Canine Mammary Tumor Cell Lines Derived from Metastatic Foci Show Increased RAD51 Expression but Diminished Radioresistance via p21 Inhibition
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Cell Culture
2.2. Double Thymidine Block
2.3. Western Blot Analysis
2.4. Induction of DNA Damage
2.5. MTT Assay
2.6. Tumor Cell Migration Assay
2.7. Immunostaining of RAD51 Foci and Microscopy
2.8. cDNA Cloning, Sequencing, and Structure Analysis of Canine p21
2.9. Cell Cycle Analysis
2.10. Microsatellite Genotyping of CHMp and CHMm Using PCR and Amplified Fragment Length Polymorphism Analysis
2.11. p53 Tetramerization Reporter Assay
2.12. Statistical Analysis
3. Results
3.1. RAD51 Expression Was Different in Each Canine Mammary Tumor Cell Line
3.2. Cell Survival after Irradiation
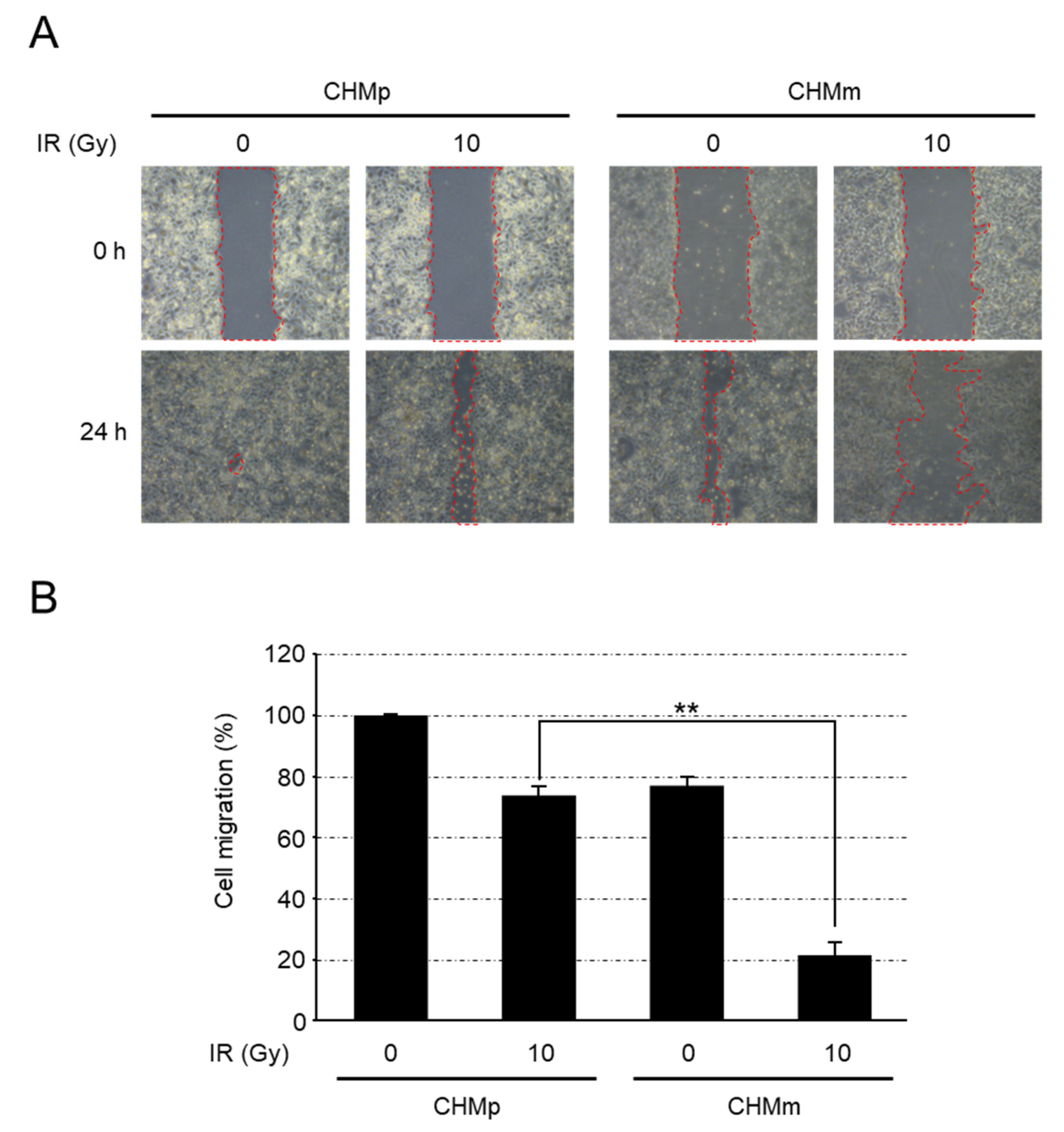
3.3. Comparison of Migration Abilities after Radiation
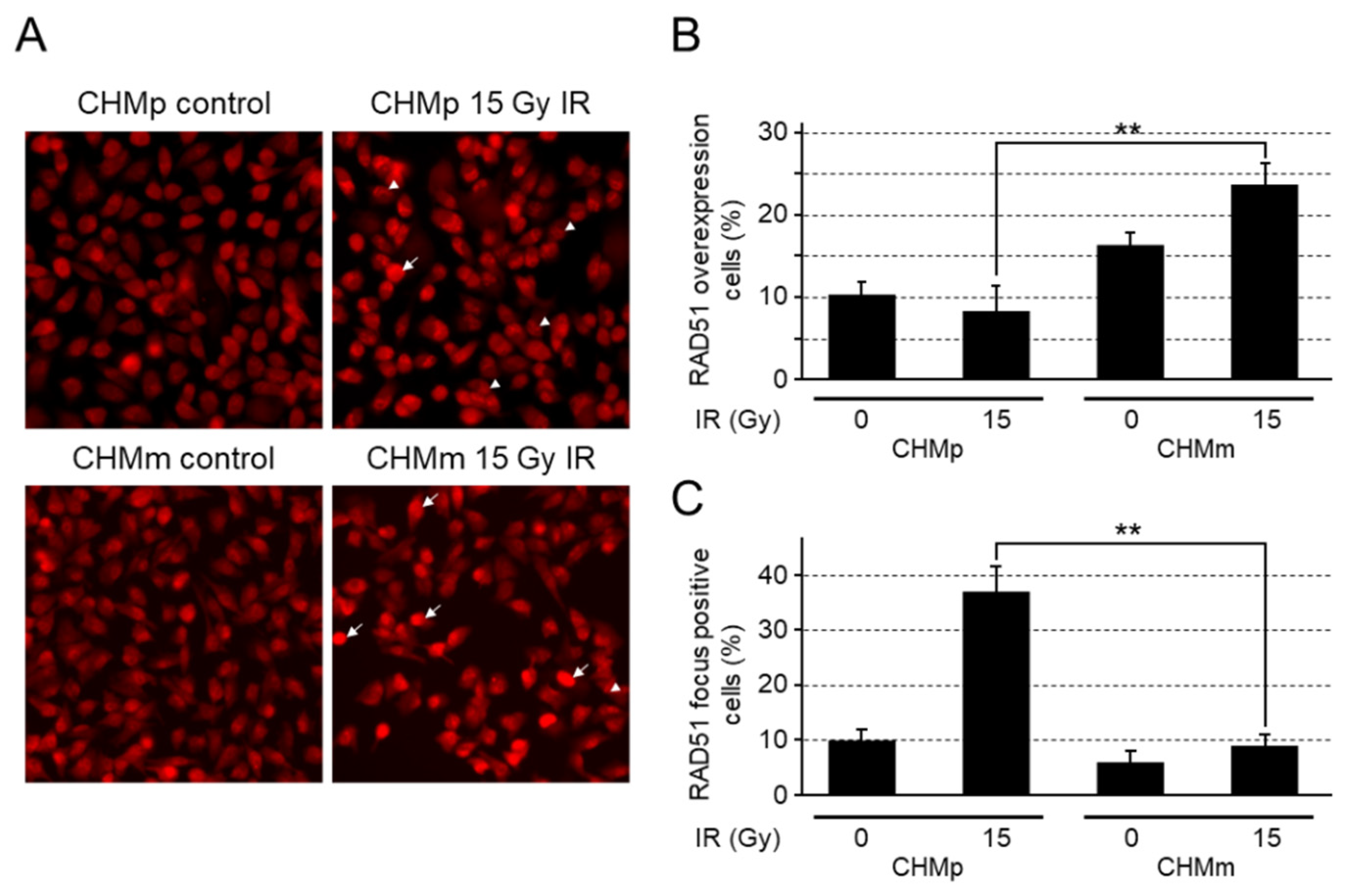
3.4. CHMm Cells Had Reduced Ability to Form RAD51 Foci after Irradiation
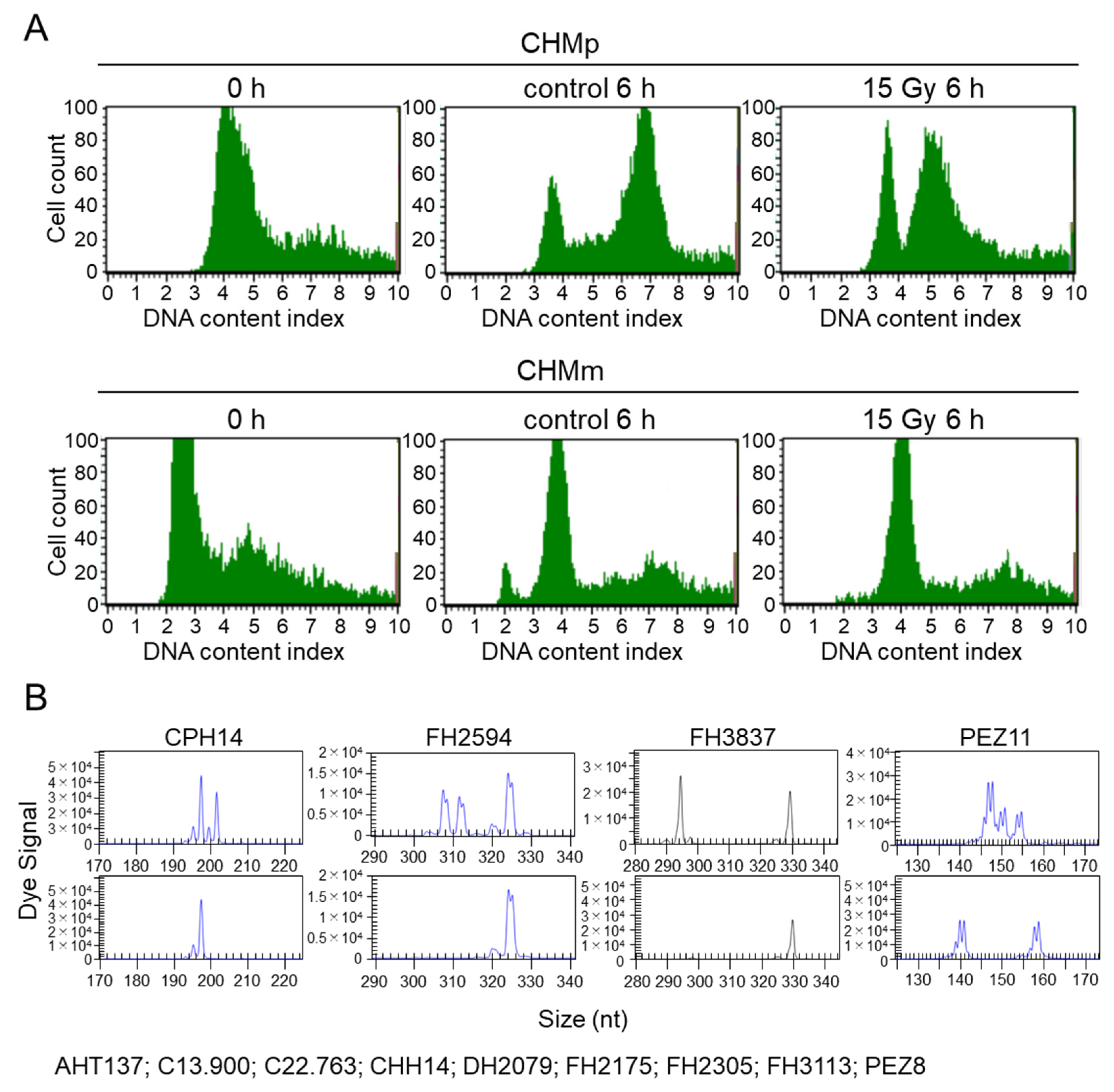
3.4.1. Irradiation Does Not Lead to Cell Cycle Arrest in CHMm
3.4.2. Analysis of Cell Identity Using Microsatellite Analysis
3.4.3. Cloning and Structural Analysis of Canine PALB2
3.4.4. Confirmation of Cross-Reactivity of Anti-Human p21 Antibodies against Canine p21
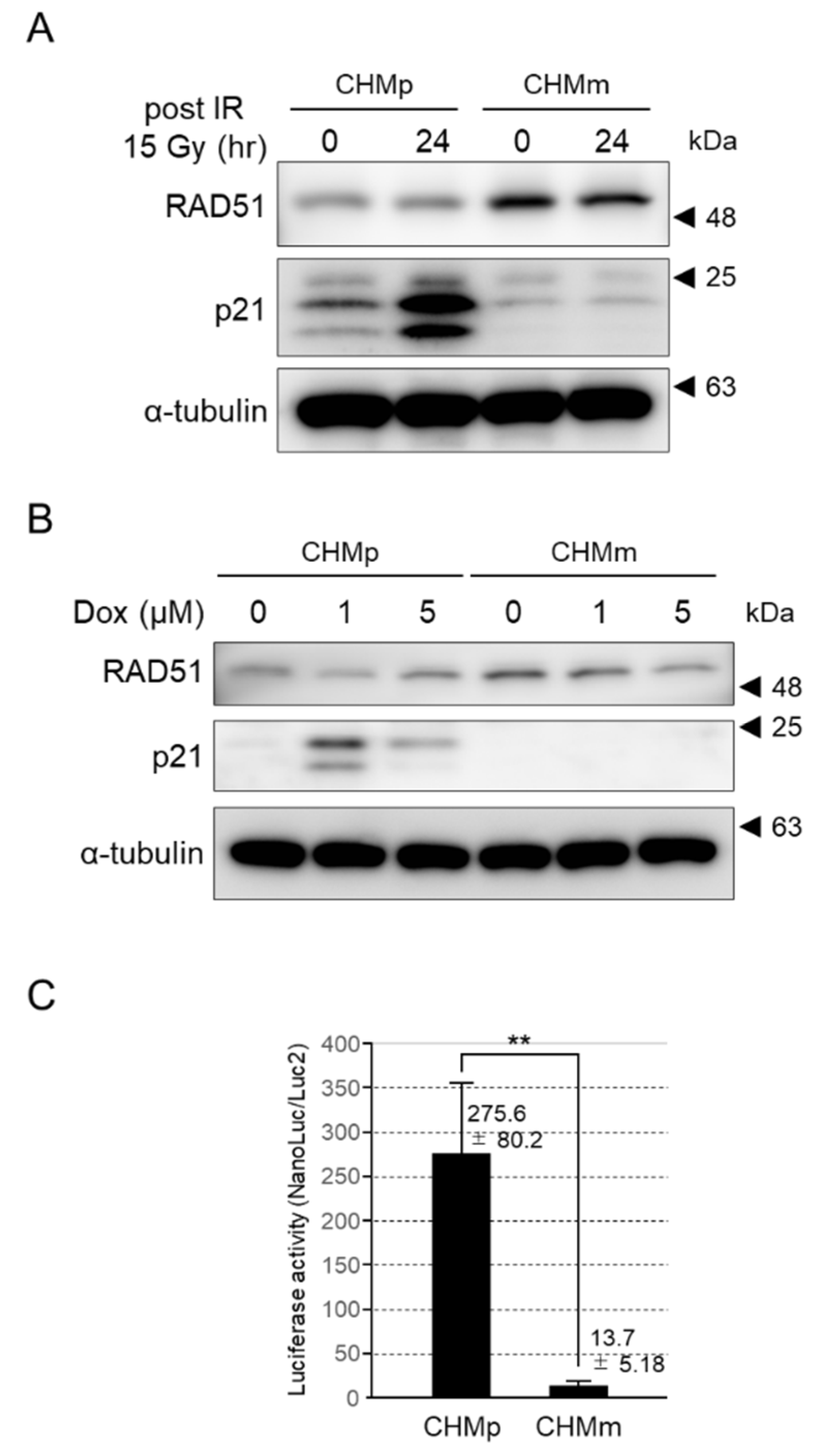
3.4.5. p21 Expression Was Induced by DNA Damage in CHMp, but Not CHMm
3.4.6. The p21 Transcriptional Activation of CHMm Cells Was Significantly Reduced
