Identification of a 5-Methylcytosine Site (mC-7) That May Inhibit CXCL11 Expression and Regulate E. coli F18 Susceptibility in IPEC-J2 Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Samples
2.2. E. coli F18 Infection
2.3. RNA Isolation, cDNA Synthesis and qPCR Analysis
2.4. Western Blot Analysis
2.5. Construction of CXCL11 Overexpression IPEC-J2 Cell Line
2.6. Adhesion Level Detection of E. coli F18 to IPEC-J2 In Vitro
2.7. Methylation Detection of CpG Island in Pig CXCL11 Promoter
2.8. Detection of Double Luciferase Activity
2.9. Identification of Key Transcription Factor in Pig CXCL11 Promoter Region CpG Island
2.10. Statistical Analyses
3. Results
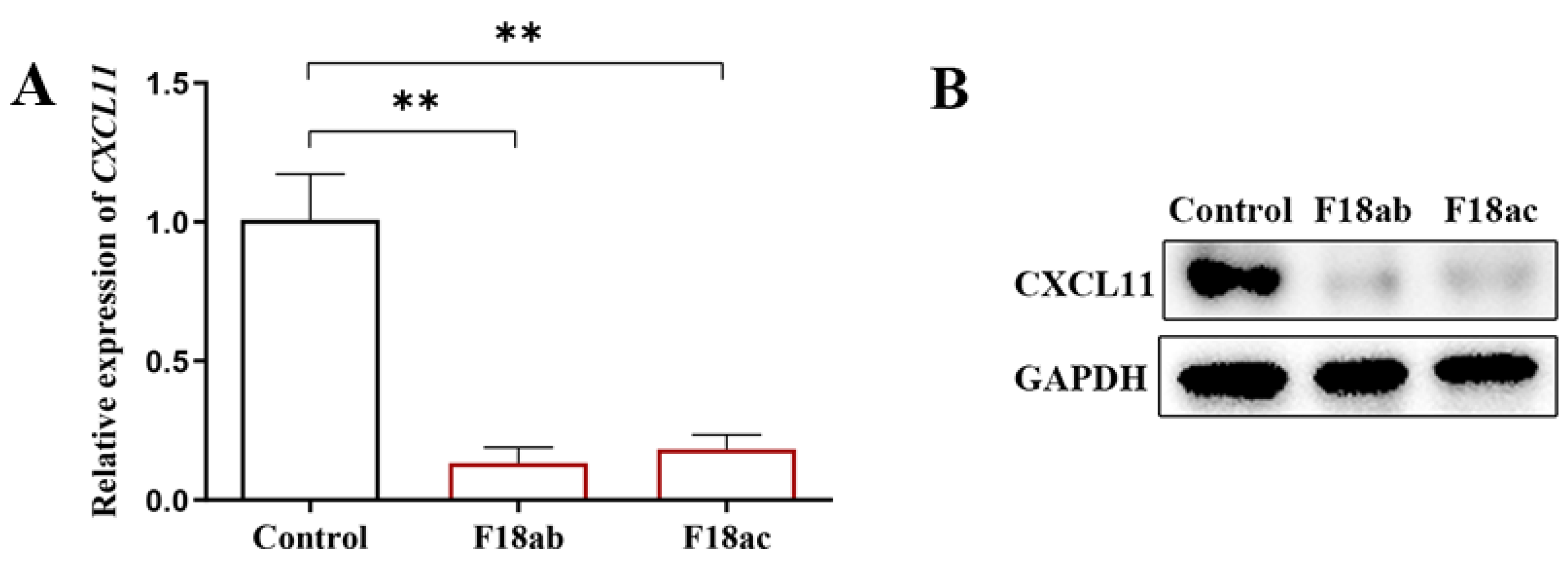
3.1. Expression Detection of CXCL11 in the E. coli F18-Infected IPEC-J2 Cells
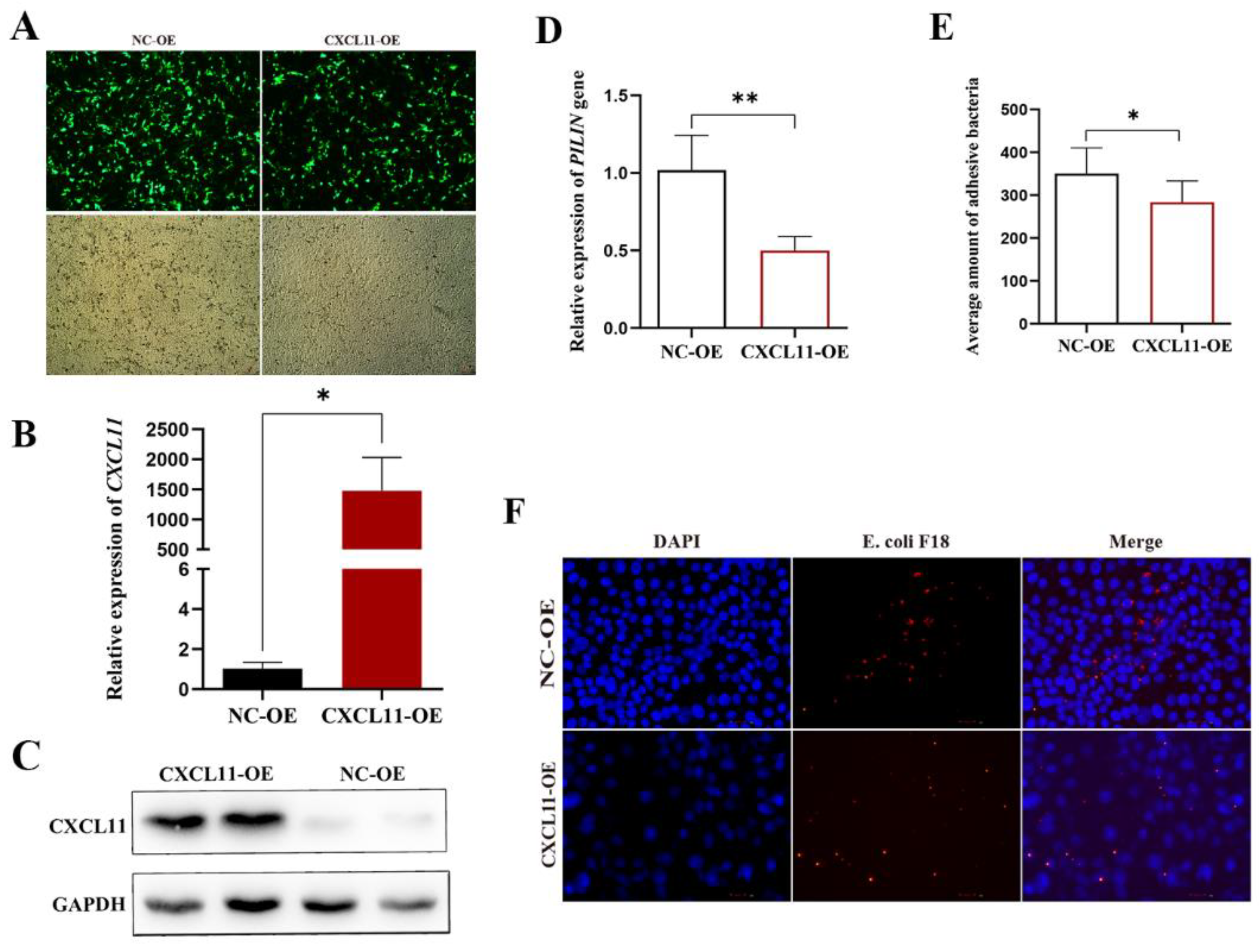
3.2. Effect of CXCL11 Overexpression on the Adhesion Ability of E. coli F18 to IPEC-J2
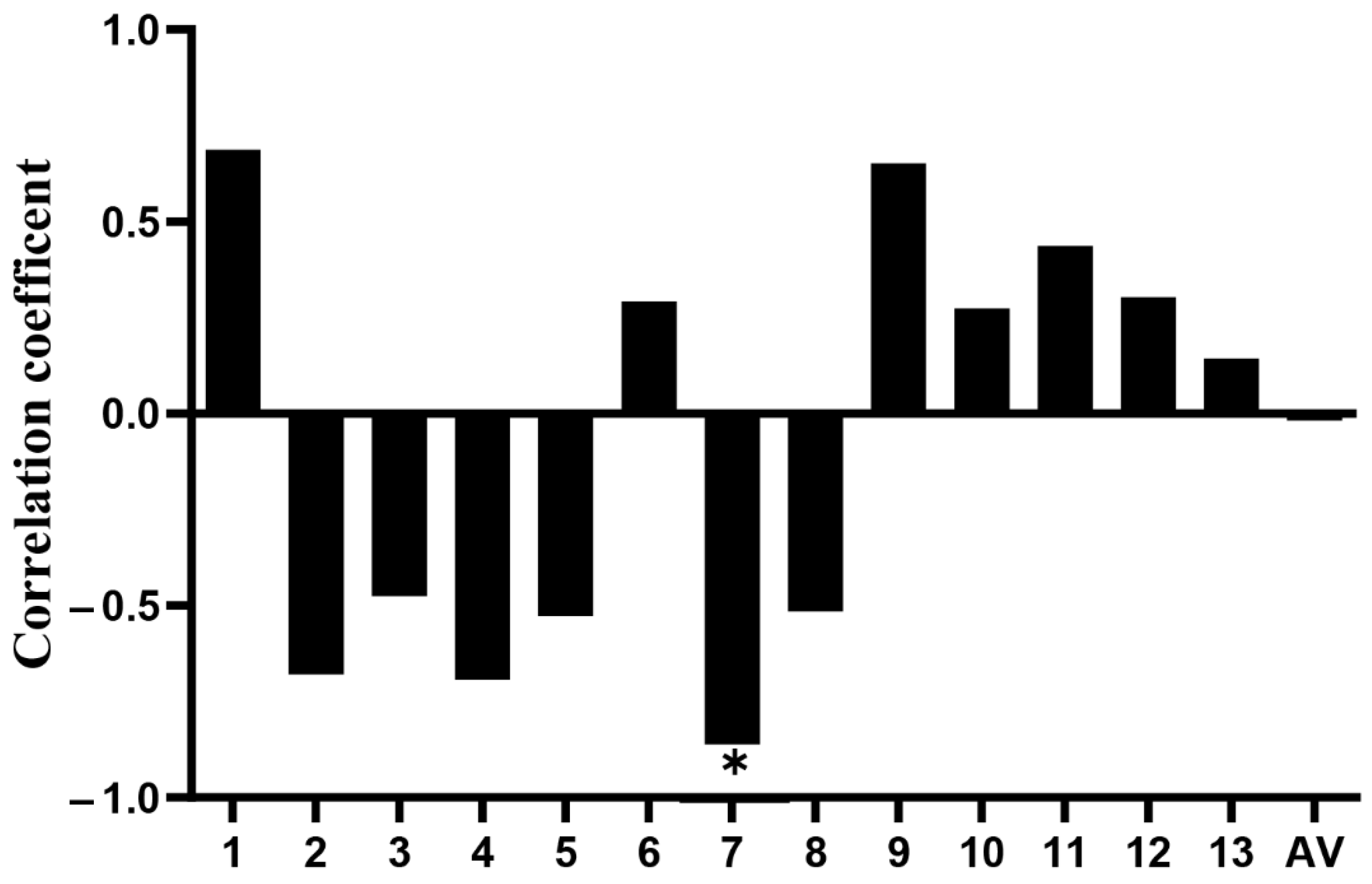
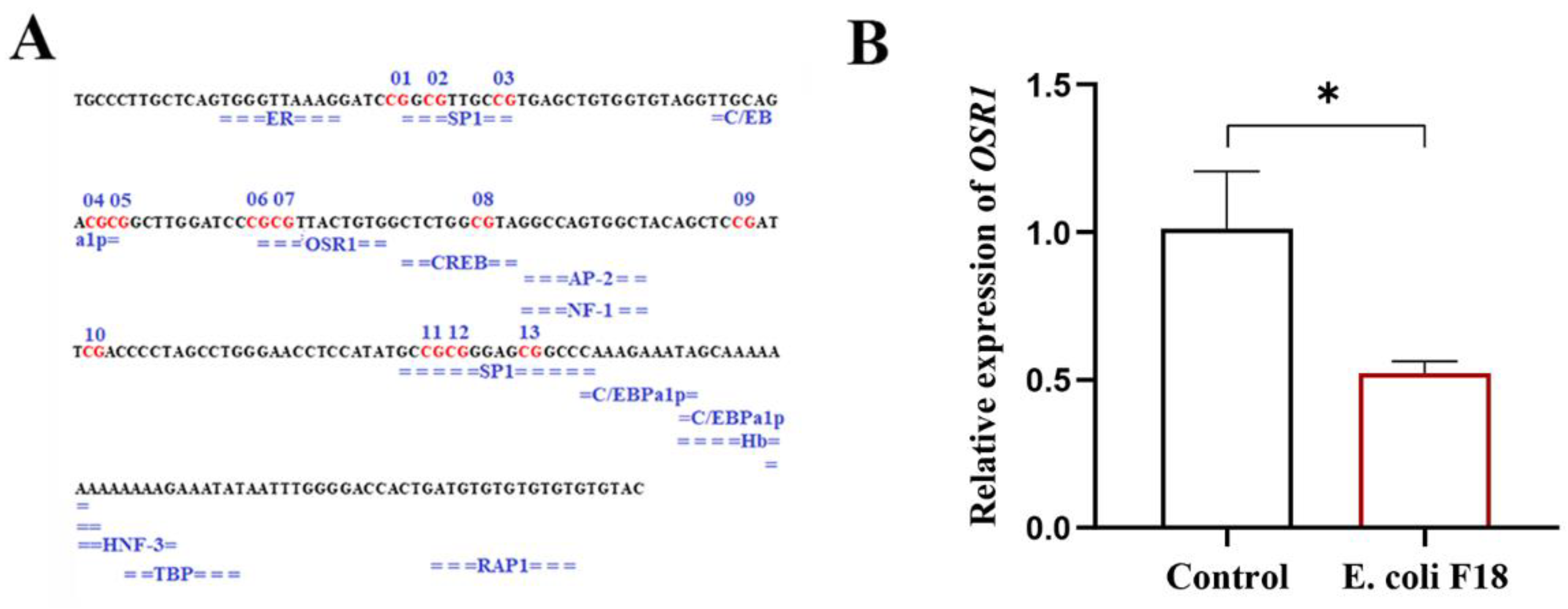
3.3. Detection of Methylation Level of CpG Island in Promoter Region of the CXCL11 Gene and Screening of Key CpG Sites
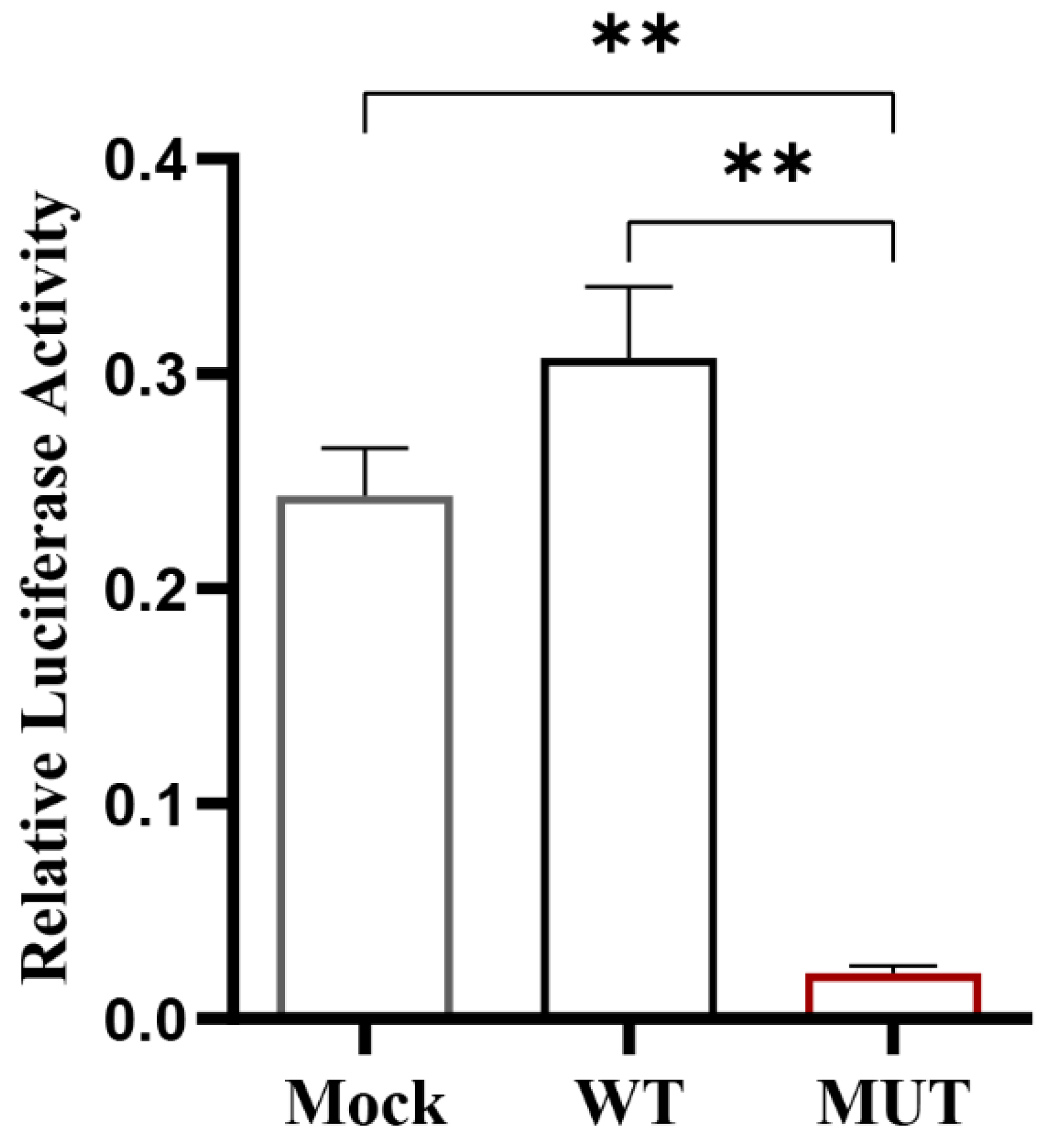
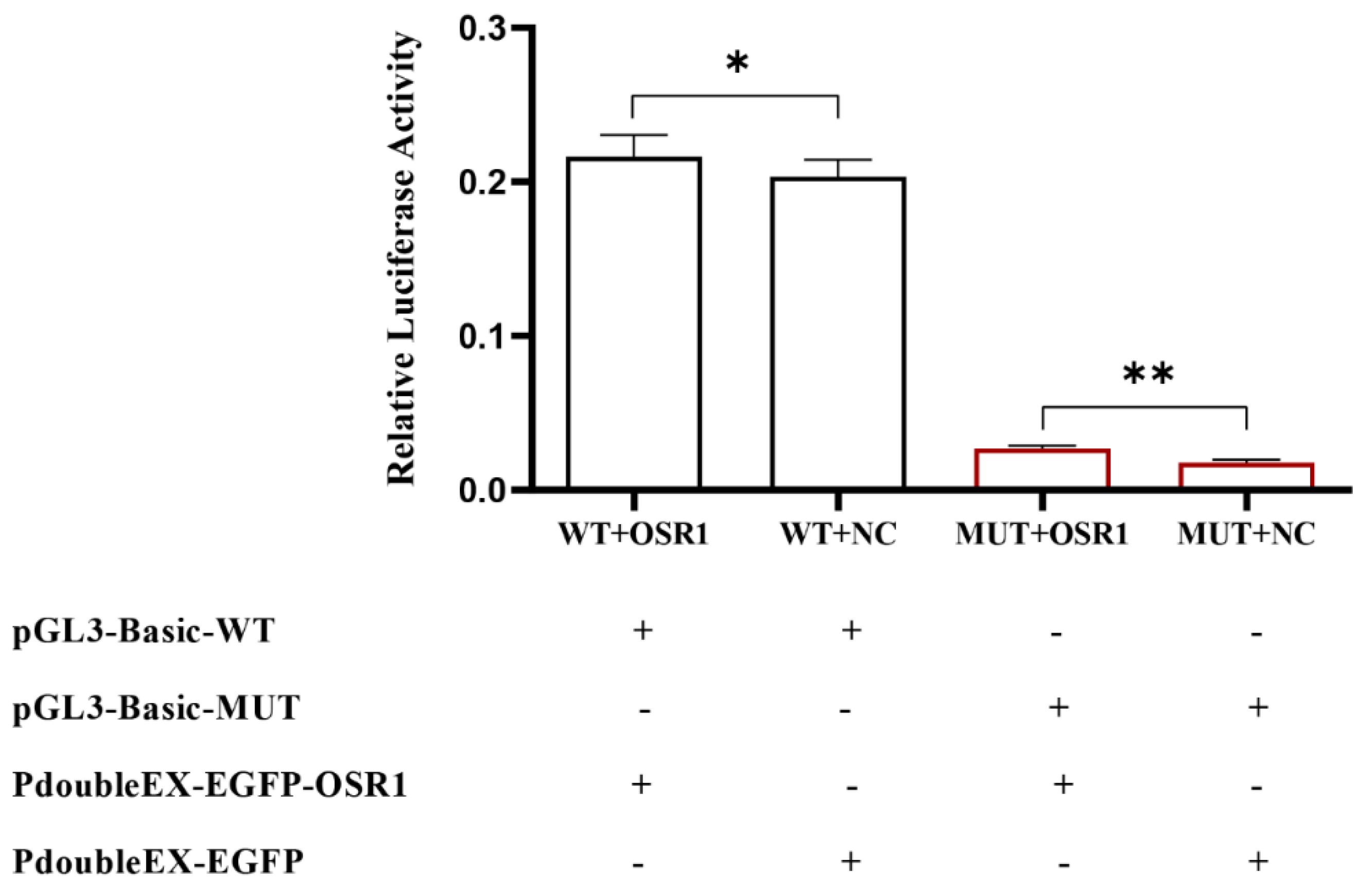
3.4. Effect of Key CpG Site Mutation on CXCL11 Transcription Activity
3.5. Evaluation of Key TFs in CpG Island of Pig CXCL11 Promoter Region
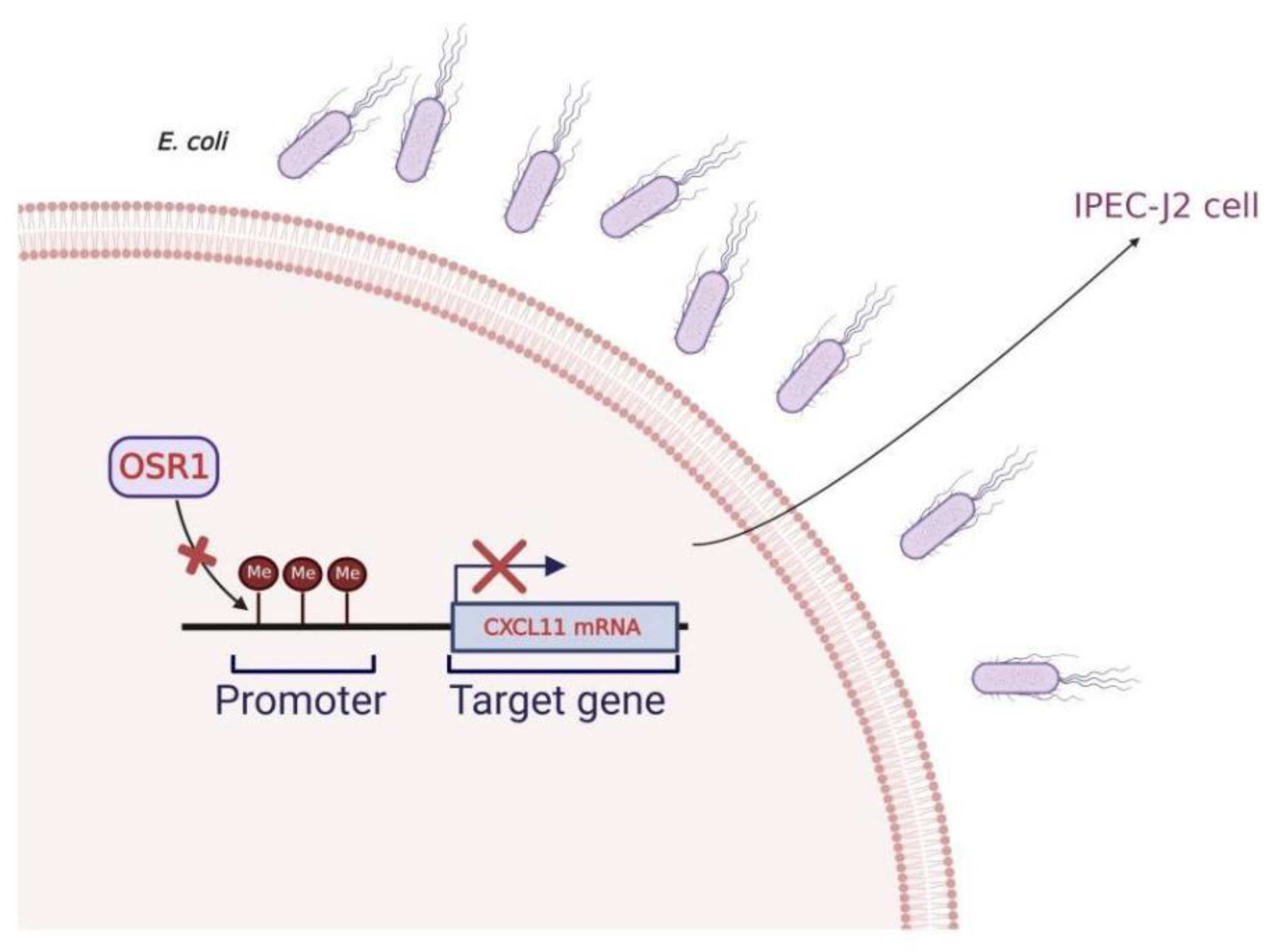
