Simple Summary
This study aims to investigate the influence of climate change on the spread of the African swine fever virus (ASFV). ASFV data in wild boar outbreak locations were sampled and investigated using the Maxent model, with WorldClim bioclimatic data as the predictor variables. The future impacts of climate change on ASFV distribution were scoped with Representative Concentration Pathways (RCP) scenarios for 2050 and 2070. The results show that the precipitation of the driest month (Bio14) and annual mean temperature (Bio1) were contributable factors and indicate a higher possibility of spreading ASFV in the future. The Maxent model was best fitted with an area under curve (AUC) value of 0.99. The proposed Maxent model and the results of this study can be potentially applied to predict disease risks associated with climate change and provide guidance for prevention management.
Abstract
Climate change is an inevitable and urgent issue in the current world. African swine fever virus (ASFV) is a re-emerging viral animal disease. This study investigates the quantitative association between climate change and the potential spread of ASFV to a global extent. ASFV in wild boar outbreak locations recorded from 1 January 2019 to 29 July 2022 were sampled and investigated using the ecological distribution tool, the Maxent model, with WorldClim bioclimatic data as the predictor variables. The future impacts of climate change on ASFV distribution based on the model were scoped with Representative Concentration Pathways (RCP 2.6, 4.5, 6.0, and 8.5) scenarios of Coupled Model Intercomparison Project 5 (CMIP5) bioclimatic data for 2050 and 2070. The results show that precipitation of the driest month (Bio14) was the highest contributor, and annual mean temperature (Bio1) was obtained as the highest permutation importance variable on the spread of ASFV. Based on the analyzed scenarios, we found that the future climate is favourable for ASFV disease; only quantitative ratios are different and directly associated with climate change. The current study could be a reference material for wildlife health management, climate change issues, and World Health Organization sustainability goal 13: climate action.
1. Introduction
Monitoring, modelling, and managing (3Ms) are the pillars for understanding specific contexts, challenges, and bottlenecks in epidemiology [1]. African swine fever virus (ASFV) is a re-emerging viral animal disease [2] that is a large double-stranded DNA virus in the Asfarviridae family and the causative agent of African swine fever (ASF) with high mortality (100%) rates in pigs [3]. Outbreaks of ASFV have been continuously monitored and analyzed with different modelling approaches and management strategies [4,5]. Since there is no effective vaccine or treatment, vector control and preventive measures are currently the only options for mitigating ASFV outbreaks [6]. Climate change generates more uncertainty and greater susceptibility, and understanding impacts under different climatic scenarios supports setting suitable mitigation actions [7].
The impact of climate change on animal and human health is a highly topical concern, with extensive debates and speculations frequently forecasting the worst [8,9,10]. The World Health Organization (WHO) has concluded that climatic changes have occurred since the mid-1970s [11], which is the shifting of climate patterns particularly caused by greenhouse gases emitted from the natural system as forest fires, earthquakes and volcanoes, and human activities [12,13]. Multiple organizations and institutions have collaborated to provide independent and global monitoring mechanisms to track climate change issues [14,15]. Researchers have contributed different strategies and models for adopting and mitigating climate change [16].
Climate change threatens biodiversity, mainly habitat loss, natural disasters, human-wildlife conflict, and species extinction [17,18,19] and escalates the risk of infectious disease outbreaks [20,21]. The spread of diseases has challenged ecologists to understand the host-parasite interactions and driving factors [22]. Previous studies reported that human and animal pathogenic viruses such as West Nile virus (WNV), herpes simplex (HSV), rabies, Chikungunya virus (CHIKV), novel coronavirus infections (COVID-19 or SARS-CoV-2), Rift Valley fever virus (RVFV), African swine fever virus (ASFV), and Bluetongue virus (BTV) have emerged as a result of climate-induced changes in vegetation, human activities [23,24,25], and a variety of contributory determinants [26,27,28]. However, significant research and models for analyzing the relationship of climate change to disease vector populations and epidemiology still require more research for proper control [29].
Long-term studies of climate factors may reveal the association between ASF and climate change [7]. Similarly, it is imperative to know the distribution pattern but hard to determine for environmental management. Various modelling approaches have been used on theoretical ecology, on climate change and conservation policy impact, and for planning purposes [30]. In recent years, the maximum entropy model (Maxent) has been the most popular and broadly used algorithm for analyzing distribution with presence-only data [30,31]. The spatial distribution of pests and some diseases using the Maxent model has been extensively studied [32,33,34,35,36,37,38,39,40,41]. However, the climate impact on wildlife disease ASFV spread in the global scenario is still limited.
Pathogenic diseases can be exacerbated by climate change [42], and bioclimatic indices are used to examine the risk of suffering from heat stress [43]. We hypothesized that multiple bioclimatic scenarios could illustrate the possible risk of ASFV. This study examines the current and future distribution of ASFV globally using the Maxent model [44] and ASFV in wild boar outbreaks locations recorded from 1 January 2019 to 29 July 2022 with 19 bioclimatic predictors. The predictive ASFV risk for 2050 and 2070 in different greenhouse gas concentration trajectories and representative concentration pathway (RCP) [45] scenarios have been reported. These findings will aid in demonstrating the spread of ASFV, identifying hidden high-risk areas, and improving the efficacy of wildlife disease management.
