Detection and Characterization of a Novel Picornavirus in European Badger (Meles meles)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Molecular Investigation for PVs
2.3. Oxford Nanopore Technologies (ONT) Sequencing
2.4. Virus Isolation
2.5. RT-PCR for Sakobuviruses
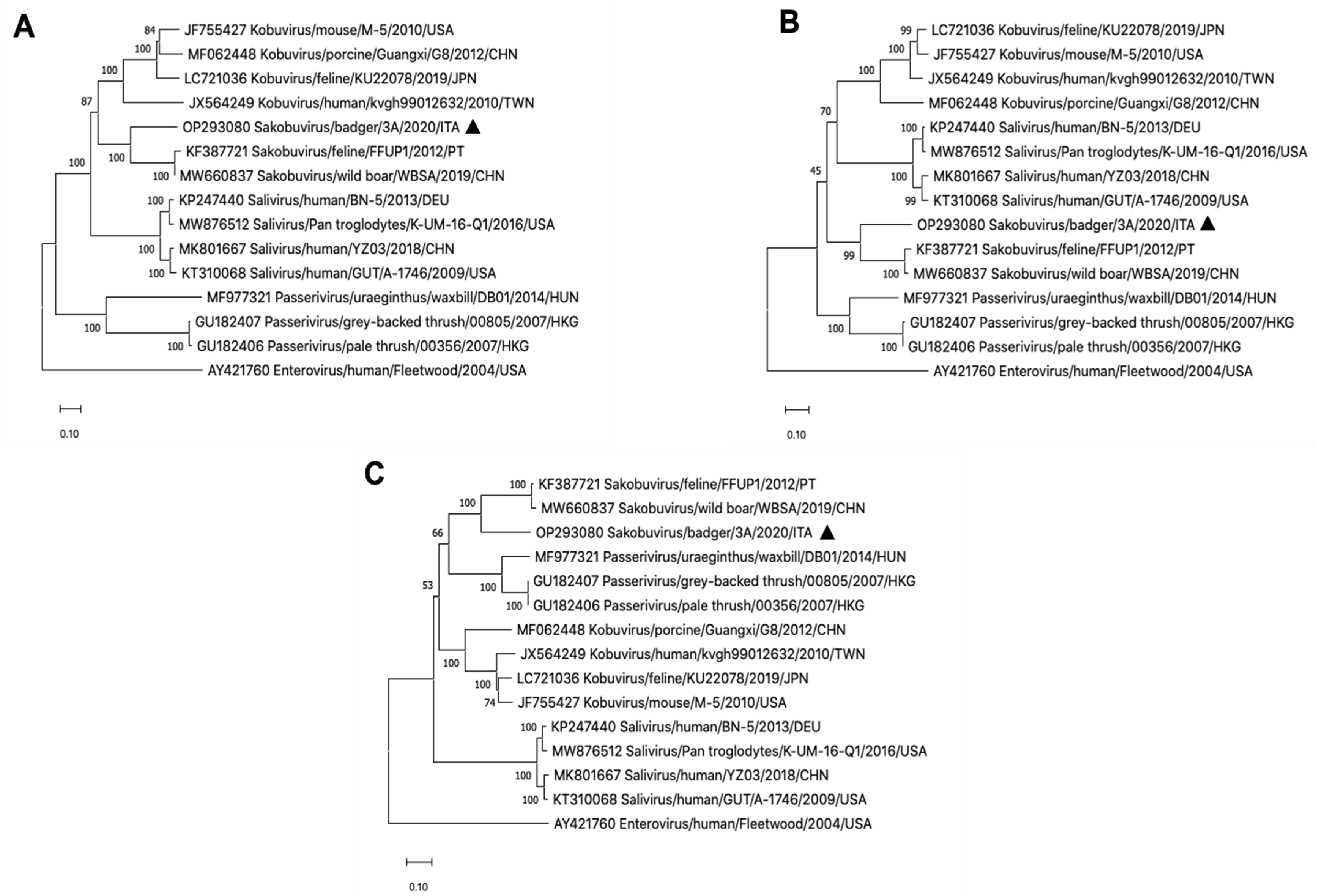
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zell, R.; Knowles, N.J.; Simmonds, P. A proposed division of the family Picornaviridae into subfamilies based on phylogenetic relationships and functional genomic organization. Arch. Virol. 2021, 166, 2927–2935. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Simmonds, P.; Dubovi, E.J.; Qaisar, N.; Henriquez, J.A.; Medina, J.; Shields, S.; Lipkin, W.I. Characterization of a canine homolog of human Aichivirus. J. Virol. 2011, 85, 11520–11525. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.; Woo, P.C.; Yip, C.C.; Choi, G.K.; Wu, Y.; Bai, R.; Fan, R.Y.; Lai, K.K.; Chan, K.H.; Yuen, K.Y. Identification of a novel feline picornavirus from the domestic cat. J. Virol. 2012, 86, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Lau, S.K.; Choi, G.K.; Huang, Y.; Teng, J.L.; Tsoi, H.W.; Tse, H.; Yeung, M.L.; Chan, K.H.; Jin, D.Y.; et al. Natural occurrence and characterization of two internal ribosome entry site elements in a novel virus, canine picodicistrovirus, in the picornavirus-like superfamily. J. Virol. 2012, 86, 2797–2808. [Google Scholar] [CrossRef]
- Woo, P.C.; Lau, S.K.; Choi, G.K.; Yip, C.C.; Huang, Y.; Tsoi, H.W.; Yuen, K.Y. Complete genome sequence of a novel picornavirus, canine picornavirus, discovered in dogs. J. Virol. 2012, 86, 3402–3403. [Google Scholar] [CrossRef]
- Chung, J.Y.; Kim, S.H.; Kim, Y.H.; Lee, M.H.; Lee, K.K.; Oem, J.K. Detection and genetic characterization of feline kobuviruses. Virus Genes 2013, 47, 559–562. [Google Scholar] [CrossRef]
- Di Martino, B.; Di Profio, F.; Melegari, I.; Robetto, S.; Di Felice, E.; Orusa, R.; Marsilio, F. Molecular evidence of kobuviruses in free-ranging red foxes (Vulpes vulpes). Arch. Virol. 2014, 159, 1803–1806. [Google Scholar] [CrossRef]
- Ng, T.F.; Mesquita, J.R.; Nascimento, M.S.; Kondov, N.O.; Wong, W.; Reuter, G.; Knowles, N.J.; Vega, E.; Esona, M.D.; Deng, X.; et al. Feline fecal virome reveals novel and prevalent enteric viruses. Vet. Microbiol. 2014, 171, 102–111. [Google Scholar] [CrossRef]
- Olarte-Castillo, X.A.; Heeger, F.; Mazzoni, C.J.; Greenwood, A.D.; Fyumagwa, R.; Moehlman, P.D.; Hofer, H.; East, M.L. Molecular characterization of canine kobuvirus in wild carnivores and the domestic dog in Africa. Virology 2015, 477, 89–97. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Choi, G.K.Y.; Huang, Y.; Sivakumar, S.; Tsoi, H.W.; Yip, C.C.Y.; Jose, S.V.; Bai, R.; Wong, E.Y.M.; et al. Molecular epidemiology of canine picornavirus in Hong Kong and Dubai and proposal of a novel genus in Picornaviridae. Infect. Genet. Evol. 2016, 41, 191–200. [Google Scholar] [CrossRef]