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lu, T.; Seo, H.; Moxley, R.A.; Zhang, W. Mapping the neutralizing epitopes of F18 fimbrial adhesin subunit FedF of enterotoxigenic Escherichia coli (ETEC). Vet. Microbiol. 2019, 230, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Frydendahl, K.; Kåre Jensen, T.; Strodl Andersen, J.; Fredholm, M.; Evans, G. Association between the porcine Escherichia coli F18 receptor genotype and phenotype and susceptibility to colonisation and postweaning diarrhoea caused by E. coli O138:F18. Vet. Microbiol. 2003, 93, 39–51. [Google Scholar] [PubMed]
- Brzozowski, B.; Mazur-Bialy, A.; Pajdo, R.; Kwiecien, S.; Bilski, J.; Zwolinska-Wcislo, M.; Mach, T.; Brzozowski, T. Mechanisms by which Stress Affects the Experimental and Clinical Inflammatory Bowel Disease (IBD): Role of Brain-Gut Axis. Curr. Neuropharmacol. 2016, 14, 892–900. [Google Scholar] [CrossRef] [PubMed]
- Baumgart, D.C.; Dignass, A.U. Intestinal barrier function. Curr. Opin. Clin. Nutr. Metab. Care 2002, 5, 685–694. [Google Scholar] [CrossRef] [PubMed]
- Alonso, C.; Vicario, M.; Pigrau, M.; Lobo, B.; Santos, J. Intestinal Barrier Function and the Brain-Gut Axis. Adv. Exp. Med. Biol. 2014, 817, 73–113. [Google Scholar]
- Daudelin, J.F.; Lessard, M.; Beaudoin, F.; Nadeau, E.; Bissonnette, N.; Boutin, Y.; Brousseau, J.P.; Lauzon, K.; Fairbrother, J.M. Administration of probiotics influences F4 (K88)-positive enterotoxigenic Escherichia coli attachment and intestinal cytokine expression in weaned pigs. Vet. Res. 2011, 42, 69. [Google Scholar] [CrossRef]
- Ren, W.; Yin, J.; Xiao, H.; Chen, S.; Liu, G.; Tan, B.; Li, N.; Peng, Y.; Li, T.; Zeng, B.; et al. Intestinal Microbiota-Derived GABA Mediates Interleukin-17 Expression during Enterotoxigenic Escherichia coli Infection. Front. Immunol. 2017, 7, 685. [Google Scholar]
