Molecular Detection and Characterization of Blastocystis sp. and Enterocytozoon bieneusi in Cattle in Northern Spain
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. DNA Extraction and Purification
2.3. Molecular Detection of Blastocystis sp.
2.4. Molecular Detection of Enterocytozoon bieneusi
2.5. Sanger Sequencing and Sequence Analysis
2.6. Blastocystis Subtype Identification Using Next Generation Amplicon Sequencing
2.7. Statistical Analysis
3. Results
3.1. Detection of Blastocystis sp. and E. bieneusi by PCR
3.2. Molecular Characterization of Blastocystis sp. and E. bieneusi by Sanger Sequencing
3.3. Blastocystis Subtype Identification by Next Generation Amplicon Sequencing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stensvold, C.R. Thinking Blastocystis out of the box. Trends Parasitol. 2012, 28, 305. [Google Scholar] [CrossRef] [PubMed]
- Ajjampur, S.S.; Tan, K.S. Pathogenic mechanisms in Blastocystis spp. Interpreting results from in vitro and in vivo studies. Parasitol. Int. 2016, 65, 772–779. [Google Scholar] [CrossRef]
- Scanlan, P.D.; Stensvold, C.R.; Rajilić-Stojanović, M.; Heilig, H.G.; De Vos, W.M.; O’Toole, P.W.; Cotter, P.D. The microbial eukaryote Blastocystis is a prevalent and diverse member of the healthy human gut microbiota. FEMS Microbiol. Ecol. 2014, 90, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Feng, Y.; Santin, M. Host specificity of Enterocytozoon bieneusi and public health implications. Trends Parasitol. 2019, 35, 436–451. [Google Scholar] [CrossRef]
- Matos, O.; Lobo, M.L.; Xiao, L. Epidemiology of Enterocytozoon bieneusi infection in humans. J. Parasitol. Res. 2012, 2012, 981424. [Google Scholar] [CrossRef] [PubMed]
- Weiss, L.M.; Becnel, J.J. Microsporidia: Pathogens of Opportunity: First Edition; Wiley Blackwell: Hoboken, NJ, USA, 2014. [Google Scholar]
- Li, W.; Feng, Y.; Xiao, L. Diagnosis and molecular typing of Enterocytozoon bieneusi: The significant role of domestic animals in transmission of human microsporidiosis. Res. Vet. Sci. 2020, 133, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.S. New insights on classification, identification, and clinical relevance of Blastocystis spp. Clin. Microbiol. Rev. 2008, 21, 639–665. [Google Scholar] [CrossRef] [PubMed]
- Koloren, Z.; Gulabi, B.B.; Karanis, P. Molecular identification of Blastocystis sp. subtypes in water samples collected from Black sea, Turkey. Acta Trop. 2018, 180, 58–68. [Google Scholar] [CrossRef]
- Li, W.; Xiao, L. Ecological and public health significance of Enterocytozoon bieneusi. One Health 2021, 12, 100209. [Google Scholar] [CrossRef]
- Stensvold, C.R.; Clark, C.G. Pre-empting Pandora’s box: Blastocystis subtypes revisited. Trends Parasitol. 2020, 36, 229–232. [Google Scholar] [CrossRef]
- Maloney, J.G.; Santin, M. Mind the gap: New full-length sequences of Blastocystis subtypes generated via Oxford Nanopore Minion sequencing allow for comparisons between full-length and partial sequences of the small subunit of the ribosomal RNA gene. Microorganisms 2021, 9, 997. [Google Scholar] [CrossRef] [PubMed]
- Maloney, J.G.; Jang, Y.; Molokin, A.; George, N.S.; Santin, M. Wide genetic diversity of Blastocystis in white-tailed deer (Odocoileus virginianus) from Maryland, USA. Microorganisms 2021, 9, 1343. [Google Scholar] [CrossRef]
- Alfellani, M.A.; Stensvold, C.R.; Vidal-Lapiedra, A.; Onuoha, E.S.; Fagbenro-Beyioku, A.F.; Clark, C.G. Variable geographic distribution of Blastocystis subtypes and its potential implications. Acta Trop. 2013, 126, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Alfellani, M.A.; Jacob, A.S.; Perea, N.O.; Krecek, R.C.; Taner-Mulla, D.; Verweij, J.J.; Levecke, B.; Tannich, E.; Clark, C.G.; Stensvold, C.R. Diversity and distribution of Blastocystis sp. subtypes in non-human primates. Parasitology 2013, 140, 966–971. [Google Scholar] [CrossRef]
- Badparva, E.; Sadraee, J.; Kheirandish, F. Genetic diversity of Blastocystis isolated from cattle in Khorramabad, Iran. Jundishapur J. Microbiol. 2015, 8, e14810. [Google Scholar] [CrossRef]
