 | Kuchta et al., 2017 | ||
| Bothridium pithonis | Czech Republic | Chondropython viridis | KY552803 | KY552838 | Kuchta et al., 2017 | |
| Vietnam | Xenopeltis unicolor | KY552804 | KY552839 | Kuchta et al., 2017 | ||
| Dibothriocephalus nihonkaiensis | Japan | Homo sapiens | AB512013 | LC312467 | Yanagida et al., 2021 Yamasaki et al., 2021 | |
| Dibothriocephalus latus | Russia | Gymnocephalus cernuus | DQ925309 | DQ925326 | Brabec et al., 2016 | |
| Dibothriocephalus dendriticus | USA | Larus hyperboreus | KY552779 | KY552814 | Kuchta et al., 2017 | |
| United Kingdom | Coregonus lavaretus | KY552778 | KY552812 | Kuchta et al., 2017 | ||
| Dibothriocephalus ditremus | United Kingdom | Salvelinus alpinus | KY552780 | KY552813 | Kuchta et al., 2017 | |
| USA | Oncorhynchus tshawytscha | KY552787 | KY552815 | Kuchta et al., 2017 | ||
| Diphyllobothrium scoticum | Australia | Mirounga leonina | KY552777 | KY552811 | Kuchta et al., 2017 | |
| Diphyllobothrium dendriticum | USA | Larus hyperboreus | KY552779 | KY552814 | Kuchta et al., 2017 | |
| Diphyllobothrium schistochilos | Norway | Pusa hispida | KY552782 | KY552821 | Kuchta et al., 2017 | |
| Diphyllobothrium tetrapterum | USA | Callorhinus ursinus | KY552786 | KY552826 | Kuchta et al., 2017 | |
| Diphyllobothrium cordatum | USA | Erignathus barbatus | KY552788 | KY552882 | Kuchta et al., 2017 | |
| Diphyllobothrium lanceolatum | USA | Erignathus barbatus | KY552789 | KY552823 | Kuchta et al., 2017 | |
| Diphyllobothrium stemmacephalum | USA | Lagenorhynchus acutus | AF124459 | AF286943 | Kuchta et al., 2017 | |
| Diphyllobothrium balaenopterae | Japan | Homo sapiens | KY552792 | KY552824 | Kuchta et al., 2017 | |
| Duthiersia fimbriata | Ghana | Varanus exanthematicus | AF267290 | DQ925328 | Kodedova et al., 2001 Brabec et al., 2006 | |
| Duthiersia expansa | Vietnam | Varanus salvator | KY552806 | KY552840 | ||
| Ligula intestinalis | USA | Oncorhynchus tshawytscha | KY552783 | KY552818 | Kuchta et al., 2017 | |
| Ligula intestinalis | Czech Republic | Podiceps cristatus | KY552785 | KY552819 | Kuchta et al., 2017 | |
| Ligula pavlovskii | Ukraine | Neogobius fluviatilis | KY552784 | KY552820 | Kuchta et al., 2017 | |
| Probothriocephalus alaini | Atlantic Ocean | Xenodermichthys copei | KR780925 | KR780881 | Brabec et al., 2015 | |
| Pyramicocephalus phocarum | Norway | Myoxocephalus scorpius | KY552790 | KY552827 | Kuchta et al., 2017 | |
| Norway | Pollachius virens | KY552791 | KY552828 | Kuchta et al., 2017 | ||
| Schistocephalus solidus | Poland | Gasterosteus aculeatus | KY552797 | KY552832 | Kuchta et al., 2017 | |
| Norway | Gasterosteus aculeatus | KY552798 | KY552833 | Kuchta et al., 2017 | ||
| Schistocephalus pungitii | Germany | Pungitius pungitius | KY552799 | KY552834 | Kuchta et al., 2017 | |
| Haplobothrium globuliforme | Canada | Amia calva | AF124458 | AF286926 | Olson et al., 1999 Olson et al., 2001 | |
References
- Hughes, A.J.; Biggs, B.A. Parasitic worms of the central nervous system: An Australian perspective. Intern. Med. J. 2002, 32, 541–553. [Google Scholar] [CrossRef]