- Wang, X.; Hardwidge Philip, R.; McCormick, B.A. Enterotoxigenic Escherichia coli Prevents Host NF-κB Activation by Targeting IκBα Polyubiquitination. Infect. Immun. 2012, 80, 4417–4425. [Google Scholar] [CrossRef]
- Ren, W.; Yin, J.; Duan, J.; Liu, G.; Zhu, X.; Chen, S.; Li, T.; Wang, S.; Tang, Y.; Hardwidge, P.R. Mouse intestinal innate immune responses altered by enterotoxigenic Escherichia coli (ETEC) infection. Microbes Infect. 2014, 16, 954–961. [Google Scholar] [CrossRef]
- Wang, G.; Geisbrecht, B.V.; Rueter, C.; Hardwidge, P.R. Enterotoxigenic Escherichia coli Flagellin Inhibits TNF-Induced NF-κB Activation in Intestinal Epithelial Cells. Pathogens 2017, 6, 18. [Google Scholar] [CrossRef]
- Wu, Z.; Liu, Y.; Dong, W.; Zhu, G.-q.; Wu, S.; Bao, W. CD14 in the TLRs signaling pathway is associated with the resistance to E. coli F18 in Chinese domestic weaned piglets. Sci. Rep. 2016, 6, 24611. [Google Scholar] [CrossRef] [PubMed]
- Rani, M.R.S.; Foster, G.R.; Leung, S.; Leaman, D.; Stark, G.R.; Ransohoff, R.M. Characterization of β-R1, a Gene That Is Selectively Induced by Interferon β (IFN-β) Compared with IFN-α*. J. Biol. Chem. 1996, 271, 22878–22884. [Google Scholar] [CrossRef] [PubMed]
- Tokunaga, R.; Zhang, W.; Naseem, M.; Puccini, A.; Berger, M.D.; Soni, S.; McSkane, M.; Baba, H.; Lenz, H.-J. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation—A target for novel cancer therapy. Cancer Treat. Rev. 2018, 63, 40–47. [Google Scholar] [PubMed]