- Ramírez, J.D.; Sánchez, L.V.; Bautista, D.C.; Corredor, A.F.; Flórez, A.C.; Stensvold, C.R. Blastocystis subtypes detected in humans and animals from Colombia. Infect. Genet. Evol. 2014, 22, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Noradilah, S.A.; Anuar, T.S.; Moktar, N.; Lee, I.L.; Salleh, F.M.; Manap, S.n.A.A.; Mohtar, n.S.H.M.; Azrul, S.M.; Abdullah, W.O.; Nordin, A.; et al. Molecular epidemiology of Blastocystis sp. in animals reared by the aborigines during wet and dry seasons in rural communities, Pahang, Malaysia. Southeast Asian J. Trop. Med. Public Health 2017, 48, 1151–1160. [Google Scholar] [CrossRef]
- Ma, L.; Qiao, H.; Wang, H.; Li, S.; Zhai, P.; Huang, J.; Guo, Y. Molecular prevalence and subtypes of Blastocystis sp. in primates in northern China. Transbound. Emerg. Dis. 2020, 67, 2789–2796. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.G.; van der Giezen, M.; Alfellani, M.A.; Stensvold, C.R. Recent developments in Blastocystis research. Adv. Parasitol. 2013, 82, 1–32. [Google Scholar] [PubMed]
- Ramírez, J.D.; Sánchez, A.; Hernández, C.; Flórez, C.; Bernal, M.C.; Giraldo, J.C.; Reyes, P.; López, M.C.; García, L.; Cooper, P.J.; et al. Geographic distribution of human Blastocystis subtypes in South America. Infect. Genet. Evol. 2016, 41, 32–35. [Google Scholar] [CrossRef]
- Khaled, S.; Gantois, N.; Ly, A.T.; Senghor, S.; Even, G.; Dautel, E.; Dejager, R.; Sawant, M.; Baydoun, M.; Benamrouz-Vanneste, S.; et al. Prevalence and subtype distribution of Blastocystis sp. in Senegalese school children. Microorganisms 2020, 8, 1408. [Google Scholar] [CrossRef]
- Alfellani, M.A.; Taner-Mulla, D.; Jacob, A.S.; Imeede, C.A.; Yoshikawa, H.; Stensvold, C.R.; Clark, C.G. Genetic diversity of Blastocystis in livestock and zoo animals. Protist 2013, 164, 497–509. [Google Scholar] [CrossRef]
- Maloney, J.G.; Molokin, A.; da Cunha, M.J.R.; Cury, M.C.; Santin, M. Blastocystis subtype distribution in domestic and captive wild bird species from Brazil using next generation amplicon sequencing. Parasite Epidemiol. Control 2020, 9, e00138. [Google Scholar] [CrossRef] [PubMed]
- Maloney, J.G.; da Cunha, M.J.R.; Molokin, A.; Cury, M.C.; Santin, M. Next-generation sequencing reveals wide genetic diversity of Blastocystis subtypes in chickens including potentially zoonotic subtypes. Parasitol. Res. 2021, 120, 2219–2231. [Google Scholar] [CrossRef] [PubMed]
- Santin, M. Enterocytozoon bieneusi. In Biology of Foodborne Parasites; Xiao, L., Ryan, U., Feng, Y., Eds.; CRC Press: Boca Raton, FL, USA, 2015; pp. 149–174. [Google Scholar]
- Galván, A.L.; Sánchez, A.M.; Valentín, M.A.; Henriques-Gil, N.; Izquierdo, F.; Fenoy, S.; del Aguila, C. First cases of microsporidiosis in transplant recipients in Spain and review of the literature. J. Clin. Microbiol. 2011, 49, 1301–1306. [Google Scholar] [CrossRef]
- Paulos, S.; Köster, P.C.; de Lucio, A.; Hernández-de-Mingo, M.; Cardona, G.A.; Fernández-Crespo, J.C.; Stensvold, C.R.; Carmena, D. Occurrence and subtype distribution of Blastocystis sp. in humans, dogs and cats sharing household in northern Spain and assessment of zoonotic transmission risk. Zoonoses Public Health 2018, 65, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Muadica, A.S.; Köster, P.C.; Dashti, A.; Bailo, B.; Hernández-de-Mingo, M.; Reh, L.; Balasegaram, S.; Verlander, N.Q.; Ruiz Chércoles, E.; Carmena, D. Molecular diversity of Giardia duodenalis, Cryptosporidium spp. and Blastocystis sp. in asymptomatic school children in Leganés, Madrid (Spain). Microorganisms 2020, 8, 466. [Google Scholar] [CrossRef]
- Rivero-Juarez, A.; Dashti, A.; López-López, P.; Muadica, A.S.; Risalde, M.L.A.; Köster, P.C.; Machuca, I.; Bailo, B.; de Mingo, M.H.; Dacal, E.; et al. Protist enteroparasites in wild boar (Sus scrofa ferus) and black Iberian pig (Sus scrofa domesticus) in southern Spain: A protective effect on hepatitis E acquisition? Parasit. Vectors 2020, 13, 281. [Google Scholar] [CrossRef]
