Prediction of the Effect of Methylation in the Promoter Region of ZP2 Gene on Egg Production in Jinghai Yellow Chickens
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection
2.3. Primer Synthesis and Quantitative Real-Time PCR (qRT-PCR)
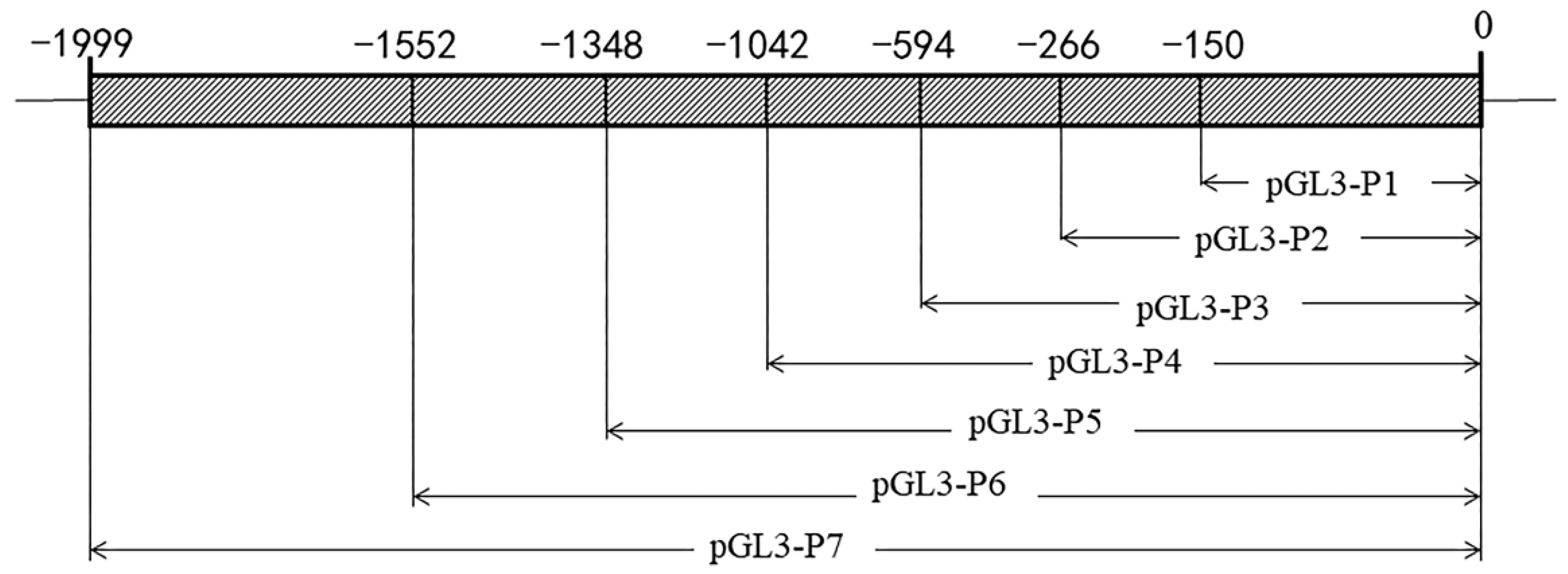
2.4. ZP2 Gene Core Promoter Region Prediction and Deletion Vector Construction
2.5. Cell Transfection and Determination of Luciferase Activity
2.6. DNA Extraction from Ovarian Tissue and Sulfite Treatment
2.7. CpG Island Analysis of Promoter Region of ZP2 Gene and BSP Primer Design
2.8. PCR Amplification and Product Recovery
2.9. BSP Sequencing
2.10. Statistical Analysis
3. Results
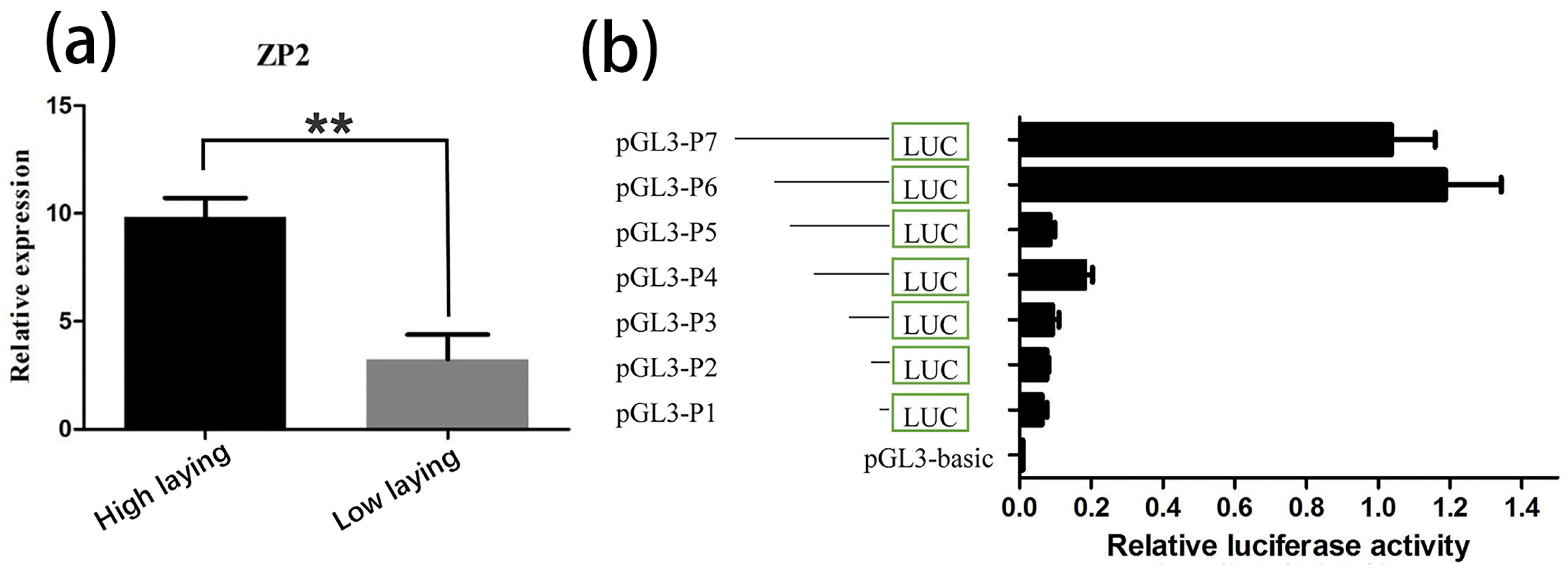
3.1. Differential Expression Analysis of ZP2 Gene in Ovarian Tissues
3.2. Prediction of the Core Promoter Region of ZP2 Gene and Assay of ZP2 Activity in DF-1 Cells Transfected with Deletion Vector
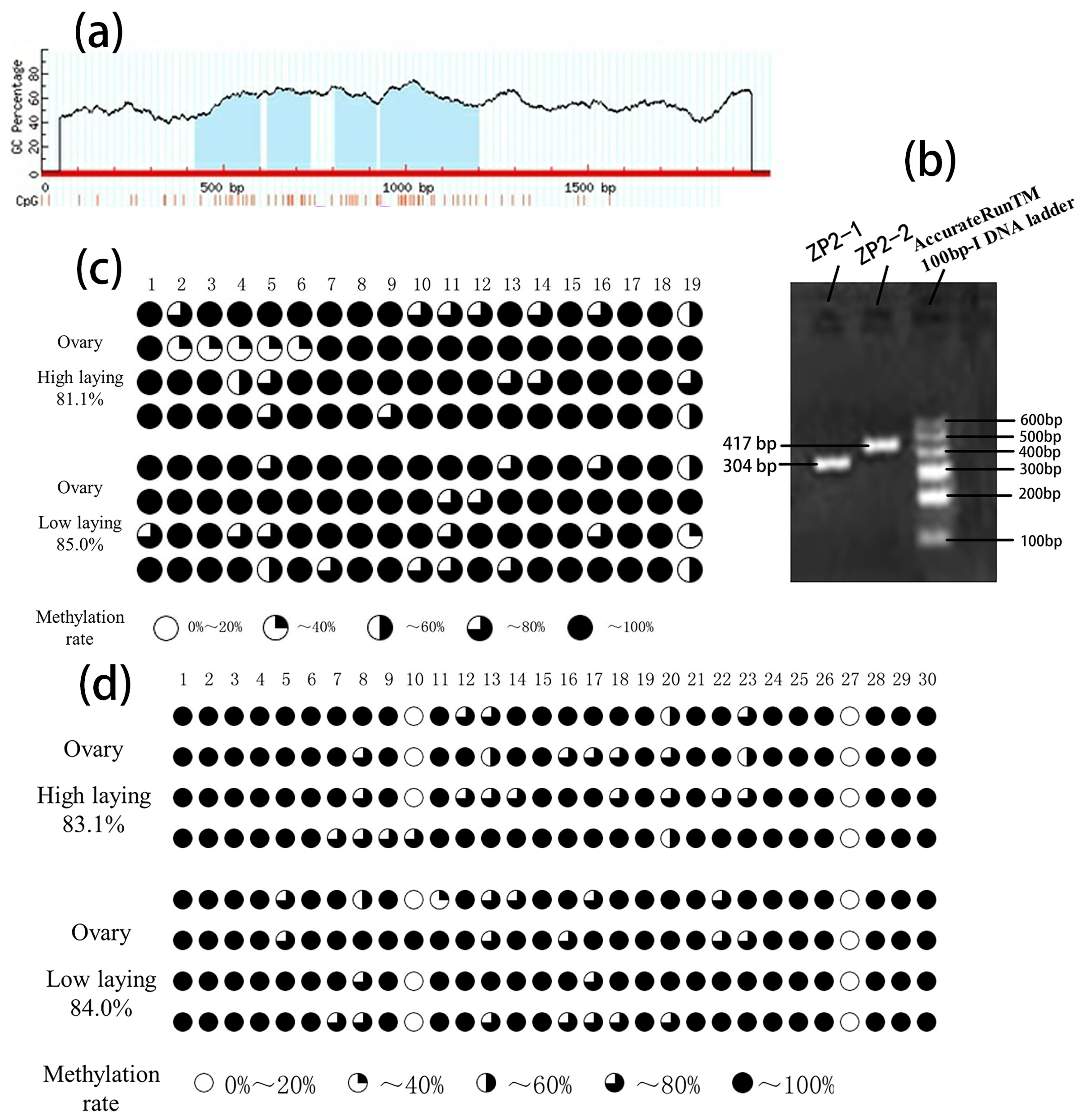
3.3. ZP2 Gene CpG Island Prediction and PCR Amplification
3.4. BSP Sequencing Results of CpG Island Methylation of the ZP2 Gene
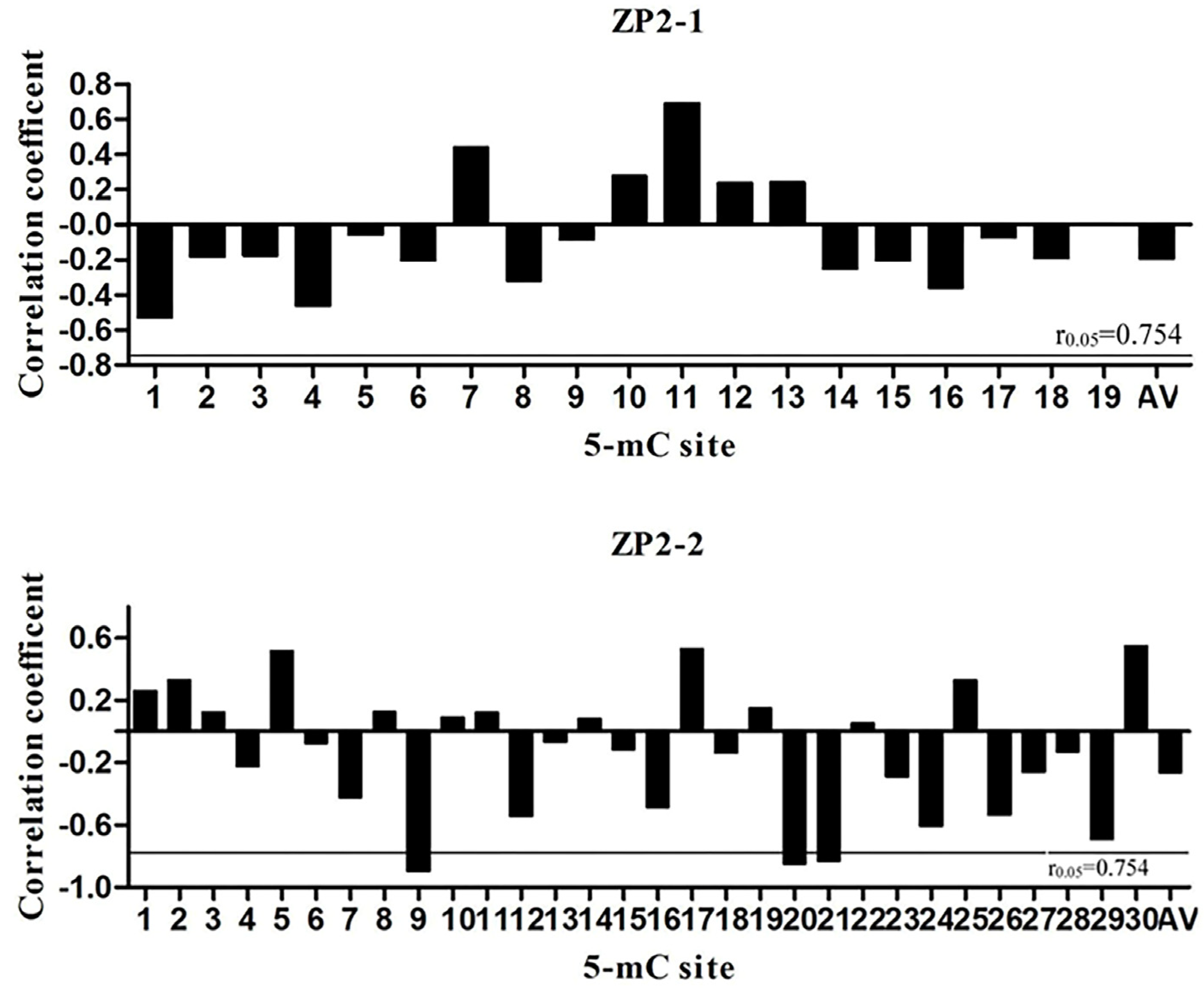
3.5. Pearson Correlation Analysis between Methylation Level and mRNA Expression
3.6. Prediction of the Potential Transcription Factor Binding Sites of the ZP2 Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Goto, T.; Tsudzuki, M. Genetic Mapping of Quantitative Trait Loci for Egg Production and Egg Quality Traits in Chickens: A Review. J. Poult. Sci. 2017, 54, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Jaworska-Adamu, J.; Krawczyk, A.; Rycerz, K. Zona pellucida in the process of fertilization and mammal embryonal development. Med. Weter 2022, 78, 121–125. [Google Scholar] [CrossRef]