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shinohara, A.; Ogawa, H.; Matsuda, Y.; Ushio, N.; Ikeo, K.; Ogawa, T. Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nat. Genet. 1993, 4, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Bhat, K.P.; Cortez, D. RPA and RAD51: Fork reversal, fork protection, and genome stability. Nat. Struct. Mol. Biol. 2018, 25, 446–453. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; McCorvie, T.J.; Yates, L.A.; Zhang, X. Structural basis of homologous recombination. Cell. Mol. Life Sci. 2020, 77, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Schipani, F.; Manerba, M.; Marotta, R.; Poppi, L.; Gennari, A.; Rinaldi, F.; Armirotti, A.; Farabegoli, F.; Roberti, M.; Di Stefano, G.; et al. The mechanistic understanding of RAD51 defibrillation: A critical step in BRCA2-mediated DNA repair by homologous recombination. Int. J. Mol. Sci. 2022, 23, 8338. [Google Scholar] [CrossRef]
- Richardson, C. RAD51, genomic stability, and tumorigenesis. Cancer Lett. 2005, 218, 127–139. [Google Scholar] [CrossRef]
- Chen, J.J.; Silver, D.; Cantor, S.; Livingston, D.M.; Scully, R. BRCA1, BRCA2, and Rad51 operate in a common DNA damage response pathway. Cancer Res. 1999, 59, 1752s–1756s. [Google Scholar]
- Tarsounas, M.; Davies, D.; West, S.C. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene 2003, 22, 1115–1123. [Google Scholar] [CrossRef]
- Sliwinski, T.; Krupa, R.; Majsterek, I.; Rykala, J.; Kolacinska, A.; Morawiec, Z.; Drzewoski, J.; Zadrozny, M.; Blasiak, J. Polymorphisms of the BRCA2 and RAD51 genes in breast cancer. Breast Cancer Res. Treat. 2005, 94, 105–109. [Google Scholar] [CrossRef]