- Chheda, Z.S.; Sharma, R.K.; Jala, V.R.; Luster, A.D.; Haribabu, B. Chemoattractant Receptors BLT1 and CXCR3 Regulate Antitumor Immunity by Facilitating CD8+T Cell Migration into Tumors. J. Immunol. 2016, 197, 2016. [Google Scholar]
- Sinigaglia, F.; D’Ambrosio, D.; Panina-Bordignon, P.; Rogge, L. Regulation of the IL-12/IL-12R axis: A critical step in T-helper cell differentiation and effector function. Immunol. Rev. 1999, 170, 65–72. [Google Scholar] [CrossRef]
- Shinkai, H.; Tanaka, M.; Morozumi, T.; Eguchi-Ogawa, T.; Okumura, N.; Muneta, Y.; Awata, T.; Uenishi, H. Biased distribution of single nucleotide polymorphisms (SNPs) in porcine Toll-like receptor 1 (TLR1), TLR2, TLR4, TLR5, and TLR6 genes. Immunogenetics 2006, 58, 324–330. [Google Scholar] [CrossRef]
- Xanthou, G.; Duchesnes, C.E.; Williams, T.J.; Pease, J.E. CCR3 functional responses are regulated by both CXCR3 and its ligands CXCL9, CXCL10 and CXCL11. Eur. J. Immunol. 2003, 33, 2241–2250. [Google Scholar] [CrossRef]
- Wang, H.; Feng, H.; Sun, J.; Zhou, Y.; Zhu, G.; Wu, S.; Bao, W. Age-associated changes in DNA methylation and expression of the TNFα gene in pigs. Genes Genet. Syst. 2018, 93, 191–198. [Google Scholar] [CrossRef]
- Saito, Y.; Nakaoka, T.; Sakai, K.; Muramatsu, T.; Toshimitsu, K.; Kimura, M.; Kanai, T.; Sato, T.; Saito, H. Inhibition of DNA Methylation Suppresses Intestinal Tumor Organoids by Inducing an Anti-Viral Response. Sci. Rep. 2016, 6, 25311. [Google Scholar]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar]
- Wang, X.; Kadarmideen, H.N. An Epigenome-Wide DNA Methylation Map of Testis in Pigs for Study of Complex Traits. Front. Genet. 2019, 10, 405. [Google Scholar] [CrossRef]