2. Materials and Methods
2.1. Data
2.1.1. Presence Data
ASF was first detected in East Africa in Kenya in the early 1900s, later spread to Europe in the late 1950s and started to spread in Asian countries in 2018 [2,46]. Wild boars freely move in the forest and are more susceptible to ASFV than domestic pigs [47]. Therefore, we sampled the global ASFV wild boar outbreaks data from 1 January 2019 to 29 July 2022 mined from the Food and Agriculture Organization (FAO) of the United Nations data portal https://empres-i.apps.fao.org/epidemiology (accessed on 30 July 2022) [48] for Maxent analysis. The portal is the FAO’s updated global health intelligence and early warning platform to improve forecasting and allow countries to track the spread of the virus and the risk of new outbreaks in different livestock species. Initially, we obtained a total of 17,828 coordinates, and after removing the repeated outbreak locations, 16,186 points were analyzed in this study.
2.1.2. Climatic Variables
Nineteen historical bioclimatic feature raster data, most commonly used over a long-time frame (from 1970 to 2000, considered as current data released in January 2020), were extracted from the WorldClim dataset [49] using the ‘getData’ function of the ‘Raster’ library in Rstuio [50,51]. Similarly, future bio variables of 2050 and 2070 on different RCPs (RCP 2.6, 4.5, 6.0, and 8.5) [45] were downloaded from the Coupled Model Intercomparison Project Phase 5 (CMIP5) [52] application programming interface (API) in Rstuio [50,51]. The corresponding current 19 bioclimatic features [49] (climatic data) of each outbreak location were extracted, and collinearity problems [53] were checked with the threshold of correlation coefficient 0.8. After checking the collinearity test using a variable infection factor (VIF), ten features (Bio1, Bio2, Bio3, Bio7, Bio8, Bio9, Bio10, Bio12, Bio14, and Bio15) were selected and used for further analysis (bold text in Table 1). Bio5, Bio6, and Bio7 were the highest VIF coefficients (Inf) variables and the lowest of Bio8 (6.17).

Table 1.
The bioclimatic features, their codes, VIF values and units.
2.2. Modelling Approach and Evaluation
Maxent is the density estimation, where probability distribution over a set of data of the survey area is examined from the outbreak locations and environmental variables based on the Bayesian rule (Equation (1)). The presence data in set X is considered as 1 and 0 for absence as response variable Y, and the distribution π (X) is the conditional probability P(X|Y = 1) [54].
In this study, we used the Maxent model suggested by Maxent [44] using the ‘dismo’ library in Rstudio [51]. The presence data were grouped randomly for 75% training (12,949 points) and 25% testing (3237 points), and the model was trained with the training dataset for 5000 replications. Maxent contracts presence against pseudo-absence background data that need to be greater than 10,000 points for higher samples [55]. Therefore, we used 10,000 random background locations to evaluate the model with a testing data set. Current bioclimatic variables have been used as environmental predictors for training and testing the model.
The model’s performance was evaluated using the area under the curve (AUC) [44], Cohen’s Kappa [56], and True skill statistics (TSS) [57] values in measuring the distribution of ASFV outbreak locations. AUC values range between 0 and 1; 0.9–1.0 was considered excellent, 0.8–0.9 good, 0.7–0.8 fair, and 0.7 poor [58]. Cohen’s Kappa values range between −1 and +1, whereas 0.80–1.0 suggests excellent, 0.60–0.80 as substantial, 0.40–0.60 as moderate, 0.20–0.40 as fair, 0.01–0.20 as non to slight, and as no agreement [59]. TSS values range from 1 to +1, with +1 indicating perfect agreement and a value less than or equal to zero, indicating the performance is not superior to random [57].
The percentage contribution and permutation importance [60] of the ASFV suitability have been evaluated from the optimal fitted model. The model was further used to predict the possible distribution of ASFV worldwide under different RCP scenarios. The average complementary log-log (cloglog) output values [61] for each future RCP and current distribution maps were compared, and possible influences due to climate change were reported. Finally, since entropy provides average information about the event, the higher the entropy value, the greater the suitability and higher probability of diffusion [44,62]; we concluded the influences of climatic change on spreading ASFV. The detailed study flow is illustrated in Figure 1.
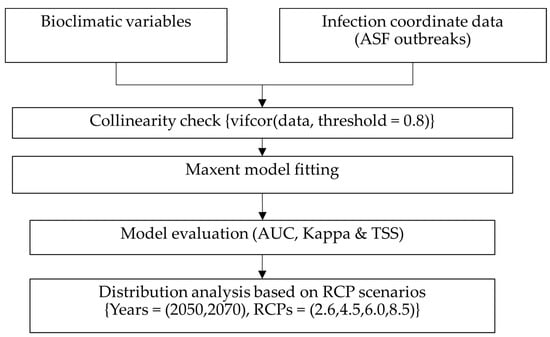
Figure 1.
Study flow.