- Sharan, S.K.; Morimatsu, M.; Albrecht, U.; Lim, D.S.; Regel, E.; Dinh, C.; Sands, A.; Eichele, G.; Hasty, P.; Bradley, A. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 1997, 386, 804–810. [Google Scholar] [CrossRef]
- Qin, J.; Huang, T.; Wang, J.; Xu, L.; Dang, Q.; Xu, X.; Liu, H.; Liu, Z.; Shao, C.; Zhang, X. RAD51 is essential for spermatogenesis and male fertility in mice. Cell Death Discov. 2022, 8, 118. [Google Scholar] [CrossRef]
- Sleeckx, N.; de Rooster, H.; Veldhuis Kroeze, E.J.; Van Ginneken, C.; Van Brantegem, L. Canine mammary tumours, an overview. Reprod. Domest. Anim. 2011, 46, 1112–1131. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, Y.; Morimatsu, M.; Ochiai, K.; Nagano, M.; Yamane, Y.; Tomizawa, N.; Sasaki, N.; Hashizume, K. Insertion/deletion polymorphism in the BRCA2 nuclear localization signal. Biomed. Res. 2005, 26, 109–116. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rivera, P.; Melin, M.; Biagi, T.; Fall, T.; Haggstrom, J.; Lindblad-Toh, K.; von Euler, H. Mammary tumor development in dogs is associated with BRCA1 and BRCA2. Cancer Res. 2009, 69, 8770–8774. [Google Scholar] [CrossRef]
- Hsu, W.L.; Huang, Y.H.; Chang, T.J.; Wong, M.L.; Chang, S.C. Single nucleotide variation in exon 11 of canine BRCA2 in healthy and cancerous mammary tissue. Vet. J. 2010, 184, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Ozmen, O.; Kul, S.; Risvanli, A.; Ozalp, G.; Sabuncu, A.; Kul, O. Somatic SNPs of the BRCA2 gene at the fragments encoding RAD51 binding sites of canine mammary tumors. Vet. Comp. Oncol. 2017, 15, 1479–1486. [Google Scholar] [CrossRef] [PubMed]