- Chuang-Ju, L.; Qi-Wei, W.; Xi-Hua, C.; Li, Z.; Hong, C.; Fang, G.; Jian-Fang, G. Molecular characterization and expression pattern of three zona pellucida 3 genes in the Chinese sturgeon, Acipenser sinensis. Fish Physiol. Biochem. 2011, 37, 471–484. [Google Scholar] [CrossRef]
- Gupta, S.K. Zona pellucida glycoproteins: Relevance in fertility and development of contraceptive vaccines. Am. J. Reprod. Immunol. 2022. [Google Scholar] [CrossRef]
- Gupta, S.K. Human Zona Pellucida Glycoproteins: Binding Characteristics With Human Spermatozoa and Induction of Acrosome Reaction. Front. Cell Dev. Biol. 2021, 9, 619868. [Google Scholar] [CrossRef]
- Gupta, S.K.; Chakravarty, S.; Suraj, K.; Bansal, P.; Ganguly, A.; Jain, M.K.; Bhandari, B. Structural and functional attributes of zona pellucida glycoproteins. Soc. Reprod. Fertil. Suppl. 2007, 63, 203–216. [Google Scholar] [PubMed]
- Goudet, G.; Mugnier, S.; Callebaut, I.; Monget, P. Phylogenetic analysis and identification of pseudogenes reveal a progressive loss of zona pellucida genes during evolution of vertebrates. Biol. Reprod. 2008, 78, 796–806. [Google Scholar] [CrossRef]
- Kiefer, S.M.; Saling, P. Proteolytic processing of human zona pellucida proteins. Biol. Reprod. 2002, 66, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Bleil, J.D.; Wassarman, P.M. Mammalian sperm-egg interaction: Identification of a glycoprotein in mouse egg zonae pellucidae possessing receptor activity for sperm. Cell 1980, 20, 873–882. [Google Scholar] [CrossRef]
- Waclawek, M.; Foisner, R.; Nimpf, J.; Schneider, W.J. The chicken homologue of zona pellucida protein-3 is synthesized by granulosa cells. Biol. Reprod. 1998, 59, 1230–1239. [Google Scholar] [CrossRef][Green Version]