- Oda, F.H.; Borteiro, C.; da Graça, R.J.; Tavares, L.E.R.; Crampet, A.; Guerra, V.; Lima, F.S.; Bellay, S.; Karling, L.C.; Castro, O.; et al. Parasitism by larval tapeworms genus Spirometra in South American amphibians and reptiles: New records from Brazil and Uruguay, and a review of current knowledge in the region. Acta Trop. 2016, 164, 150–164. [Google Scholar] [CrossRef]
- Hong, D.; Xie, H.; Wan, H.; An, N.; Xu, C.; Zhang, J. Efficacy comparison between long-term high-dose praziquantel and surgical therapy for cerebral sparganosis: A multicenter retrospective cohort study. PLoS Negl. Trop. Dis. 2016, 12, e0006918. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Li, W.; Hua, L.; Gong, S.; Xiao, J.; Hou, F.; Ge, Y.; Yang, G.D. Spirometra (Pseudophyllidea, Diphyllobothriidae) severely infecting wild-caught snakes from food markets in Guangzhou and Shenzhen, Guangdong, China: Implications for public health. Sci. World J. 2014, 874014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Li, M.W.; Wang, Z.D.; Zhao, G.H.; Zhu, X.Q. Human sparganosis, a neglected food borne zoonosis. Lancet Infect. Dis. 2015, 15, 1226–1235. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, H.; Cui, J.; Jiang, P.; Lin, M.L.; Zhang, Y.L.; Liu, R.D.; Wang, Z.Q. The phylogenetic diversity of Spirometra erinaceieuropaei isolates from southwest China revealed by multi genes. Acta Trop. 2016, 156, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Kuchta, R.; Kołodziej-Sobocińska, M.; Brabec, J.; Młocicki, D.; Sałamatin, R.; Scholz, T. Sparganosis (Spirometra) in Europe in the Molecular Era. Clin. Infect. Dis. 2021, 72, 882–890. [Google Scholar] [CrossRef]
- Anantaphruti, M.T.; Nawa, Y.; Vanvanitchai, Y. Human sparganosis in Thailand: An overview. Acta Trop. 2011, 118, 171–176. [Google Scholar] [CrossRef]
- Li, M.W.; Song, H.Q.; Li, C.; Lin, H.Y.; Xie, W.T.; Lin, R.Q.; Zhu, X.Q. Sparganosis in mainland China. Int. J. Infect. Dis. 2011, 15, e154–e156. [Google Scholar] [CrossRef] [Green Version]
- Bennett, H.M.; Mok, H.P.; Gkrania-Klotsas, E.; Tsai, I.J.; Stanley, E.J.; Antoun, N.M.; Coghlan, A.; Harsha, B.; Berriman, M. The genome of the sparganosis tapeworm Spirometra erinaceieuropaei isolated from the biopsy of a migrating brain lesion. Genome Biol. 2014, 15, 510. [Google Scholar] [CrossRef]
- Zhang, X.; Hong, X.; Liu, S.N.; Jiang, P.; Zhao, S.C.; Sun, C.X.; Wang, Z.Q.; Cui, J. Large-scale survey of a neglected agent of sparganosis Spirometra erinaceieuropaei (Cestoda: Diphyllobothriidae) in wild frogs in China. PLoS Negl. Trop. Dis. 2020, 14, e0008019. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Tang, H.; Abuzeid, A.M.I.; Tan, L.; Wang, A.; Wan, X.; Zhang, H.J.; Liu, Y.S.; Li, G.Q. Protein phosphorylation networks in spargana of Spirometra erinaceieuropaei revealed by phosphoproteomic analysis. Parasit. Vectors 2020, 13, 248. [Google Scholar] [CrossRef]
- Eom, K.S.; Park, H.; Lee, D.; Choe, S.; Kim, K.H.; Jeon, H.K. Mitochondrial Genome Sequences of Spirometra erinaceieuropaei and S. decipiens (Cestoidea: Diphyllobothriidae). Korean J. Parasitol. 2015, 53, 455–463. [Google Scholar] [CrossRef]
- Jeon, H.K.; Park, H.; Lee, D.; Choe, S.; Kim, K.H.; Sohn, W.M.; Eom, S. Genetic Identification of Spirometra decipiens Plerocercoids in Terrestrial Snakes from Korea and China. Korean J. Parasitol. 2016, 54, 181–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamasaki, H.; Sanpool, O.; Rodpai, R.; Sadaow, L.; Laummaunwai, P.; Un, M.; Thanchomnang, T.; Laymanivong, S.; Aung, W.P.P.; Intapan, P.M.; et al. Spirometra species from Asia: Genetic diversity and taxonomic challenges. Parasitol. Int. 2021, 80, 102181. [Google Scholar] [CrossRef]