- Di Giacomo, D.; Di Domenico, M.; Defourny, S.V.P.; Malatesta, D.; Di Teodoro, G.; Martino, M.; Viola, A.; D’Alterio, N.; Camma, C.; Modesto, P.; et al. Validation of AmpliSeq NGS panel for BRCA1 and BRCA2 variant detection in canine formalin-fixed paraffin-embedded mammary tumors. Life 2022, 12, 851. [Google Scholar] [CrossRef]
- Yoshikawa, Y.; Ochiai, K.; Morimatsu, M.; Suzuki, Y.; Wada, S.; Taoda, T.; Iwai, S.; Chikazawa, S.; Orino, K.; Watanabe, K. Effects of the missense mutations in canine BRCA2 on BRC repeat 3 functions and comparative analyses between canine and human BRC repeat 3. PLoS ONE 2012, 7, e45833. [Google Scholar] [CrossRef]
- Klopfleisch, R.; Gruber, A.D. Increased expression of BRCA2 and RAD51 in lymph node metastases of canine mammary adenocarcinomas. Vet. Pathol. 2009, 46, 416–422. [Google Scholar] [CrossRef]
- Klopfleisch, R.; Schutze, M.; Gruber, A.D. RAD51 protein expression is increased in canine mammary carcinomas. Vet. Pathol. 2010, 47, 98–101. [Google Scholar] [CrossRef]
- Hu, J.; Zhang, Z.; Zhao, L.; Li, L.; Zuo, W.; Han, L. High expression of RAD51 promotes DNA damage repair and survival in KRAS-mutant lung cancer cells. BMB Rep. 2019, 52, 151–156. [Google Scholar] [CrossRef]
- Song, J.; Cui, D.; Wang, J.; Qin, J.; Wang, S.; Wang, Z.; Zhai, X.; Ma, H.; Ma, D.; Liu, Y.; et al. Overexpression of HMGA1 confers radioresistance by transactivating RAD51 in cholangiocarcinoma. Cell Death Discov. 2021, 7, 322. [Google Scholar] [CrossRef] [PubMed]
- Wiegmans, A.P.; Al-Ejeh, F.; Chee, N.; Yap, P.Y.; Gorski, J.J.; Da Silva, L.; Bolderson, E.; Chenevix-Trench, G.; Anderson, R.; Simpson, P.T.; et al. Rad51 supports triple negative breast cancer metastasis. Oncotarget 2014, 5, 3261–3272. [Google Scholar] [CrossRef] [PubMed]