- Takeuchi, Y.; Nishimura, K.; Aoki, N.; Adachi, T.; Sato, C.; Kitajima, K.; Matsuda, T. A 42-kDa glycoprotein from chicken egg-envelope, an avian homolog of the ZPC family glycoproteins in mammalian Zona pellucida. Its first identification, cDNA cloning and granulosa cell-specific expression. Eur. J. Biochem. 1999, 260, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Bausek, N.; Waclawek, M.; Schneider, W.J.; Wohlrab, F. The major chicken egg envelope protein ZP1 is different from ZPB and is synthesized in the liver. J. Biol. Chem. 2000, 275, 28866–28872. [Google Scholar] [CrossRef] [PubMed]
- Okumura, H.; Kohno, Y.; Iwata, Y.; Mori, H.; Aoki, N.; Sato, C.; Kitajima, K.; Nadano, D.; Matsuda, T. A newly identified zona pellucida glycoprotein, ZPD, and dimeric ZP1 of chicken egg envelope are involved in sperm activation on sperm-egg interaction. Biochem. J. 2004, 384, 191–199. [Google Scholar] [CrossRef]
- Kinoshita, M.; Rodler, D.; Sugiura, K.; Matsushima, K.; Kansaku, N.; Tahara, K.; Tsukada, A.; Ono, H.; Yoshimura, T.; Yoshizaki, N.; et al. Zona pellucida protein ZP2 is expressed in the oocyte of Japanese quail (Coturnix japonica). Reproduction 2010, 139, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Serizawa, M.; Kinoshita, M.; Rodler, D.; Tsukada, A.; Ono, H.; Yoshimura, T.; Kansaku, N.; Sasanami, T. Oocytic expression of zona pellucida protein ZP4 in Japanese quail (Coturnix japonica). Anim. Sci. J. 2011, 82, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Rodler, D.; Sasanami, T.; Sinowatz, F. Assembly of the inner perivitelline layer, a homolog of the mammalian zona pellucida: An immunohistochemical and ultrastructural study. Cells Tissues Organs 2012, 195, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Nishio, S.; Kohno, Y.; Iwata, Y.; Arai, M.; Okumura, H.; Oshima, K.; Nadano, D.; Matsuda, T. Glycosylated chicken ZP2 accumulates in the egg coat of immature oocytes and remains localized to the germinal disc region of mature eggs. Biol. Reprod. 2014, 91, 107. [Google Scholar] [CrossRef] [PubMed]