- Dashti, A.; Santín, M.; Cano, L.; de Lucio, A.; Bailo, B.; de Mingo, M.H.; Köster, P.C.; Fernández-Basterra, J.A.; Aramburu-Aguirre, J.; López-Molina, N.; et al. Occurrence and genetic diversity of Enterocytozoon bieneusi (Microsporidia) in owned and sheltered dogs and cats in Northern Spain. Parasitol. Res. 2019, 118, 2979–2987. [Google Scholar] [CrossRef]
- Köster, P.C.; Dashti, A.; Bailo, B.; Muadica, A.S.; Maloney, J.G.; Santín, M.; Chicharro, C.; Migueláñez, S.; Nieto, F.J.; Cano-Terriza, D.; et al. Occurrence and genetic diversity of protist parasites in captive non-human primates, zookeepers, and free-living sympatric rats in the Córdoba Zoo Conservation Centre, Southern Spain. Animals 2021, 11, 700. [Google Scholar] [CrossRef]
- Martínez-Padilla, A.; Caballero-Gómez, J.; Magnet, Á.; Gómez-Guillamón, F.; Izquierdo, F.; Camacho-Sillero, L.; Jiménez-Ruiz, S.; Del Águila, C.; García-Bocanegra, I. Zoonotic Microsporidia in wild lagomorphs in Southern Spain. Animals 2020, 10, 2218. [Google Scholar] [CrossRef]
- Santín, M.; Calero-Bernal, R.; Carmena, D.; Mateo, M.; Balseiro, A.; Barral, M.; Lima Barbero, J.F.; Habela, M.Á. Molecular characterization of Enterocytozoon bieneusi in wild carnivores in Spain. J. Eukaryot. Microbiol. 2018, 65, 468–474. [Google Scholar] [CrossRef]
- Quílez, J.; Sánchez-Acedo, C.; Clavel, A.; Causapé, A.C. Occurrence of Blastocystis sp. in cattle in Aragón, northeastern Spain. Parasitol. Res. 1995, 81, 703–705. [Google Scholar] [CrossRef]
- Stensvold, C.R.; Alfellani, M.A.; Nørskov-Lauritsen, S.; Prip, K.; Victory, E.L.; Maddox, C.; Nielsen, H.V.; Clark, C.G. Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int. J. Parasitol. 2009, 39, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, P.; Zhao, X.; Xu, H.; Wu, W.; Wang, Y.; Guo, Y.; Wang, L.; Feng, Y.; Xiao, L. Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Vet. Parasitol. 2015, 207, 220–227. [Google Scholar] [CrossRef]
- da Silva Fiuza, V.R.; Lopes, C.W.; de Oliveira, F.C.; Fayer, R.; Santin, M. New findings of Enterocytozoon bieneusi in beef and dairy cattle in Brazil. Vet. Parasitol. 2016, 216, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.T.; Wang, R.J.; Ren, G.J.; Yu, Z.Q.; Zhang, L.X.; Zhang, S.Y.; Lu, H.; Peng, X.Q.; Zhao, G.H. Multilocus genotyping of Giardia duodenalis and Enterocytozoon bieneusi in dairy and native beef (Qinchuan) calves in Shaanxi province, northwestern China. Parasitol. Res. 2016, 115, 1355–1361. [Google Scholar] [CrossRef]
- Tang, C.; Cai, M.; Wang, L.; Guo, Y.; Li, N.; Feng, Y.; Xiao, L. Genetic diversity within dominant Enterocytozoon bieneusi genotypes in pre-weaned calves. Parasit. Vectors. 2018, 11, 170. [Google Scholar] [CrossRef] [PubMed]
- Maloney, J.G.; Lombard, J.E.; Urie, N.J.; Shivley, C.B.; Santin, M. Zoonotic and genetically diverse subtypes of Blastocystis in US pre-weaned dairy heifer calves. Parasitol. Res. 2019, 118, 575–582. [Google Scholar] [CrossRef]
- Scicluna, S.M.; Tawari, B.; Clark, C.G. DNA barcoding of Blastocystis. Protist 2006, 157, 77–85. [Google Scholar] [CrossRef]
- Buckholt, M.A.; Lee, J.H.; Tzipori, S. Prevalence of Enterocytozoon bieneusi in swine: An 18-month survey at a slaughterhouse in Massachusetts. Appl. Environ. Microbiol. 2002, 68, 2595–2599. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Maloney, J.G.; Molokin, A.; Santin, M. Next generation amplicon sequencing improves detection of Blastocystis mixed subtype infections. Infect. Genet. Evol. 2019, 73, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Santín, M.; Gómez-Muñoz, M.T.; Solano-Aguilar, G.; Fayer, R. Development of a new PCR protocol to detect and subtype Blastocystis spp. from humans and animals. Parasitol. Res. 2011, 109, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B. BBMap Download. SourceForge.net. 2014. Available online: https://sourceforge.net/projects/bbmap/ (accessed on 27 April 2020).