- Zhang, X.; Sun, W.; He, L.; Wang, L.; Qiu, K.; Yin, J. Global DNA methylation pattern involved in the modulation of differentiation potential of adipogenic and myogenic precursors in skeletal muscle of pigs. Stem Cell Res. Ther. 2020, 11, 536. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.H.; Gan, L.N.; Bao, W.B.; Wu, S.L.; Qin, W.U.; Zi, C.; Zhu, G.Q. Use of Fluorescence Quantitative Polymerase Chain Reaction (PCR) for the Detection of Escherichia coli Adhesion to Pig Intestinal Epithelial Cells. Pol. J. Vet. Sci. 2016, 19, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wang, L.; Zhou, Y.; Miao, Z. Prevalence and characterization of virulence genes in Escherichia coli isolated from piglets suffering post-weaning diarrhoea in Shandong Province, China. Vet. Med. Sci. 2020, 6, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Luise, D.; Lauridsen, C.; Bosi, P.; Trevisi, P. Methodology and application of Escherichia coli F4 and F18 encoding infection models in post-weaning pigs. J. Anim. Sci. Biotechnol. 2019, 10, 53. [Google Scholar] [CrossRef]
- Stirm, S.; Orskov, F. K88, an Episome-determined Protein Antigen of Escherichia coli. Nature 1966, 209, 507–508. [Google Scholar] [CrossRef]
- Cui, D.; Xu, X. DNA Methyltransferases, DNA Methylation, and Age-Associated Cognitive Function. Int. J. Mol. Sci. 2018, 19, 1315. [Google Scholar] [CrossRef]
- Miller, C.A.; Sweatt, J.D. Covalent Modification of DNA Regulates Memory Formation. Neuron 2007, 53, 857–869. [Google Scholar] [CrossRef]
- Tirado-Magallanes, R.; Rebbani, K.; Lim, R.; Pradhan, S.; Benoukraf, T. Whole genome DNA methylation: Beyond genes silencing. Oncotarget 2016, 8, 5629–5637. [Google Scholar] [CrossRef]