- Berlin, S.; Qu, L.; Ellegren, H. Adaptive Evolution of Gamete-Recognition Proteins in Birds. JMolE 2008, 67, 488–496. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wang, Y.; Liu, Z.; Guo, X.; Sun, Y.; Kang, L.; Jiang, Y. Transcriptomic and proteomic analyses of ovarian follicles reveal the role of VLDLR in chicken follicle selection. BMC Genom. 2020, 21, 486. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Su, B.; Li, W.H.; Zhao, Z. CpG island density and its correlations with genomic features in mammalian genomes. Genome Biol. 2008, 9, R79. [Google Scholar] [CrossRef] [PubMed]
- Sarda, S.; Hannenhalli, S. Orphan CpG islands as alternative promoters. Transcription 2018, 9, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Li, N.; Hu, X.; Li, J.; Du, Z.; Chen, L.; Yin, G.; Duan, J.; Zhang, H.; Zhao, Y.; et al. Genome-wide mapping of DNA methylation in chicken. PLoS ONE 2011, 6, e19428. [Google Scholar]
- Burt, D.; Pourquie, O. Genetics. Chicken genome--science nuggets to come soon. Science 2003, 300, 1669. [Google Scholar] [CrossRef]
- Smith, J.; Sen, S.; Weeks, R.J.; Eccles, M.R.; Chatterjee, A. Promoter DNA Hypermethylation and Paradoxical Gene Activation. Trends Cancer 2020, 6, 392–406. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.Y.; Hsu, S.D.; Huang, H.Y.; Sun, Y.M.; Chou, C.H.; Weng, S.L.; Huang, H.D. MethHC: A database of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 2015, 43, D856–D861. [Google Scholar] [CrossRef]
- Sin, H.S.; Barski, A.; Zhang, F.; Kartashov, A.V.; Nussenzweig, A.; Chen, J.; Andreassen, P.R.; Namekawa, S.H. RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids. Genes Dev. 2012, 26, 2737–2748. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.X. Studies on Genetic Diversity and Genetic Effect of the Myostatin Gene on Growth and Reproductive Traits of Bian Chicken. Ph.D. Thesis, Yangzhou University, Yangzhou, China, 2010. [Google Scholar]