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PEER J. 2016, 4, e2584. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinformatics 2009, 10, 421. [Google Scholar] [CrossRef]
- Quilez, J.; Clavel, A.; Sanchez-Acedo, C.; Causape, A.C. Detection of Blastocystis sp. in pigs in Aragon (Spain). Vet. Parasitol. 1995, 56, 345–348. [Google Scholar] [CrossRef]
- Ponce Gordo, F.; Herrera, S.; Castro, A.T.; García Durán, B.; Martínez Díaz, R.A. Parasites from farmed ostriches (Struthio camelus) and rheas (Rhea americana) in Europe. Vet. Parasitol. 2002, 107, 137–160. [Google Scholar] [CrossRef]
- Pérez Cordón, G.; Hitos Prados, A.; Romero, D.; Sánchez Moreno, M.; Pontes, A.; Osuna, A.; Rosales, M.J. Intestinal parasitism in the animals of the zoological garden “Peña Escrita” (Almuñecar, Spain). Vet. Parasitol. 2008, 156, 302–309. [Google Scholar] [CrossRef]
- Cordón, G.P.; Prados, A.H.; Romero, D.; Moreno, M.S.; Pontes, A.; Osuna, A.; Rosales, M.J. Intestinal and haematic parasitism in the birds of the Almuñecar (Granada, Spain) ornithological garden. Vet. Parasitol. 2009, 165, 361–366. [Google Scholar] [CrossRef]
- Soledad Gómez, M.; Gracenea, M.; Montoliu, I.; Feliu, C.; Monleon, A.; Fernandez, J.; Enseñat, C. Intestinal parasitism--protozoa and helminths--in primates at the Barcelona Zoo. J. Med. Primatol. 1996, 25, 419–423. [Google Scholar] [CrossRef]
- Zhu, W.; Tao, W.; Gong, B.; Yang, H.; Li, Y.; Song, M.; Lu, Y.; Li, W. First report of Blastocystis infections in cattle in China. Vet. Parasitol. 2017, 246, 38–42. [Google Scholar] [CrossRef]
- Shams, M.; Shamsi, L.; Sadrebazzaz, A.; Asghari, A.; Badali, R.; Omidian, M.; Hassanipour, S. A systematic review and meta-analysis on the global prevalence and subtypes distribution of Blastocystis sp. infection in cattle: A zoonotic concern. Comp. Immunol. Microbiol. Infect. Dis. 2021, 76, 101650. [Google Scholar] [CrossRef]
- Maloney, J.G.; Molokin, A.; Santin, M. Use of Oxford Nanopore MinION to generate full-length sequences of the Blastocystis small subunit (SSU) rRNA gene. Parasit. Vectors 2020, 13, 595. [Google Scholar] [CrossRef]
- Calero-Bernal, R.; Santín, M.; Maloney, J.G.; Martín-Pérez, M.; Habela, M.A.; Fernández-García, J.L.; Figueiredo, A.; Nájera, F.; Palacios, M.J.; Mateo, M.; et al. Blastocystis sp. subtype diversity in wild carnivore species from Spain. J. Eukaryot. Microbiol. 2020, 67, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Hublin, J.S.Y.; Maloney, J.G.; Santin, M. Blastocystis in domesticated and wild mammals and birds. Res. Vet. Sci. 2021, 135, 260–282. [Google Scholar] [CrossRef]
- Yan, Y.; Su, S.; Ye, J.; Lai, X.; Lai, R.; Liao, H.; Chen, G.; Zhang, R.; Hou, Z.; Luo, X. Blastocystis sp. subtype 5: A possibly zoonotic genotype. Parasitol. Res. 2007, 101, 1527–1532. [Google Scholar] [CrossRef]