- Uyama, R.; Nakagawa, T.; Hong, S.H.; Mochizuki, M.; Nishimura, R.; Sasaki, N. Establishment of four pairs of canine mammary tumour cell lines derived from primary and metastatic origin and their E-cadherin expression. Vet. Comp. Oncol. 2006, 4, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Michishita, M.; Akiyoshi, R.; Yoshimura, H.; Katsumoto, T.; Ichikawa, H.; Ohkusu-Tsukada, K.; Nakagawa, T.; Sasaki, N.; Takahashi, K. Characterization of spheres derived from canine mammary gland adenocarcinoma cell lines. Res. Vet. Sci. 2011, 91, 254–260. [Google Scholar] [CrossRef]
- Yoshitake, R.; Saeki, K.; Watanabe, M.; Nakaoka, N.; Ong, S.M.; Hanafusa, M.; Choisunirachon, N.; Fujita, N.; Nishimura, R.; Nakagawa, T. Molecular investigation of the direct anti-tumour effects of nonsteroidal anti-inflammatory drugs in a panel of canine cancer cell lines. Vet. J. 2017, 221, 38–47. [Google Scholar] [CrossRef]
- Ochiai, K.; Azakami, D.; Morimatsu, M.; Hirama, H.; Kawakami, S.; Nakagawa, T.; Michishita, M.; Egusa, A.S.; Sasaki, T.; Watanabe, M.; et al. Endogenous Leu332Gln mutation in p53 disrupts the tetramerization ability in a canine mammary gland tumor cell line. Oncol. Rep. 2018, 40, 488–494. [Google Scholar] [CrossRef]
- Wada, S.; Van Khoa, T.; Kobayashi, Y.; Funayama, T.; Ogihara, K.; Ueno, S.; Ito, N. Prediction of cellular radiosensitivity from DNA damage induced by gamma-rays and carbon ion irradiation in canine tumor cells. J. Vet. Med. Sci. 2005, 67, 1089–1095. [Google Scholar] [CrossRef][Green Version]