3. Results and Analysis
The Maxent model was fitted with ASFV outbreak point data and ten climatic predictors conducted after the collinearity test. Based on the performance measures that we considered in the study, we could say the examined model performed excellent prediction through AUC (0.99) and TSS (0.77) and substantial agreement on Kappa (0.60) tests. Two variable contribution measures, relative percentage contribution and permutation importance [60] with ten bioclimatic features, were investigated from the optimal model (Table 2). Bio14 highly contributed (49.3%) to the spread of ASFV, followed by Bio1 (34%), Bio9 (5.5%), Bio12 (3.1%), and, the lowest, Bio8 (0.2%). Similarly, the highest permutation was obtained for Bio1 (29.4%), followed by Bio2 (23.8%), Bio14 (17%), and a negligible (0%) effect by Bio8.

Table 2.
Variable contribution for ASFV distribution.
Based on the observed model, the potential global distribution of ASFV with presence occurrence data and current bioclimatic variables can be seen in Figure 2. The Maxent output entropy values with current climatic conditions in the world ranged between 0 and 0.82 (mean 0.06), and the potential distribution area is in the northern hemisphere.
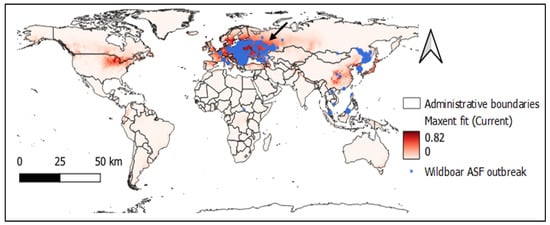
Figure 2.
Maxent predicted map with current bioclimatic features and wild boar ASF outbreak locations (black arrow points to Europe as the recent hotspot of ASFV).
We have examined the impact of future climate conditions on the suitability of ASFV from the above optimal Maxent model. The probable impact maps with different scenarios for the years 2050 and 2070 were captured in Figure 3. For each scenario (RCP 2.6_2050 (Figure 3a), RCP 2.6_2070 (Figure 3b), RCP 4.5_ 2050 (Figure 3c), RCP 4.5_ 2070 (Figure 3d), RCP 6.0 _ 2050 (Figure 3e), RCP 6.0 _2070 (Figure 3f), RCP 8.5_ 2050 (Figure 3g), and RCP 8.5_ 2070 (Figure 3h)), the Maxent entropy value ranges slightly decreased compared to the current scenario but increased the suitability zones.
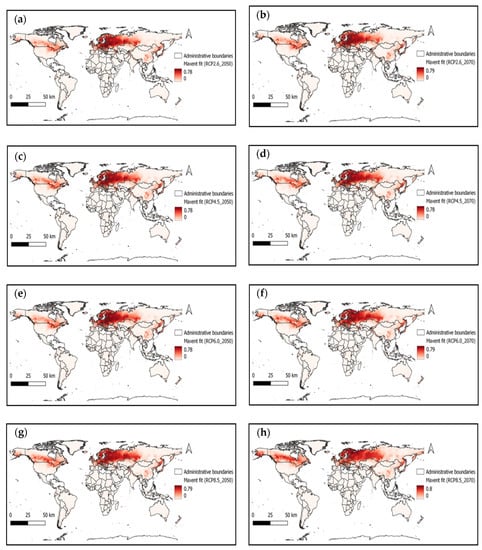
Figure 3.
ASFV suitability map with different (a) RCP 2.6_2050, (b) RCP 2.6_2070, (c) RCP 4.5_ 2050, (d) RCP 4.5_ 2070, (e) RCP 6.0 _ 2050, (f) RCP 6.0 _2070, (g) RCP 8.5_ 2050, and (h) RCP 8.5_ 2070 scenarios.
We further examine the quantitative influence of climate changes on the suitability of ASFV based on the average Maxent entropy value and error as their standard deviation (Figure 4). The average suitability index in 2050 for RCPs 2.6, 4.5, 6.0, and 8.0 increased by 38.56, 46.03, 44.78, and 53.61 percent over the current index, respectively. The highest risk of ASFV in 2050 can be seen in RCP 8.5, followed by RCPs 4.5, 6.0, and 2.6. Similarly, the highest risk of ASFV in 2070 compared with the current index can be seen in RCP 8.5 (73.96%) followed by RCPs 6.0 (55.66%), 4.5 (50.75%), and 2.6 (40.96%). Overall, we can see that risk of ASFV is higher than in the current climatic conditions, and the RCP 6.0 (44.78%) scenario is a relatively lower risk than RCP 4.5 (46.03%) in 2050. With these changes in bioclimatic values under different scenarios, ASFV suitability indices have been changed, which infers evidence of climate change’s impact on the spread of ASFV.
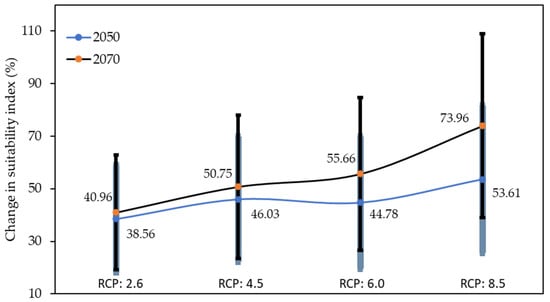
Figure 4.
Average change (percent) of ASFV suitability index and standard deviation (error bars) in different scenarios from current year to 2050 and 2070.