- Li, L.H.; Zhou, X.N.; Du, Z.W.; Wang, X.Z.; Wang, L.B.; Jiang, J.Y.; Yoshikawa, H.; Steinmann, P.; Utzinger, J.; Wu, Z.; et al. Molecular epidemiology of human Blastocystis in a village in Yunnan province, China. Parasitol. Int. 2007, 56, 281–286. [Google Scholar] [CrossRef]
- Wang, W.; Owen, H.; Traub, R.J.; Cuttell, L.; Inpankaew, T.; Bielefeldt-Ohmann, H. Molecular epidemiology of Blastocystis in pigs and their in-contact humans in Southeast Queensland, Australia, and Cambodia. Vet. Parasitol. 2014, 203, 264–269. [Google Scholar] [CrossRef]
- del Aguila, C.; Izquierdo, F.; Navajas, R.; Pieniazek, N.J.; Miró, G.; Alonso, A.I.; Da Silva, A.J.; Fenoy, S. Enterocytozoon bieneusi in animals: Rabbits and dogs as new hosts. J. Eukaryot. Microbiol. 1999, 46, 8S–9S. [Google Scholar] [PubMed]
- Galván-Díaz, A.L.; Magnet, A.; Fenoy, S.; Henriques-Gil, N.; Haro, M.; Ponce-Gordo, F.; Miró, G.; Del Águila, C.; Izquierdo, F. Microsporidia Detection and Genotyping Study of Human Pathogenic E. bieneusi in Animals from Spain. PLoS ONE 2014, 9, e92289. [Google Scholar] [CrossRef] [PubMed]
- Haro, M.; Henriques-Gil, N.; Fenoy, S.; Izquierdo, F.; Alonso, F.; Del Aguila, C. Detection and genotyping of Enterocytozoon bieneusi in pigeons. J. Eukaryot. Microbiol. 2006, 53, S58–S60. [Google Scholar] [CrossRef] [PubMed]
- Lores, B.; del Aguila, C.; Arias, C. Enterocytozoon bieneusi (microsporidia) in faecal samples from domestic animals from Galicia, Spain. Mem. Inst. Oswaldo Cruz 2002, 97, 941–945. [Google Scholar] [CrossRef]
- del Aguila, C.; Navajas, R.; Gurbindo, D.; Ramos, J.T.; Mellado, M.J.; Fenoy, S.; Muñoz Fernandez, M.A.; Subirats, M.; Ruiz, J.; Pieniazek, N.J. Microsporidiosis in HIV-positive children in Madrid (Spain). J. Eukaryot. Microbiol. 1997, 44, 84S–85S. [Google Scholar] [CrossRef]
- del Aguila, C.; Soriano, V.; Navajas, R.; Subirats, M.; Fenoy, S.; Valencia, E.; Baquero, M.; Pieniazek, N.J. Species identification of intestinal microsporidiosis in HIV-positive patients using the polymerase chain reaction. Enferm. Infecc. Microbiol. Clin. 1997, 15, 456–461. [Google Scholar]
- López-Vélez, R.; Turrientes, M.C.; Garrón, C.; Montilla, P.; Navajas, R.; Fenoy, S.; del Aguila, C. Microsporidiosis in travelers with diarrhea from the tropics. J. Travel Med. 1999, 6, 223–227. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lores, B.; López-Miragaya, I.; Arias, C.; Fenoy, S.; Torres, J.; del Aguila, C. Intestinal microsporidiosis due to Enterocytozoon bieneusi in elderly human immunodeficiency virus--negative patients from Vigo, Spain. Clin. Infect. Dis. 2002, 34, 918–921. [Google Scholar] [CrossRef]