- Jones, P.A. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012, 13, 484–492. [Google Scholar] [PubMed]
- Dai, C.; Yang, L.; Jin, J.; Wang, H.; Wu, S.; Bao, W. Regulation and Molecular Mechanism of TLR5 on Resistance to Escherichia coli F18 in Weaned Piglets. Animals 2019, 9, 735. [Google Scholar] [CrossRef]
- Sun, L.; Wang, J.; Yin, X.; Sun, S.; Zi, C.; Zhu, G.; Wu, S.; Bao, W. Identification of a 5-Methylcytosine Site that may Regulate C/EBPβ Binding and Determine Tissue-Specific Expression of the BPI Gene in Piglets. Sci. Rep. 2016, 6, 28506. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yin, X.; Sun, L.; Sun, S.; Zi, C.; Zhu, G.; Wu, S.; Bao, W. Correlation between BPI Gene Upstream CpG Island Methylation and mRNA Expression in Piglets. Int. J. Mol. Sci. 2014, 15, 10989–10998. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Sun, L.; Xia, R.; Sun, S.; Zhu, G.; Wu, S.; Bao, W. Correlation between the methylation of the FUT1 promoter region and FUT1 expression in the duodenum of piglets from newborn to weaning. 3 Biotech 2017, 7, 247. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Feng, H.; Cao, Y.; Huang, Y.; Dai, C.; Wu, S.; Bao, W. New Insight into the Molecular Mechanism of the FUT2 Regulating Escherichia coli F18 Resistance in Weaned Piglets. Int. J. Mol. Sci. 2018, 19, 3301. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, C.; Tang, H.; Yu, Y.; Zhang, Q. Combined Analysis of DNA Methylome and Transcriptome Reveal Novel Candidate Genes Related to Porcine Escherichia coli F4ab/ac-Induced Diarrhea. Front. Cell. Infect. Microbiol. 2020, 10, 250. [Google Scholar] [CrossRef]