4. Discussion
Climate change is today’s central concern that must be mitigated for a sustainable ecosystem [63]. Climate change increases cross-species viral sharing [64]. ASF is a socio-economic burden on biodiversity and food security. Swine industries worldwide have the biggest threat from ASF [65]. Analyzing this threat linking climate change and its potential distribution could be a supportive reference for wildlife and pig industry management. Identifying possible factors and modelling approaches could support the control of epidemic spread. Suitability modelling is a powerful tool for analyzing the potential distribution that can help to design ecology and epidemic management policies [1,44]. Ecological niche models have been broadly used for risk analysis in epidemiology [66].
This study analyses the possible impact of climate changes on the suitability of ASFV through the commonly used ecological distribution model, Maxent [30]. Maxent deals with presence-only data [54]; 16,186 locations of ASFV in wild boar outbreaks were analyzed with current [49] and predicted CMIP5 [52] RCP scenarios’ bioclimatic data. Initially, we checked the collinearity problem with the current 19 bioclimatic variables and presence data and obtained ten eligible bio variables for the Maxent model. The model was fitted with outbreak data and ten bioclimatic variables and predicted with an optimal model for current and future climatic data. The observed model had AUC (0.99), TSS (0.77), and Kappa (0.60), which confirmed that Maxent could effectively estimate the probability of risk of ASF.
Climatic and environmental factors influence ASFV. The number of ASFV infections in pigs decrease in summer temperatures below 16 °C, and outbreaks are higher in locations above −1 °C [67]. Our study investigated the impact and influenceable climatic factors of ASFV spread and probable future scenarios for 2050 and 2070 with different RCP conditions. We found that the precipitation of the driest month (Bio 14) and annual mean temperature (Bio 1) were the primary contributors to ASFV distribution. For each future scenario, average suitability indexes were found to be higher than the current climatic conditions (see Figure 3 and Figure 4).
The future climatic RCP scenarios have different trends. RCP 2.6 is the straight pathway where carbon emission will decline by 2020 and reach zero by 2100; RCP 4.5 is moderate: emission will peak about 2040 and then decline. Similarly, the emission rate is high, peaking around 2080 in RCP 6.0, declining, and continuously increasing through the 21st century in 8.5 [68]. Bioclimatic factors are climate change-induced variables derived from temperature and precipitation and are extensively used for ecological modelling [49]. Observing the current occurrence locations, future distributions can be predicted using data-driven models [36,54]. Based on the available two-period bioclimatic data of different RCP pathways by 2050 and 2070 and data-driven maxent estimations, a higher risk of ASFV was observed with climate change.
This study illustrates climate change in the spread of ASFV with bioclimatic scenarios and the Maxent model; however, there are significant limitations. The impact of climate change on ASFV is complex, and outbreak locations could be driven by multiple factors that make general predictions impossible. The spread of disease is influenced by various factors such as movement behaviour, the habitat and population density of reservoir and novel host populations [69], climate and land use changes [70], and anthropogenic factors [71], but the current study only examined the climatic covariates. Detailed studies considering disease-driving factors, including domestic pig cases, would present better output. In the future, more variables with heterogeneous effects on ASFV distribution could be analyzed. The economic value of one health approach impacting disease spread [72] also needs research to control climate change and disease as ASFV diffusion. The current study used the Maxent model to investigate the effect of climatic change; comparison, validation with ensemble models [73], and hybrid data-driven-mechanistic models [1,74] could be the better-represented model that warrants further investigation. The changes in bioclimatic variable data influenced entropy values, and the study advocated that climate change influences the spread of ASFV: further investigation and validation must be undertaken.
5. Conclusions
In this study, we analyzed the ASFV in wild boar outbreak locations to fit the Maxent model with ten bioclimatic variables after removing nine variables that created the collinearity problems and examined the possible impact of climatic variables for 2050 and 2070 with multiple RCP scenarios. The results show climatic changes are influencing the spread of ASFV, and the future climate is favourable for ASFV spread: only quantitative ratios are different. Based on the observed model with climatic variables, the spread of wild disease ASFV is directly associated with climate changes. There is no vaccine to eradicate ASF, so improving biosafety measures is currently the best option to prevent ASFV [75,76]. We have emphasized the importance of ASFV control management in conjunction with climate change. Mitigation of climatic change supporting the WHO sustainability goal 13 (Climate action) [77] is to be strictly implemented to control the risk of ASFV. WHO and OIE have recommended operational strategies for maintaining essential human and animal health services that must be assessed and reported [78]. The implication of distribution model predictions can be a supportive tool to deal with the future distribution of wildlife diseases like ASFV. The current study could be a reference to global health concerns.