- Reese, M.G. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 2001, 26, 51–56. [Google Scholar] [CrossRef]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The Ultimate qPCR Experiment: Producing Publication Quality, Reproducible Data the First Time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef]
- Kanamori, A.; Naruse, K.; Mitani, H.; Shima, A.; Hori, H. Genomic organization of ZP domain containing egg envelope genes in medaka (Oryzias latipes). Gene 2003, 305, 35–45. [Google Scholar] [CrossRef]
- Xie, Y.; Nie, C.C.; Cao, Z.W. Expression of mouse ZP2 mRNA in the granulosa cells of follicles. J. Jinan Univ. (Nat. Sci. Med. Ed.) 2010, 31, 116–119. [Google Scholar]
- Hudson, N.O.; Buck-Koehntop, B.A. Zinc Finger Readers of Methylated DNA. Molecules 2018, 23, 2555. [Google Scholar] [CrossRef] [PubMed]
- Kubo, N.; Ishii, H.; Xiong, X.; Bianco, S.; Meitinger, F.; Hu, R.; Hocker, J.D.; Conte, M.; Gorkin, D.; Yu, M.; et al. Promoter-proximal CTCF binding promotes distal enhancer-dependent gene activation. Nat. Struct. Mol. Biol. 2021, 28, 152–161. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.G.; Srinivasan, K.; Dai, Z.; Duan, W.; Druhan, L.J.; Ding, H.; Yee, L.; Villalona-Calero, M.A.; Plass, C.; Otterson, G.A. Methylation of adjacent CpG sites affects Sp1/Sp3 binding and activity in the p21(Cip1) promoter. Mol. Cell Biol. 2003, 23, 4056–4065. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, M.V.C.; Bourc’his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef]