- Zhang, X.; Duan, J.Y.; Shi, Y.L.; Jiang, P.; Zeng, D.J.; Wang, Z.Q.; Cui, J. Comparative mitochondrial genomics among Spirometra (Cestoda: Diphyllobothriidae) and the molecular phylogeny of related tapeworms. Mol. Phylogenet. Evol. 2017, 117, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Brabec, J.; Kuchta, R.; Scholz, T. Paraphyly of the Pseudophyllidea (Platyhelminthes: Cestoda): Circumscription of monophyletic clades based on phylogenetic analysis of ribosomal RNA. Int. J. Parasitol. 2006, 36, 1535–1541. [Google Scholar] [CrossRef]
- Hernández-Orts, J.S.; Scholz, T.; Brabec, J.; Kuzmina, T.; Kuchta, R. High morphological plasticity and global geographical distribution of the Pacific broad tapeworm Adenocephalus pacificus (syn. Diphyllobothrium pacificum): Molecular and morphological survey. Acta Trop. 2015, 149, 168–178. [Google Scholar] [CrossRef]
- Tan, L.; Wang, A.B.; Zheng, S.Q.; Zhang, X.L.; Huang, C.J.; Liu, W. Molecular characterization and phylogenetic analysis of Taenia multiceps from China. Acta Parasitol. 2018, 63, 721–727. [Google Scholar] [CrossRef] [PubMed]
- Olson, P.D.; Caira, J.N. Evolution of the major lineages of tapeworms (Platyhelminthes: Cestoidea) inferred from 18S ribosomal DNA and elongation factor-1alpha. J. Parasitol. 1999, 85, 1134–1159. [Google Scholar] [CrossRef] [Green Version]
- Waeschenbach, A.; Brabec, J.; Scholz, T.; Littlewood, D.T.J.; Kuchta, R. The catholic taste of broad tapeworms-multiple routes to human infection. Int. J. Parasitol. 2017, 47, 831–843. [Google Scholar] [CrossRef]
- Avcioglu, H.; Yildirim, A.; Duzlu, O.; Inci, A.; Terim, K.A.K.; Balkaya, I. Prevalence and molecular characterization of bovine coenurosis from Eastern Anatolian region of Turkey. Vet. Parasitol. 2011, 176, 59–64. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, X. DAMBE7, New and Improved Tools for Data Analysis in Molecular Biology and Evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Methods Mol. Biol. 2000, 132, 71–91. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5, a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Duan, J.Y.; Wang, Z.Q.; Jiang, P.; Liu, R.D.; Cui, J. Using the small subunit of nuclear ribosomal DNA to reveal the phylogenetic position of the plerocercoid larvae of Spirometra tapeworms. Exp. Parasitol. 2017, 175, 1–7. [Google Scholar] [CrossRef]
- Hao, G.Y. Sequencing and Phylogenetic Analysis of Mitochondrial Cytb Gene and nad4 Gene of Cysticercus cellulosae Xichang lsolates. Prog. Vet. Med. 2015, 36, 59–63. [Google Scholar]
- Hao, G.Y.; He, X.Q.; Wang, R.K. Genetic Variation Analysis of Mitochondrial cox1 and nad4 Genes of Ascaridia galli. China Anim. Husb. Vet. Med. 2017, 44, 113–122. [Google Scholar]
- Okino, T.; Yamasaki, H.; Yamamoto, Y.; Fukuma, Y.; Kurebayashi, J.; Sanuki, F.; Moriya, T.; Ushirogawa, H.; Saito, M. A case of human breast sparganosis diagnosed as Spirometra Type I by molecular analysis in Japan. Parasitol. Int. 2021, 84, 102383. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).