- Norby, E.E.; Jarman, R.G.; Keiser, P.B.; Binn, L.N.; Hang, J. Genome Sequence of a Novel Canine Picornavirus Isolated from an American Foxhound. Genome Announc. 2017, 5, e00338-17. [Google Scholar] [CrossRef] [PubMed]
- Melegari, I.; Sarchese, V.; Di Profio, F.; Robetto, S.; Carella, E.; Bermudez Sanchez, S.; Orusa, R.; Martella, V.; Marsilio, F.; Di Martino, B. First molecular identification of kobuviruses in wolves (Canis lupus) in Italy. Arch. Virol. 2018, 163, 509–513. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Huang, M.; Chen, X.; Sun, Y.; Huang, J.; Hu, R.; Li, S. Identification and genome characterization of a novel feline picornavirus proposed in the Hunnivirus genus. Infect. Genet. Evol. 2019, 71, 47–50. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, Y.; Li, H.; Chen, Z.; Zhou, J.; Liu, G.; Wang, Y. Genomic characterization and phylogenetic analysis of a new canine picornavirus variant in the mainland of China. Virus Res. 2021, 296, 198351. [Google Scholar] [CrossRef] [PubMed]
- Zell, R. Picornaviridae-the ever-growing virus family. Arch. Virol. 2018, 163, 299–317. [Google Scholar] [CrossRef]
- Reyes, G.R.; Kim, J.P. Sequence-independent, single-primer amplification (SISPA) of complex DNA populations. Mol. Cell. Probes 1991, 5, 473–481. [Google Scholar] [CrossRef]
- Allander, T.; Tammi, M.T.; Eriksson, M.; Bjerkner, A.; Tiveljung-Lindell, A.; Andersson, B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA 2005, 102, 12891–12896. [Google Scholar] [CrossRef]
- Djikeng, A.; Halpin, R.; Kuzmickas, R.; Depasse, J.; Feldblyum, J.; Sengamalay, N.; Afonso, C.; Zhang, X.; Anderson, N.G.; Ghedin, E.; et al. Viral genome sequencing by random priming methods. BMC Genom. 2008, 9, 5. [Google Scholar] [CrossRef]
- Vilsker, M.; Moosa, Y.; Nooij, S.; Fonseca, V.; Ghysens, Y.; Dumon, K.; Pauwels, R.; Alcantara, L.C.; Vanden Eynden, E.; Vandamme, A.M.; et al. Genome Detective: An automated system for virus identification from high-throughput sequencing data. Bioinformatics 2019, 35, 871–873. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kluge, M.; Campos, F.S.; Tavares, M.; de Amorim, D.B.; Valdez, F.P.; Giongo, A.; Roehe, P.M.; Franco, A.C. Metagenomic Survey of Viral Diversity Obtained from Feces of Subantarctic and South American Fur Seals. PLoS ONE 2016, 11, e0151921. [Google Scholar] [CrossRef] [PubMed]
- Kozak, M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 1986, 44, 283–292. [Google Scholar] [CrossRef]
- Li, L.; Pesavento, P.A.; Shan, T.; Leutenegger, C.M.; Wang, C.; Delwart, E. Viruses in diarrhoeic dogs include novel kobuviruses and sapoviruses. J. Gen. Virol. 2011, 92, 2534–2541. [Google Scholar] [CrossRef]