- Mikeska, T.; Bock, C.; El-Maarri, O.; Hübner, A.; Ehrentraut, D.; Schramm, J.; Felsberg, J.; Kahl, P.; Büttner, R.; Pietsch, T.; et al. Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J. Mol. Diagn. 2007, 9, 368–381. [Google Scholar] [CrossRef]
- Xie, X.L.; Yu, Y.; Yuan, Z.F.; Yang, J.; Ma, P.P.; Li, D.C.; Yu, S.K.; An, F.; Feng, X.J.; Zhang, Y. Comparative analysis on content and distribution of CpG sites in milk production traits and mastitis-related genes in dairy cattle. Yi Chuan 2012, 34, 437–444. [Google Scholar] [CrossRef]
- Mahboob Morshed, J.Y.; Sano, S.; Nishijima, K.; Iijima, M.K.a.S. Biochemical analysis of chicken ovalbumin promoter. Anim. Cell Technol. Basic Appl. Asp. 2006, 14, 301–307. [Google Scholar]
- Gilmour, J.; O’Connor, L.; Middleton, C.P.; Keane, P.; Gillemans, N.; Cazier, J.B.; Philipsen, S.; Bonifer, C. Robust hematopoietic specification requires the ubiquitous Sp1 and Sp3 transcription factors. Epigenetics Chromatin 2019, 12, 33. [Google Scholar] [CrossRef]
- Peng, F.; Zhou, Y.; Wang, J.; Guo, B.; Wei, Y.; Deng, H.; Wu, Z.; Zhang, C.; Shi, K.; Li, Y.; et al. The transcription factor Sp1 modulates RNA polymerase III gene transcription by controlling BRF1 and GTF3C2 expression in human cells. J. Biol. Chem. 2020, 295, 4617–4630. [Google Scholar] [CrossRef]
- Xu, X.W.; Pan, C.W.; Yang, X.M.; Zhou, L.; Zheng, Z.Q.; Li, D.C. SP1 reduces autophagic flux through activating p62 in gastric cancer cells. Mol. Med. Rep. 2018, 17, 4633–4638. [Google Scholar] [CrossRef]
- Isomura, H.; Stinski, M.F.; Kudoh, A.; Daikoku, T.; Shirata, N.; Tsurumi, T. Two Sp1/Sp3 binding sites in the major immediate-early proximal enhancer of human cytomegalovirus have a significant role in viral replication. J. Virol. 2005, 79, 9597–9607. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, Y.; Struhl, K. Different SP1 binding dynamics at individual genomic loci in human cells. Proc. Natl. Acad. Sci. USA 2021, 118, e2113579118. [Google Scholar] [CrossRef] [PubMed]