Author Contributions
Conceptualization, S.T., G.-S.J. and Y.O.; Data curation, T.-S.K. and D.-H.L.; Formal analysis, S.T. and T.D.; Investigation, D.-H.L., and G.-S.J.; Methodology, T.D. and D.-H.L.; Project administration, S.T., G.-S.J. and Y.O.; Resources, S.T., T.-S.K., D.-H.L., G.-S.J. and Y.O.; Software, S.T. and T.D.; Supervision, G.-S.J. and Y.O.; Validation, G.-S.J. and Y.O.; Visualization, G.-S.J. and Y.O.; Writing—original draft, S.T., T.D., T.-S.K. and D.-H.L.; Writing—review & editing, S.T., T.D., T.-S.K., D.-H.L., G.-S.J. and Y.O. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through the Animal Disease Management Technology Advancement Support Program, funded by the Ministry of Agriculture, Food and Rural Affairs (MAFRA) (Project No. 122013-2), and by Yeungnam University research grant, 2019.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Alamo, T.; Reina, D.G.; Millán Gata, P.; Preciado, V.M.; Giordano, G. Data-Driven Methods for Present and Future Pandemics: Monitoring, Modelling and Managing. Annu. Rev. Control 2021, 52, 448–464. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cordón, P.J.; Montoya, M.; Reis, A.L.; Dixon, L.K. African Swine Fever: A Re-Emerging Viral Disease Threatening the Global Pig Industry. Vet. J. 2018, 233, 41–48. [Google Scholar] [CrossRef]
- Galindo, I.; Alonso, C. African Swine Fever Virus: A Review. Viruses 2017, 9, 103. [Google Scholar] [CrossRef]
- Plavšic, B.; Rozstalnyy, A.; Park, J.Y.; Guberti, V.; Depner, K.R.; Torres, G. Strategic Challenges to Global Control of African Swine Fever. In Proceedings of the General Sessions on the World Assembly of the Delegates of the OIE, Paris, France, 26–31 May 2019; Volume 33, pp. 26–31. [Google Scholar]
- Ezanno, P.; Picault, S.; Bareille, S.; Beaunée, G.; Boender, G.J.; Dankwa, E.A.; Deslandes, F.; Donnelly, C.A.; Hagenaars, T.J.; Hayes, S.; et al. The African Swine Fever Modelling Challenge: Model Comparison and Lessons Learnt. Epidemics 2022, 40, 100615. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Park, B.; Kang, H.E. Control Measures to African Swine Fever Outbreak: Active Response in South Korea, Preparation for the Future, and Cooperation. J. Vet. Sci. 2021, 22, e13. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, H.; Schulz, K.; Conraths, F.J.; Sauter-Louis, C. A Review of Environmental Risk Factors for African Swine Fever in European Wild Boar. Animals 2021, 11, 2692. [Google Scholar] [CrossRef]
- De La Rocque, S. Climate Change: Impact on the Epidemiology and Control of Animal Diseases. OIE Rev. Sci. Tech. 2008, 27, 303–308. [Google Scholar]
- Abbass, K.; Qasim, M.Z.; Song, H.; Murshed, M.; Mahmood, H.; Younis, I. A Review of the Global Climate Change Impacts, Adaptation, and Sustainable Mitigation Measures. Environ. Sci. Pollut. Res. 2022, 29, 42539–42559. [Google Scholar] [CrossRef]
- WHO. Climate Change and Health Fact Sheets on Sustainable Development Goals: Health Targets; WHO: Geneva, Switzerland, 2018; Volume 143. [Google Scholar]
- Patz, J.A.; Olson, S.H. Climate Change and Health: Global to Local Influences on Disease Risk. Ann. Trop. Med. Parasitol. 2006, 100, 535–549. [Google Scholar] [CrossRef]
- Yue, X.L.; Gao, Q.X. Contributions of Natural Systems and Human Activity to Greenhouse Gas Emissions. Adv. Clim. Change Res. 2018, 9, 243–252. [Google Scholar] [CrossRef]
- Masson-Delmotte, V.; Zhai, P.; Chen, Y.; Goldfarb, L.; Gomis, M.I.; Matthews, J.B.R.; Berger, S.; Huang, M.; Yelekçi, O.; Yu, R.; et al. (Eds.) Climate Change 2021: The Physical Science Basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2021. [Google Scholar]
- Caroprese, M.; Bradford, B.J.; Rhoads, R.P. Impact of Climate Change on Immune Responses in Agricultural Animals. Front. Vet. Sci. 2021, 8, 844. [Google Scholar] [CrossRef] [PubMed]
- Watts, N.; Amann, M.; Arnell, N.; Ayeb-Karlsson, S.; Beagley, J.; Belesova, K.; Boykoff, M.; Byass, P.; Cai, W.; Campbell-Lendrum, D.; et al. The 2020 Report of The Lancet Countdown on Health and Climate Change: Responding to Converging Crises. Lancet 2021, 397, 129–170. [Google Scholar] [CrossRef]
- Fawzy, S.; Osman, A.I.; Doran, J.; Rooney, D.W. Strategies for Mitigation of Climate Change: A Review. Environ. Chem. Lett. 2020, 18, 2069–2094. [Google Scholar] [CrossRef]
- IPCC Climate Change: A Threat to Human Wellbeing and Health of the Planet. 2022. Available online: https://www.ipcc.ch/2022/02/28/pr-wgii-ar6 (accessed on 31 August 2022).