- Abreu-Acosta, N.; Lorenzo-Morales, J.; Leal-Guio, Y.; Coronado-Alvarez, N.; Foronda, P.; Alcoba-Florez, J.; Izquierdo, F.; Batista-Díaz, N.; Del Aguila, C.; Valladares, B. Enterocytozoon bieneusi (microsporidia) in clinical samples from immunocompetent individuals in Tenerife, Canary Islands, Spain. Trans. R. Soc. Trop. Med. Hyg. 2005, 99, 855. [Google Scholar] [CrossRef]
- Galván, A.L.; Magnet, A.; Izquierdo, F.; Fenoy, S.; Rueda, C.; Fernández Vadillo, C.; Henriques-Gil, N.; del Aguila, C. Molecular characterization of human-pathogenic microsporidia and Cyclospora cayetanensis isolated from various water sources in Spain: A year-long longitudinal study. Appl. Environ. Microbiol. 2013, 79, 449–459. [Google Scholar] [CrossRef]
- Ruan, Y.; Xu, X.; He, Q.; Li, L.; Guo, J.; Bao, J.; Pan, G.; Li, T.; Zhou, Z. The largest meta-analysis on the global prevalence of microsporidia in mammals, avian and water provides insights into the epidemic features of these ubiquitous pathogens. Parasit. Vectors 2021, 14, 186. [Google Scholar] [CrossRef]
- Del Coco, V.F.; Córdoba, M.A.; Bilbao, G.; de Almeida Castro, P.; Basualdo, J.A.; Santín, M. First report of Enterocytozoon bieneusi from dairy cattle in Argentina. Vet. Parasitol. 2014, 199, 112–115. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Z.; Su, Y.; Liang, X.; Sun, X.; Peng, S.; Lu, H.; Jiang, N.; Yin, J.; Xiang, M.; et al. Identification and genotyping of Enterocytozoon bieneusi in China. J. Clin. Microbiol. 2011, 49, 2006–2008. [Google Scholar] [CrossRef]
- Hwang, S.; Shin, S.U.; Kim, S.; Ryu, J.H.; Choi, K.S. Zoonotic potential of Enterocytozoon bieneusi in pre-weaned Korean native calves. Parasit. Vectors 2020, 13, 300. [Google Scholar] [CrossRef] [PubMed]
- Abu Samra, N.; Thompson, P.N.; Jori, F.; Zhang, H.; Xiao, L. Enterocytozoon bieneusi at the wildlife/livestock interface of the Kruger National Park, South Africa. Vet. Parasitol. 2012, 190, 587–590. [Google Scholar]
- Santín, M.; Fayer, R. A longitudinal study of Enterocytozoon bieneusi in dairy cattle. Parasitol. Res. 2009, 105, 141–144. [Google Scholar] [CrossRef]
- Ma, J.; Cai, J.; Ma, J.; Feng, Y.; Xiao, L. Enterocytozoon bieneusi genotypes in yaks (Bos grunniens) and their public health potential. J. Eukaryot. Microbiol. 2015, 62, 21–25. [Google Scholar] [CrossRef]
- Zhao, A.; Zhang, Y.; Wang, W.; Jing, B.; Xing, J.; Tao, D.; Zhao, W.; Qi, M. Enterocytozoon bieneusi in donkeys from Xinjiang, China: Prevalence, molecular characterization and the assessment of zoonotic risk. BMC Vet. Res. 2020, 16, 196. [Google Scholar] [CrossRef]
- Karim, M.R.; Dong, H.; Li, T.; Yu, F.; Li, D.; Zhang, L.; Li, J.; Wang, R.; Li, S.; Li, X.; et al. Predomination and new genotypes of Enterocytozoon bieneusi in captive nonhuman primates in zoos in China: High genetic diversity and zoonotic significance. PLoS ONE 2015, 10, e0117991. [Google Scholar] [CrossRef]