- Tanabe, A.; Deguchi, T.; Sato, T.; Nemoto, Y.; Maruo, T.; Madarame, H.; Shida, T.; Naya, Y.; Ogihara, K.; Sahara, H. Radioresistance of cancer stem-like cell derived from canine tumours. Vet. Comp. Oncol. 2016, 14, e93–e101. [Google Scholar] [CrossRef]
- Maeda, M.; Ochiai, K.; Michishita, M.; Morimatsu, M.; Sakai, H.; Kinoshita, N.; Sakaue, M.; Onozawa, E.; Azakami, D.; Yamamoto, M.; et al. In vitro anticancer effects of alpelisib against PIK3CAmutated canine hemangiosarcoma cell lines. Oncol. Rep. 2022, 47, 84. [Google Scholar] [CrossRef]
- Blum, M.; Chang, H.Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Imagawa, T.; Terai, T.; Yamada, Y.; Kamada, R.; Sakaguchi, K. Evaluation of transcriptional activity of p53 in individual living mammalian cells. Anal. Biochem. 2009, 387, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Thumser-Henner, P.; Nytko, K.J.; Rohrer Bley, C. Mutations of BRCA2 in canine mammary tumors and their targeting potential in clinical therapy. BMC Vet. Res. 2020, 16, 30. [Google Scholar] [CrossRef] [PubMed]
- Valdivia, G.; Alonso-Diez, A.; Perez-Alenza, D.; Pena, L. From conventional to precision therapy in canine mammary cancer: A comprehensive review. Front. Vet. Sci. 2021, 8, 623800. [Google Scholar] [CrossRef] [PubMed]
- Shah, C.; Bauer-Nilsen, K.; McNulty, R.H.; Vicini, F. Novel radiation therapy approaches for breast cancer treatment. Semin. Oncol. 2020, 47, 209–216. [Google Scholar] [CrossRef]
- Vispe, S.; Cazaux, C.; Lesca, C.; Defais, M. Overexpression of Rad51 protein stimulates homologous recombination and increases resistance of mammalian cells to ionizing radiation. Nucleic Acids Res. 1998, 26, 2859–2864. [Google Scholar] [CrossRef]