- Scanes, C.G. Human Activity and Habitat Loss: Destruction, Fragmentation, and Degradation. In Animals and Human Society; Academic Press: Cambridge, MA, USA, 2017; pp. 451–482. ISBN 9780128052471. [Google Scholar]
- Bellard, C.; Bertelsmeier, C.; Leadley, P.; Thuiller, W.; Courchamp, F. Impacts of Climate Change on the Future of Biodiversity. Ecol. Lett. 2012, 15, 365–377. [Google Scholar] [CrossRef]
- Baker, R.E.; Mahmud, A.S.; Miller, I.F.; Rajeev, M.; Rasambainarivo, F.; Rice, B.L.; Takahashi, S.; Tatem, A.J.; Wagner, C.E.; Wang, L.F.; et al. Infectious Disease in an Era of Global Change. Nat. Rev. Microbiol. 2021, 20, 193–205. [Google Scholar] [CrossRef]
- Lafferty, K.D. The Ecology of Climate Change and Infectious Diseases. Ecology 2009, 90, 888–900. [Google Scholar] [CrossRef]
- Johnson, P.T.J.; Townsend, A.R.; Cleveland, C.C.; Glibert, P.M.; Howarth, R.W.; Mckenzie, V.J.; Rejmankova, E.; Ward, M.H. Linking Environmental Nutrient Enrichment and Disease Emergence in Humans and Wildlife. Ecol. Appl. 2010, 20, 16–29. [Google Scholar] [CrossRef]
- Gupta, S.; Rouse, B.T.; Sarangi, P.P. Did Climate Change Influence the Emergence, Transmission, and Expression of the COVID-19 Pandemic? Front. Med. 2021, 8, 769208. [Google Scholar] [CrossRef]
- Epstein, P.R. Chikungunya Fever Resurgence and Global Warming. Am. J. Trop. Med. Hyg. 2007, 76, 403–404. [Google Scholar] [CrossRef]
- Magiri, R.; Muzandu, K.; Gitau, G.; Choongo, K.; Iji, P. Impact of Climate Change on Animal Health, Emerging and Re-Emerging Diseases in Africa. In African Handbook of Climate Change Adaptation; Springer: Cham, Switzerland, 2021; pp. 1835–1851. [Google Scholar]
- Smolinski, M.S.; Hamburg, M.A.; Lederberg, J. Emerging Microbial Threats to Health in the 21st Century; National Academy of Sciences: Washington, DC, USA, 2003; Volume 398. [Google Scholar]
- Glud, H.A.; George, S.; Skovgaard, K.; Larsen, L.E. Zoonotic and Reverse Zoonotic Transmission of Viruses between Humans and Pigs. APMIS 2021, 129, 675–693. [Google Scholar] [CrossRef]
- Mishra, J.; Mishra, P.; Arora, N.K. Linkages between Environmental Issues and Zoonotic Diseases: With Reference to COVID-19 Pandemic. Environ. Sustain. 2021, 4, 455–467. [Google Scholar] [CrossRef]
- Patz, J.A.; Olson, S.H. Malaria Risk and Temperature: Influences from Global Climate Change and Local Land Use Practices. Proc. Natl. Acad. Sci. USA 2006, 103, 5635–5636. [Google Scholar] [CrossRef] [PubMed]
- Qazi, A.W.; Saqib, Z.; Zaman-ul-Haq, M. Trends in Species Distribution Modelling in Context of Rare and Endemic Plants: A Systematic Review. Ecol. Processes 2022, 11, 1–11. [Google Scholar] [CrossRef]
- Yackulic, C.B.; Chandler, R.; Zipkin, E.F.; Royle, J.A.; Nichols, J.D.; Campbell Grant, E.H.; Veran, S. Presence-Only Modelling Using MAXENT: When Can We Trust the Inferences? Methods Ecol. Evol. 2013, 4, 236–243. [Google Scholar] [CrossRef]
- Kopsco, H.L.; Smith, R.L.; Halsey, S.J. A Scoping Review of Species Distribution Modeling Methods for Tick Vectors. Front. Ecol. Evol. 2022, 10, 462. [Google Scholar] [CrossRef]
- Lee, D.S.; Choi, W.I.; Nam, Y.; Park, Y.S. Predicting Potential Occurrence of Pine Wilt Disease Based on Environmental Factors in South Korea Using Machine Learning Algorithms. Ecol. Inform. 2021, 64, 101378. [Google Scholar] [CrossRef]
- Gao, H.; Ma, J. Spatial Distribution and Risk Areas of Foot and Mouth Disease in Mainland China. Prev. Vet. Med. 2021, 189, 105311. [Google Scholar] [CrossRef]
- Alkhamis, M.A.; VanderWaal, K. Spatial and Temporal Epidemiology of Lumpy Skin Disease in the Middle East, 2012–2015. Front. Vet. Sci. 2016, 3, 19. [Google Scholar] [CrossRef]
- Ikegami, M.; Jenkins, T.A.R. Estimate Global Risks of a Forest Disease under Current and Future Climates Using Species Distribution Model and Simple Thermal Model—Pine Wilt Disease as a Model Case. For. Ecol. Manag. 2018, 409, 343–352. [Google Scholar] [CrossRef]
- Al Ruheili, A.M.; Boluwade, A.; Al Subhi, A.M. Assessing the Impact of Climate Change on the Distribution of Lime (16SRII-b) and Alfalfa (16srii-d) Phytoplasma Disease Using Maxent. Plants 2021, 10, 460. [Google Scholar] [CrossRef]
- Narouei-Khandan, H.A.; Worner, S.P.; Viljanen, S.L.H.; Van Bruggen, A.H.C.; Balestra, G.M.; Jones, E. The Potential Global Climate Suitability of Kiwifruit Bacterial Canker Disease (Pseudomonas Syringae Pv. Actinidiae (Psa)) Using Three Modelling Approaches: CLIMEX, Maxent and Multimodel Framework. Climate 2022, 10, 14. [Google Scholar] [CrossRef]