- Sak, B.; Brady, D.; Pelikánová, M.; Květoňová, D.; Rost, M.; Kostka, M.; Tolarová, V.; Hůzová, Z.; Kváč, M. Unapparent microsporidial infection among immunocompetent humans in the Czech Republic. J. Clin. Microbiol. 2011, 49, 1064–1070. [Google Scholar] [CrossRef]
- Wang, H.Y.; Qi, M.; Sun, M.F.; Li, D.F.; Wang, R.J.; Zhang, S.M.; Zhao, J.F.; Li, J.Q.; Cui, Z.H.; Chen, Y.C.; et al. Prevalence and population genetics analysis of Enterocytozoon bieneusi in dairy cattle in China. Front. Microbiol. 2019, 10, 1399. [Google Scholar] [CrossRef]
- Wang, R.; Li, N.; Jiang, W.; Guo, Y.; Wang, X.; Jin, Y.; Feng, Y.; Xiao, L. Infection patterns, clinical significance, and genetic characteristics of Enterocytozoon bieneusi and Giardia duodenalis in dairy cattle in Jiangsu, China. Parasitol. Res. 2019, 118, 3053–3060. [Google Scholar] [CrossRef]
- Gumbo, T.; Sarbah, S.; Gangaidzo, I.T.; Ortega, Y.; Sterling, C.R.; Carville, A.; Tzipori, S.; Wiest, P.M. Intestinal parasites in patients with diarrhea and human immunodeficiency virus infection in Zimbabwe. AIDS 1999, 13, 819–821. [Google Scholar] [CrossRef] [PubMed]
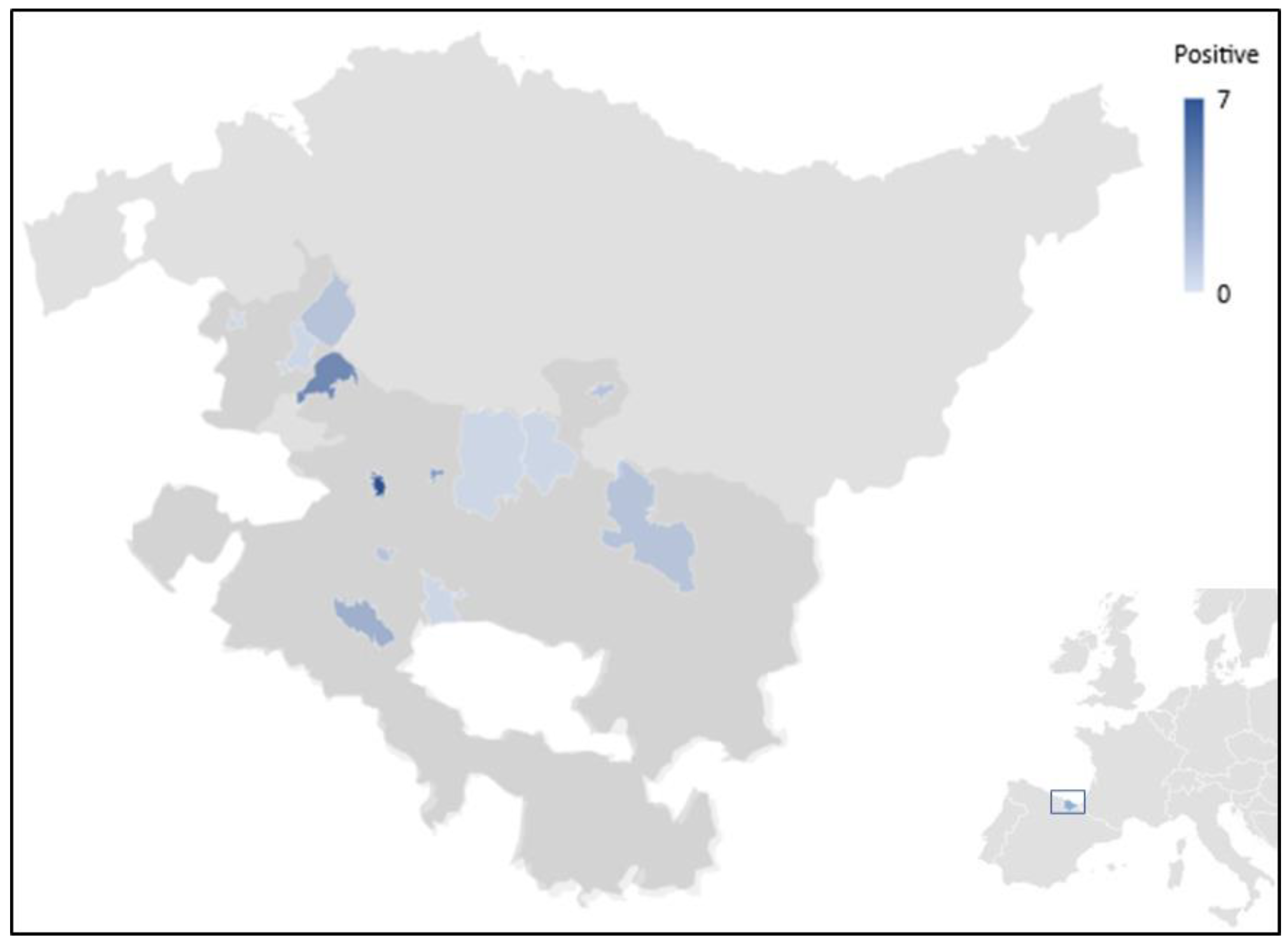
| Blastocystis sp. | Enterocytozoon bieneusi | ||||
|---|---|---|---|---|---|
| Variable | n | Positive (n) | % | Positive (n) | % |
| Production system | |||||
| Dairy | 168 | 38 | 22.6 | 2 | 1.2 |
| Beef | 65 | 21 | 32.3 | 0 | 0.0 |
| Unknown | 103 | 49 | 47.6 | 0 | 0.0 |
| County | |||||
| Añana | 11 | 6 | 54.5 | 0 | 0.0 |
| Ayala | 106 | 20 | 18.9 | 0 | 0.0 |
| Llanada Alavesa | 1 | 1 | 100 | 0 | 0.0 |
| Zuia-Gorbeialdea | 118 | 32 | 27.1 | 2 | 1.7 |
| Unknown | 100 | 49 | 49.0 | 0 | 0.0 |
| Total | 336 | 108 | 32.1 | 2 | 0.6 |
| Blastocystis sp. | Enterocytozoon bieneusi | |||
|---|---|---|---|---|
| Variable | Isolates(n) | Subtypes (n) | Isolates(n) | Genotypes (n) |
| Production system | ||||
| Dairy | 11 | ST5 (4), ST10 (7) | 2 | BEB4 (2) |
| Beef | 9 | ST5 (4), ST10 (5) | 0 | – |
| Unknown | 17 | ST5 (9), ST10 (7), ST14 (1) | 0 | – |
| Total | 37 | 2 | ||
| Subtype | Next Generation Amplicon Sequencing | Sanger Sequencing |
|---|---|---|
| ST1 | 1.9% (2/108) | Not detected |
| ST3 | 0.9% (1/108) | Not detected |
| ST5 | 13.0% (14/108) | 45.9% (17/37) |
| ST10 1 | 98.2% (106/108) | 51.4% (19/37) |
| ST14 1 | 63.0% (68/108) | 2.7% (1/37) |
| ST21 | 64.8% (70/108) | Not detected |
| ST23 | 35.2% (38/108) | Not detected |
| ST24 | 25.9% (28/108) | Not detected |
| ST25 | 86.1% (93/108) | Not detected |
| ST26 | 93.5% (101/108) | Not detected |
| Number of Subtypes in Co-Infection | Subtype Combinations | Samples (n) |
|---|---|---|
| 2 | ST10 + ST25 | 1 |