- Xu, Y.; Chen, K.; Cai, Y.; Cheng, C.; Zhang, Z.; Xu, G. Overexpression of Rad51 predicts poor prognosis and silencing of Rad51 increases chemo-sensitivity to doxorubicin in neuroblastoma. Am. J. Transl. Res. 2019, 11, 5788–5799. [Google Scholar]
- Yuan, S.S.; Chang, H.L.; Lee, E.Y. Ionizing radiation-induced Rad51 nuclear focus formation is cell cycle-regulated and defective in both ATM(−/−) and c-Abl(−/−) cells. Mutat. Res. 2003, 525, 85–92. [Google Scholar] [CrossRef]
- Demeyer, A.; Benhelli-Mokrani, H.; Chenais, B.; Weigel, P.; Fleury, F. Inhibiting homologous recombination by targeting RAD51 protein. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188597. [Google Scholar] [CrossRef]
- Daboussi, F.; Dumay, A.; Delacote, F.; Lopez, B.S. DNA double-strand break repair signalling: The case of RAD51 post-translational regulation. Cell Signal. 2002, 14, 969–975. [Google Scholar] [CrossRef]
- Laurini, E.; Marson, D.; Fermeglia, A.; Aulic, S.; Fermeglia, M.; Pricl, S. Role of Rad51 and DNA repair in cancer: A molecular perspective. Pharmacol. Ther. 2020, 208, 107492. [Google Scholar] [CrossRef]
- Engeland, K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ. 2022, 29, 946–960. [Google Scholar] [CrossRef] [PubMed]
- Thorn, C.F.; Oshiro, C.; Marsh, S.; Hernandez-Boussard, T.; McLeod, H.; Klein, T.E.; Altman, R.B. Doxorubicin pathways: Pharmacodynamics and adverse effects. Pharmacogenet. Genom. 2011, 21, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Gencel-Augusto, J.; Lozano, G. p53 tetramerization: At the center of the dominant-negative effect of mutant p53. Genes Dev. 2020, 34, 1128–1146. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shimakawa, K.; Ochiai, K.; Hirose, S.; Tanabe, E.; Michishita, M.; Sakaue, M.; Yoshikawa, Y.; Morimatsu, M.; Tajima, T.; Watanabe, M.; et al. Canine Mammary Tumor Cell Lines Derived from Metastatic Foci Show Increased RAD51 Expression but Diminished Radioresistance via p21 Inhibition. Vet. Sci. 2022, 9, 703. https://doi.org/10.3390/vetsci9120703
Shimakawa K, Ochiai K, Hirose S, Tanabe E, Michishita M, Sakaue M, Yoshikawa Y, Morimatsu M, Tajima T, Watanabe M, et al. Canine Mammary Tumor Cell Lines Derived from Metastatic Foci Show Increased RAD51 Expression but Diminished Radioresistance via p21 Inhibition. Veterinary Sciences. 2022; 9(12):703. https://doi.org/10.3390/vetsci9120703
Chicago/Turabian StyleShimakawa, Kei, Kazuhiko Ochiai, Sachi Hirose, Eri Tanabe, Masaki Michishita, Motoharu Sakaue, Yasunaga Yoshikawa, Masami Morimatsu, Tsuyoshi Tajima, Masami Watanabe, and et al. 2022. "Canine Mammary Tumor Cell Lines Derived from Metastatic Foci Show Increased RAD51 Expression but Diminished Radioresistance via p21 Inhibition" Veterinary Sciences 9, no. 12: 703. https://doi.org/10.3390/vetsci9120703
APA StyleShimakawa, K., Ochiai, K., Hirose, S., Tanabe, E., Michishita, M., Sakaue, M., Yoshikawa, Y., Morimatsu, M., Tajima, T., Watanabe, M., & Tanaka, Y. (2022). Canine Mammary Tumor Cell Lines Derived from Metastatic Foci Show Increased RAD51 Expression but Diminished Radioresistance via p21 Inhibition. Veterinary Sciences, 9(12), 703. https://doi.org/10.3390/vetsci9120703