- Pramanik, M.; Singh, P.; Dhiman, R.C. Identification of Bio-Climatic Determinants and Potential Risk Areas for Kyasanur Forest Disease in Southern India Using MaxEnt Modelling Approach. BMC Infect. Dis. 2021, 21, 1226. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Peng, W.; Liu, X.; He, G.; Cai, Y. Spatiotemporal Dynamics and Factors Driving the Distributions of Pine Wilt Disease-Damaged Forests in China. Forests 2022, 13, 261. [Google Scholar] [CrossRef]
- Musolin, D.L.; Nielsen, A.L.; Mazzoni, V.; Hwang, J.H.; Kim, S.-H.; Yoon, S.; Jung, S.; Kim, D.H.; Lee, W.-H. Evaluation of Spatial Distribution of Three Major Leptocorisa (Hemiptera: Alydidae) Pests Using MaxEnt Model. Insects 2022, 13, 750. [Google Scholar] [CrossRef]
- Mora, C.; McKenzie, T.; Gaw, I.M.; Dean, J.M.; von Hammerstein, H.; Knudson, T.A.; Setter, R.O.; Smith, C.Z.; Webster, K.M.; Patz, J.A.; et al. Over Half of Known Human Pathogenic Diseases Can Be Aggravated by Climate Change. Nat. Clim. Chang. 2022, 12, 869–875. [Google Scholar] [CrossRef]
- Neves, S.F.; Silva, M.C.F.; Miranda, J.M.; Stilwell, G.; Cortez, P.P. Predictive Models of Dairy Cow Thermal State: A Review from a Technological Perspective. Vet. Sci. 2022, 9, 416. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum Entropy Modeling of Species Geographic Distributions. Int. J. Glob. Environ. Issues 2006, 6, 231–252. [Google Scholar] [CrossRef]
- Van Vuuren, D.P.; Edmonds, J.A.; Kainuma, M.; Riahi, K.; Weyant, J. A Special Issue on the RCPs. Clim. Chang. 2011, 109, 1. [Google Scholar] [CrossRef]
- Mighell, E.; Ward, M.P. African Swine Fever Spread across Asia, 2018–2019. Transbound. Emerg. Dis. 2021, 68, 2722–2732. [Google Scholar] [CrossRef]
- Sánchez-Cordón, P.J.; Nunez, A.; Neimanis, A.; Wikström-Lassa, E.; Montoya, M.; Crooke, H.; Gavier-Widén, D. African Swine Fever: Disease Dynamics in Wild Boar Experimentally Infected with ASFV Isolates Belonging to Genotype I and II. Viruses 2019, 11, 852. [Google Scholar] [CrossRef]
- Claes, F.; Kuznetsov, D.; Liechti, R.; Von Dobschuetz, S.; Truong, B.D.; Gleizes, A.; Conversa, D.; Colonna, A.; Demaio, E.; Ramazzotto, S.; et al. The EMPRES-i Genetic Module: A Novel Tool Linking Epidemiological Outbreak Information and Genetic Characteristics of Influenza Viruses. Database 2014, 2014, bau008. [Google Scholar] [CrossRef] [PubMed]
- Fick, S.E.; Hijmans, R.J. WorldClim 2: New 1-Km Spatial Resolution Climate Surfaces for Global Land Areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Van Etten, J.; Mattiuzzi, M.; Sumner, M.; Greenberg, J.A.; Lamigueiro, O.P.; Bevan, A.; Racine, E.B.; Shortridge, A. Geographic Data Analysis and Modeling: Package “Raster”. R CRAN Proj. 2015, 2.3-40, 1–134. [Google Scholar]
- R Core Team RStudio | Open Source & Professional Software for Data Science Teams—RStudio. RStudio. 2022. Available online: http://www.rstudio.com/ (accessed on 2 April 2022).
- Taylor, K.E.; Stouffer, R.J.; Meehl, G.A. An Overview of CMIP5 and the Experiment Design. Bull. Am. Meteorol. Soc. 2012, 93, 485–498. [Google Scholar] [CrossRef]
- Dodge, Y. The Concise Encyclopedia of Statistics; Springer: New York, NY, USA, 2008; ISBN 978-0-387-31742-7. [Google Scholar]
- Phillips, S.J.; Dudík, M. Modeling of Species Distributions with Maxent: New Extensions and a Comprehensive Evaluation. Ecography 2008, 31, 161–175. [Google Scholar] [CrossRef]
- Merow, C.; Smith, M.J.; Silander, J.A. A Practical Guide to MaxEnt for Modeling Species’ Distributions: What It Does, and Why Inputs and Settings Matter. Ecography 2013, 36, 1058–1069. [Google Scholar] [CrossRef]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Allouche, O.; Tsoar, A.; Kadmon, R. Assessing the Accuracy of Species Distribution Models: Prevalence, Kappa and the True Skill Statistic (TSS). J. Appl. Ecol. 2006, 43, 1223–1232. [Google Scholar] [CrossRef]
- Swets, J.A. Measuring the Accuracy of Diagnostic Systems. Science 1988, 240, 1285–1293. [Google Scholar] [CrossRef]
- McHugh, M.L. Interrater Reliability: The Kappa Statistic. Biochem. Med. 2012, 22, 276–282. [Google Scholar] [CrossRef]
- Phillips, S.J. A Brief Tutorial on Maxent. Available online: www.cs.princeton.edu/~schapire/maxent (accessed on 31 August 2022).