- Otani, K.; Dong, Y.; Li, X.; Lu, J.; Zhang, N.; Xu, L.; Go, M.Y.Y.; Ng, E.K.W.; Arakawa, T.; Chan, F.K.L.; et al. Odd-skipped related 1 is a novel tumour suppressor gene and a potential prognostic biomarker in gastric cancer. J. Pathol. 2014, 234, 302–315. [Google Scholar] [CrossRef]
- James, R.G.; Kamei, C.N.; Wang, Q.; Jiang, R.; Schultheiss, T.M. Odd-skipped related 1 is required for development of the metanephric kidney and regulates formation and differentiation of kidney precursor cells. Development 2006, 133, 2995–3004. [Google Scholar] [CrossRef]
- Wang, Q.; Lan, Y.; Cho, E.S.; Maltby, K.M.; Jiang, R. Odd-skipped related 1 (Odd1) is an essential regulator of heart and urogenital development. Dev. Biol. 2005, 288, 582–594. [Google Scholar] [CrossRef]
- Guo, D.; Wu, B.; Yan, J.; Li, X.; Sun, H.; Zhou, D. A possible gene silencing mechanism: Hypermethylation of the Keap1 promoter abrogates binding of the transcription factor Sp1 in lung cancer cells. Biochem. Biophys. Res. Commun. 2012, 428, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Reamon-Buettner, S.M.; Borlak, J. Epigenetic Silencing of Cell Adhesion Molecule 1 in Different Cancer Progenitor Cells of Transgenic c-Myc and c-Raf Mouse Lung Tumors. Cancer Res. 2008, 68, 7587–7596. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | GenBank Accession No. | Primer Sequence | Fragment Size (bp) |
|---|---|---|---|
| CXCL11 | XM_005666774.3 | F:5′- TCAAAGCGGGAAGGTGTCTT -3′ | 301 |