- Thiel, G.; Cibelli, G. Regulation of life and death by the zinc finger transcription factor Egr-1. J. Cell Physiol. 2002, 193, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Pagel, J.I.; Deindl, E. Early growth response 1--a transcription factor in the crossfire of signal transduction cascades. Indian J. Biochem. Biophys. 2011, 48, 226–235. [Google Scholar]
- Ling, Y.; Shi, X.; Wang, Y.; Ling, X.; Li, Q. Down-regulation of thyroid hormone receptor β1 gene expression in gastric cancer involves promoter methylation. Biochem. Biophys. Res. Commun. 2014, 444, 147–152. [Google Scholar] [CrossRef]
- Ling, F.; Li, J.; Chen, Y.; Du, H.; Mei, Y.; Mo, D.; Wang, C. Cloning and characterization of the 5′-flanking region of the pig adiponectin gene. Biochem. Biophys. Res. Commun. 2009, 381, 236–240. [Google Scholar] [CrossRef]
- Head, J.A. Patterns of DNA methylation in animals: An ecotoxicological perspective. Integr. Comp. Biol. 2014, 54, 77–86. [Google Scholar] [CrossRef]

| Gene | Accession Number | Primer Sequence (5′-3′) | Product Size (bp) |
|---|---|---|---|
| ZP2 | NM_001039098.1 | F: TGATGTCTTGTGAGCACGCTGTAG R: GTGCCATTCAGCAGCAGGAG | 121 |
| β-actin | X00182 | F: CAGCCATCTTTCTTGGGTAT R: CTGTGATCTCCTTCTGCATCC | 169 |
| Fragment Name | Primer Sequence (5′-3′) | Product Size (bp) |
|---|---|---|
| pGL3-P7 | CAGCTGTGGCTTCTCTGAGCG | 1999 |
| pGL3-P6 | CTTTACTCTGTTTCCCCCCTCC | 1552 |
| pGL3-P5 | GTGCCCCTGGGAGCGTACAGA | 1348 |
| pGL3-P4 | TTCAGCTGTCAGTAGCCTGCC | 1042 |
| pGL3-P3 | TCCCCTGTGAGGGAAGGCTTG | 594 |
| pGL3-P2 | GCCTACAGGAAAGCTGGGGAG | 266 |
| pGL3-P1 | GCAGTGAGACACTGGAACAGG | 150 |
| ZPDR | TTAAGACCACCTCGTTCCACCC |
| Gene | Primer Sequence (5′-3′) | Product Size (bp) |
|---|---|---|