- Phillips, S.J.; Anderson, R.P.; Dudík, M.; Schapire, R.E.; Blair, M.E. Opening the Black Box: An Open-Source Release of Maxent. Ecography 2017, 40, 887–893. [Google Scholar] [CrossRef]
- Tang, X.; Yuan, Y.; Li, X.; Zhang, J. Maximum Entropy Modeling to Predict the Impact of Climate Change on Pine Wilt Disease in China. Front. Plant Sci. 2021, 12, 652500. [Google Scholar] [CrossRef] [PubMed]
- An, R.; Ji, M.; Zhang, S. Global Warming and Obesity: A Systematic Review. Obes. Rev. 2018, 19, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Carlson, C.J.; Albery, G.F.; Merow, C.; Trisos, C.H.; Zipfel, C.M.; Eskew, E.A.; Olival, K.J.; Ross, N.; Bansal, S. Climate Change Increases Cross-Species Viral Transmission Risk. Nature 2022, 607, 555–562. [Google Scholar] [CrossRef]
- Pig333 South Korea Reduces Wild Boar Population to Slow ASF Spread—Swine News—Pig333, Pig to Pork Community. Available online: https://www.pig333.com/latest_swine_news/south-korea-reduces-wild-boar-population-to-slow-asf-spread_17241/ (accessed on 26 April 2021).
- Escobar, L.E. Ecological Niche Modeling: An Introduction for Veterinarians and Epidemiologists. Front. Vet. Sci. 2020, 7, 713. [Google Scholar] [CrossRef]
- Ungur, A.; Cazan, C.D.; Panait, L.C.; Coroian, M.; Cătoi, C. What Is the Real Influence of Climatic and Environmental Factors in the Outbreaks of African Swine Fever? Animals 2022, 12, 781. [Google Scholar] [CrossRef]
- Van Vuuren, D.P.; Edmonds, J.; Kainuma, M.; Riahi, K.; Thomson, A.; Hibbard, K.; Hurtt, G.C.; Kram, T.; Krey, V.; Lamarque, J.F.; et al. The Representative Concentration Pathways: An Overview. Clim. Chang. 2011, 109, 5–31. [Google Scholar] [CrossRef]
- Engering, A.; Hogerwerf, L.; Slingenbergh, J. Pathogen-Host-Environment Interplay and Disease Emergence. Emerg. Microbes Infect. 2013, 2, e5. [Google Scholar] [CrossRef]
- Rakotoarinia, M.R.; Guillaume Blanchet, F.; Gravel, D.; Lapen, D.R.; Leighton, P.A.; Ogden, N.H.; Ludwig, A. Effects of Land Use and Weather on the Presence and Abundance of Mosquito-Borne Disease Vectors in a Urban and Agricultural Landscape in Eastern Ontario, Canada. PLoS ONE 2022, 17, e0262376. [Google Scholar] [CrossRef]
- Podgórski, T.; Borowik, T.; Łyjak, M.; Woźniakowski, G. Spatial Epidemiology of African Swine Fever: Host, Landscape and Anthropogenic Drivers of Disease Occurrence in Wild Boar. Prev. Vet. Med. 2020, 177, 104691. [Google Scholar] [CrossRef]
- Häsler, B.; Gilbert, W.; Jones, B.A.; Pfeiffer, D.U.; Rushton, J.; Otte, M.J. The Economic Value of One Health in Relation to the Mitigation of Zoonotic Disease Risks. In Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2012; Volume 365, pp. 127–151. [Google Scholar]
- Sharma, N.; Dev, J.; Mangla, M.; Wadhwa, V.M.; Mohanty, S.N.; Kakkar, D. A Heterogeneous Ensemble Forecasting Model for Disease Prediction. New Gener. Comput. 2021, 39, 701–715. [Google Scholar] [CrossRef] [PubMed]
- Hayes, B.H.; Andraud, M.; Salazar, L.G.; Rose, N.; Vergne, T. Mechanistic Modelling of African Swine Fever: A Systematic Review. Prev. Vet. Med. 2021, 191, 105358. [Google Scholar] [CrossRef] [PubMed]
- Denstedt, E.; Porco, A.; Hwang, J.; Nga, N.T.T.; Ngoc, P.T.B.; Chea, S.; Khammavong, K.; Milavong, P.; Sours, S.; Osbjer, K.; et al. Detection of African Swine Fever Virus in Free-Ranging Wild Boar in Southeast Asia. Transbound. Emerg. Dis. 2021, 68, 2669–2675. [Google Scholar] [CrossRef]
- Gervasi, V.; Marcon, A.; Bellini, S.; Guberti, V. Evaluation of the Efficiency of Active and Passive Surveillance in the Detection of African Swine Fever in Wild Boar. Vet. Sci. 2020, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- United Nations. Sustainable Development Goal 13: Climate Action; United Nations: New York, NY, USA, 2019. [Google Scholar]
- WHO. OIE WHO-OIE Operational Framework for Good Governance at the Human-Animal Interface: Bridging WHO and OIE Tools for the Assessment of National Capacities; WHO: Geneva, Switzerland, 2014. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).