| R:5′- TGCTTTCAGGGTGACAATCACTT -3′ | |||
| OSR1 | XM_021087818.1 | F:5′- GGTGGAGAGGGTGTTTCAGG-3′ | 166 |
| R:5′- CTCCACCATCCAGCTCCCAG-3′ | |||
| GAPDH | AF017079.1 | F: 5′-ACATCATCCCTGCTTCTACTGG-3′ | 188 |
| R: 5′-CTCGGACGCCTGCTTCAC-3′ | |||
| PILIN | M25302.1 | F: 5′-AGGCCGAACCAAAGAAGCAT-3′ | 117 |
| R: 5′-TCACCATCAGGGTTTCTGAGT-3′ | |||
| β-actin | NC_010445.3 | F: 5′-GTCGTACTCCTGCTTGCTGAT-3′ | 119 |
| R: 5′-CCTTCTCCTTCCAGATCATCGC-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, X.; Yu, L.; Huang, R.; Bao, W.; Wu, S.; Wu, Z. Identification of a 5-Methylcytosine Site (mC-7) That May Inhibit CXCL11 Expression and Regulate E. coli F18 Susceptibility in IPEC-J2 Cells. Vet. Sci. 2022, 9, 600. https://doi.org/10.3390/vetsci9110600
Shi X, Yu L, Huang R, Bao W, Wu S, Wu Z. Identification of a 5-Methylcytosine Site (mC-7) That May Inhibit CXCL11 Expression and Regulate E. coli F18 Susceptibility in IPEC-J2 Cells. Veterinary Sciences. 2022; 9(11):600. https://doi.org/10.3390/vetsci9110600
Chicago/Turabian StyleShi, Xiaoru, Luchen Yu, Rufeng Huang, Wenbin Bao, Shenglong Wu, and Zhengchang Wu. 2022. "Identification of a 5-Methylcytosine Site (mC-7) That May Inhibit CXCL11 Expression and Regulate E. coli F18 Susceptibility in IPEC-J2 Cells" Veterinary Sciences 9, no. 11: 600. https://doi.org/10.3390/vetsci9110600
APA StyleShi, X., Yu, L., Huang, R., Bao, W., Wu, S., & Wu, Z. (2022). Identification of a 5-Methylcytosine Site (mC-7) That May Inhibit CXCL11 Expression and Regulate E. coli F18 Susceptibility in IPEC-J2 Cells. Veterinary Sciences, 9(11), 600. https://doi.org/10.3390/vetsci9110600