| ZP2-1 | F: AAGTTATTTAGATTGAAAATATAAATTAGGGGAT R: CATTCATTTCCACCACAACAACAAC | 304 |
| ZP2-2 | F: GGAGTTATAGGTTTTAAGAGAGGGAGG R: CCACATTACTCCATCTATACCCAAAAAC | 417 |
| Core Region | Start (bp) | End (bp) | Primer Sequence (5′-3′) | Score (0–1) |
|---|---|---|---|---|
| 1 | −1516 | −1466 | GAATCACGGCATAAAACATCAGCCCCGGAGGGAGCGCTGCAGCGATGGGG | 0.96 |
| 2 | −249 | −198 | GGAGGGACTTTTTATAGGGGCAGGTAGTGACCAGATGGCTTTAAACTGGA | 0.95 |
| Fragment | Overall Methylation Level (%) | Number of Methylated Sites | |
|---|---|---|---|
| High Laying Group | Low Laying Group | ||
| ZP2-1 | 81.1 | 85.0 | 19 |
| ZP2-2 | 83.1 | 84.0 | 30 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Zhang, X.-Q.; Ling, X.-Z.; Zhao, X.-H.; Zhou, K.-Z.; Wang, J.-Y.; Zhang, G.-X. Prediction of the Effect of Methylation in the Promoter Region of ZP2 Gene on Egg Production in Jinghai Yellow Chickens. Vet. Sci. 2022, 9, 570. https://doi.org/10.3390/vetsci9100570
Zhang J, Zhang X-Q, Ling X-Z, Zhao X-H, Zhou K-Z, Wang J-Y, Zhang G-X. Prediction of the Effect of Methylation in the Promoter Region of ZP2 Gene on Egg Production in Jinghai Yellow Chickens. Veterinary Sciences. 2022; 9(10):570. https://doi.org/10.3390/vetsci9100570
Chicago/Turabian StyleZhang, Jin, Xiang-Qian Zhang, Xuan-Ze Ling, Xiu-Hua Zhao, Kai-Zhi Zhou, Jin-Yu Wang, and Gen-Xi Zhang. 2022. "Prediction of the Effect of Methylation in the Promoter Region of ZP2 Gene on Egg Production in Jinghai Yellow Chickens" Veterinary Sciences 9, no. 10: 570. https://doi.org/10.3390/vetsci9100570
APA StyleZhang, J., Zhang, X.-Q., Ling, X.-Z., Zhao, X.-H., Zhou, K.-Z., Wang, J.-Y., & Zhang, G.-X. (2022). Prediction of the Effect of Methylation in the Promoter Region of ZP2 Gene on Egg Production in Jinghai Yellow Chickens. Veterinary Sciences, 9(10), 570. https://doi.org/10.3390/vetsci9100570