- Di Martino, B.; Di Felice, E.; Ceci, C.; Di Profio, F.; Marsilio, F. Canine kobuviruses in diarrhoeic dogs in Italy. Vet. Microbiol. 2013, 166, 246–249. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, B.; Di Profio, F.; Melegari, I.; Marsilio, F.; Martella, V. Detection of feline kobuviruses in diarrhoeic cats, Italy. Vet. Microbiol. 2015, 176, 186–189. [Google Scholar] [CrossRef]
- Dastjerdi, A.; Benfield, C.; Everest, D.; Stidworthy, M.F.; Zell, R. Novel enteric viruses in fatal enteritis of grey squirrels. J. Gen. Virol. 2020, 101, 746–750. [Google Scholar] [CrossRef]
- Ao, Y.; Xu, J.; Duan, Z. A novel cardiovirus species identified in feces of wild Himalayan marmots. Infect. Genet. Evol. 2022, 103, 105347. [Google Scholar] [CrossRef]
- Palombieri, A.; Fruci, P.; Di Profio, F.; Sarchese, V.; Robetto, S.; Martella, V.; Di Martino, B. Detection and characterization of bopiviruses in domestic and wild ruminants. Transbound. Emerg. Dis. 2022; in press. [Google Scholar] [CrossRef]
| Primers | Target Genes | Assay | Sequence (5′ to 3′) | Position | References |
|---|---|---|---|---|---|
| 3D-for | 3DRdRp | Screening RT-PCR | GTGGGCTGCAAYCCNGA | 7438–7454 | [3] |
| 3D-rev | 3DRdRp | Screening RT-PCR | TTNAGNGCATCAAACCARA | 7544–7563 | [3] |
| FR26RV-N | -- | cDNA synthesis | GCCGGAGCTCTGCAGATATC-N6 | -- | [17] |
| FR40RV-T | Poly-(A) tail | cDNA synthesis | GCCGGAGCTCTGCAGATATC-T20 | -- | [18] |
| FR20RV | --- | SISPA | GCCGGAGCTCTGCAGATATC | -- | [17] |
| SaKoV-for | 3DRdRp | pan-SaKoV RT-PCR | GGTAGCGCGGTCGGTTGCGACCC | 6862–6884 | This study |
| SaKoV-rev | 3DRdRp | pan-SaKoV RT-PCR | CCCAGGACTGGTAGTTGTTAG | 7529–7550 | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palombieri, A.; Fruci, P.; Sarchese, V.; Robetto, S.; Orusa, R.; Arbuatti, A.; Martella, V.; Di Martino, B.; Di Profio, F. Detection and Characterization of a Novel Picornavirus in European Badger (Meles meles). Vet. Sci. 2022, 9, 645. https://doi.org/10.3390/vetsci9110645
Palombieri A, Fruci P, Sarchese V, Robetto S, Orusa R, Arbuatti A, Martella V, Di Martino B, Di Profio F. Detection and Characterization of a Novel Picornavirus in European Badger (Meles meles). Veterinary Sciences. 2022; 9(11):645. https://doi.org/10.3390/vetsci9110645
Chicago/Turabian StylePalombieri, Andrea, Paola Fruci, Vittorio Sarchese, Serena Robetto, Riccardo Orusa, Alessio Arbuatti, Vito Martella, Barbara Di Martino, and Federica Di Profio. 2022. "Detection and Characterization of a Novel Picornavirus in European Badger (Meles meles)" Veterinary Sciences 9, no. 11: 645. https://doi.org/10.3390/vetsci9110645
APA StylePalombieri, A., Fruci, P., Sarchese, V., Robetto, S., Orusa, R., Arbuatti, A., Martella, V., Di Martino, B., & Di Profio, F. (2022). Detection and Characterization of a Novel Picornavirus in European Badger (Meles meles). Veterinary Sciences, 9(11), 645. https://doi.org/10.3390/vetsci9110645