| ST25 + ST26 | 1 | |
| 3 | ST5 + ST10 + ST25 | 1 |
| ST5 + ST10 + ST14 | 1 | |
| ST10 + ST14 + ST24 | 2 | |
| ST10 + ST21 + ST26 | 1 | |
| ST10 + ST25 + ST26 | 15 | |
| ST14 + ST25 + ST26 | 1 | |
| 4 | ST5 + ST10 + ST25 + ST26 | 3 |
| ST10 + ST14 + ST21 + ST24 | 1 | |
| ST10 + ST14 + ST25 + ST26 | 5 | |
| ST10 + ST21 + ST23 + ST26 | 1 | |
| ST10 + ST21 + ST24 + ST26 | 1 | |
| ST10 + ST21 + ST25 + ST26 | 4 | |
| ST10 + ST23 + ST25 + ST26 | 4 | |
| 5 | ST5 + ST10 + ST14 + ST25 + ST26 | 1 |
| ST5 + ST10 + ST14 + ST24 + ST26 | 1 | |
| ST10 + ST14 + ST21 + ST24 + ST26 | 4 | |
| ST10 + ST14 + ST21 + ST25 + ST26 | 15 | |
| ST10 + ST21 + ST23 + ST25 + ST26 | 8 | |
| ST10 + ST14 + ST24 + ST25 + ST26 | 1 | |
| 6 | ST5 + ST10 + ST14 + ST21 + ST25 + ST26 | 4 |
| ST5 + ST10 + ST14 + ST23 + ST25 + ST26 | 1 | |
| ST10 + ST14 + ST21 + ST23 + ST24 + ST25 | 1 | |
| ST10 + ST14 + ST21 + ST23 + ST24 + ST26 | 3 | |
| ST10 + ST14 + ST21 + ST23 + ST25 + ST26 | 12 | |
| ST10 + ST14 + ST21 + ST24 + ST25 + ST26 | 5 | |
| ST10 + ST21 + ST23 + ST24 + ST25 + ST26 | 1 | |
| 7 | ST1 + ST5 + ST10 + ST14 + ST21 + ST25 + ST26 | 1 |
| ST10 + ST14 + ST21 + ST23 + ST24 + ST25 + ST26 | 7 | |
| 8 | ST3 + ST10 + ST14 + ST21 + ST23 + ST24 + ST25 + ST26 | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abarca, N.; Santín, M.; Ortega, S.; Maloney, J.G.; George, N.S.; Molokin, A.; Cardona, G.A.; Dashti, A.; Köster, P.C.; Bailo, B.; et al. Molecular Detection and Characterization of Blastocystis sp. and Enterocytozoon bieneusi in Cattle in Northern Spain. Vet. Sci. 2021, 8, 191. https://doi.org/10.3390/vetsci8090191
Abarca N, Santín M, Ortega S, Maloney JG, George NS, Molokin A, Cardona GA, Dashti A, Köster PC, Bailo B, et al. Molecular Detection and Characterization of Blastocystis sp. and Enterocytozoon bieneusi in Cattle in Northern Spain. Veterinary Sciences. 2021; 8(9):191. https://doi.org/10.3390/vetsci8090191
Chicago/Turabian StyleAbarca, Nadia, Mónica Santín, Sheila Ortega, Jenny G. Maloney, Nadja S. George, Aleksey Molokin, Guillermo A. Cardona, Alejandro Dashti, Pamela C. Köster, Begoña Bailo, and et al. 2021. "Molecular Detection and Characterization of Blastocystis sp. and Enterocytozoon bieneusi in Cattle in Northern Spain" Veterinary Sciences 8, no. 9: 191. https://doi.org/10.3390/vetsci8090191
APA StyleAbarca, N., Santín, M., Ortega, S., Maloney, J. G., George, N. S., Molokin, A., Cardona, G. A., Dashti, A., Köster, P. C., Bailo, B., Hernández-de-Mingo, M., Muadica, A. S., Calero-Bernal, R., Carmena, D., & González-Barrio, D. (2021). Molecular Detection and Characterization of Blastocystis sp. and Enterocytozoon bieneusi in Cattle in Northern Spain. Veterinary Sciences, 8(9), 191. https://doi.org/10.3390/vetsci8090191









