Predicting Dynamic Clinical Outcomes of the Chemotherapy for Canine Lymphoma Patients Using a Machine Learning Model
Abstract
1. Introduction
2. Materials and Methods
2.1. Case Selection
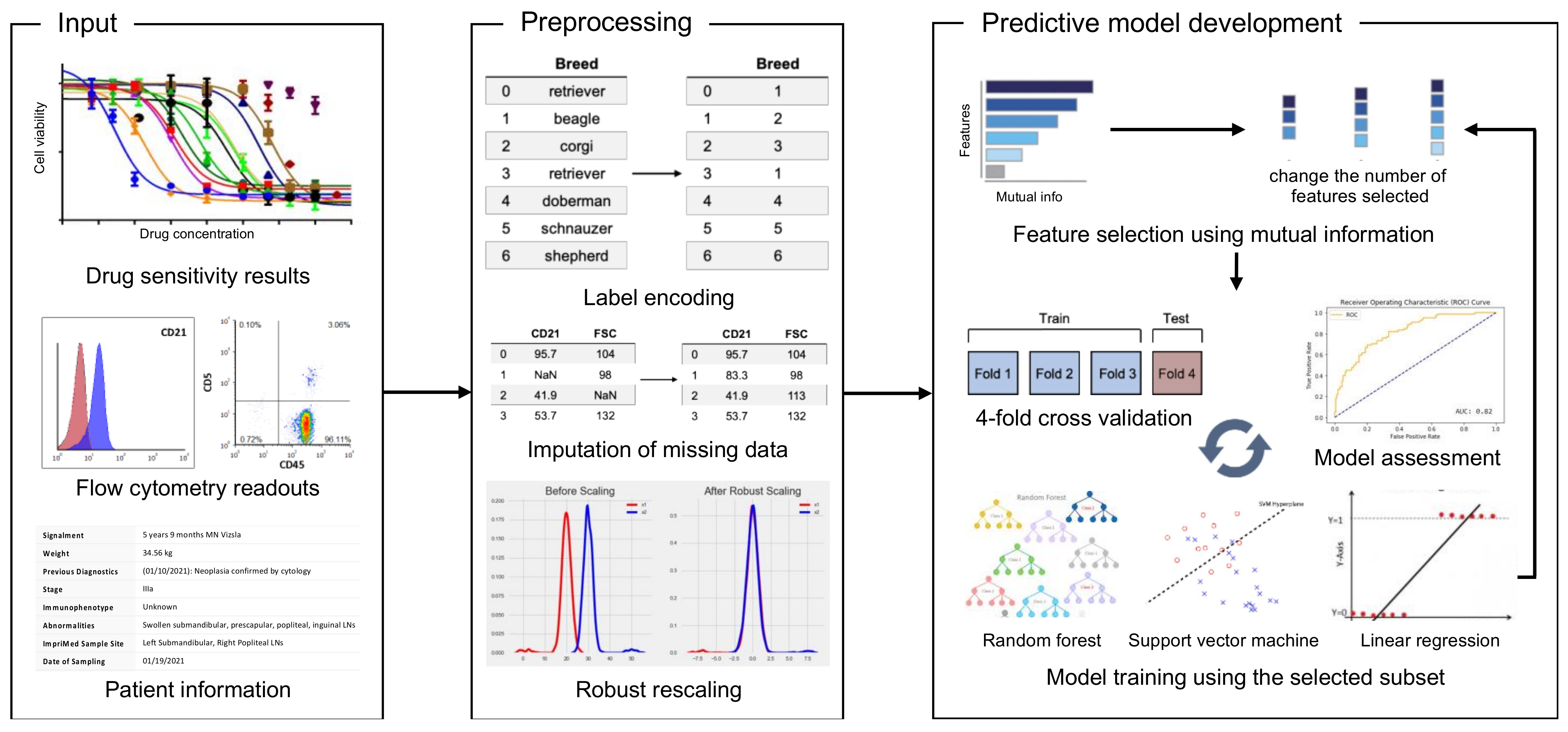
2.2. The Development of the Predictive ML Models
2.3. Survival Analysis
2.4. Model Performance Assessment
3. Results
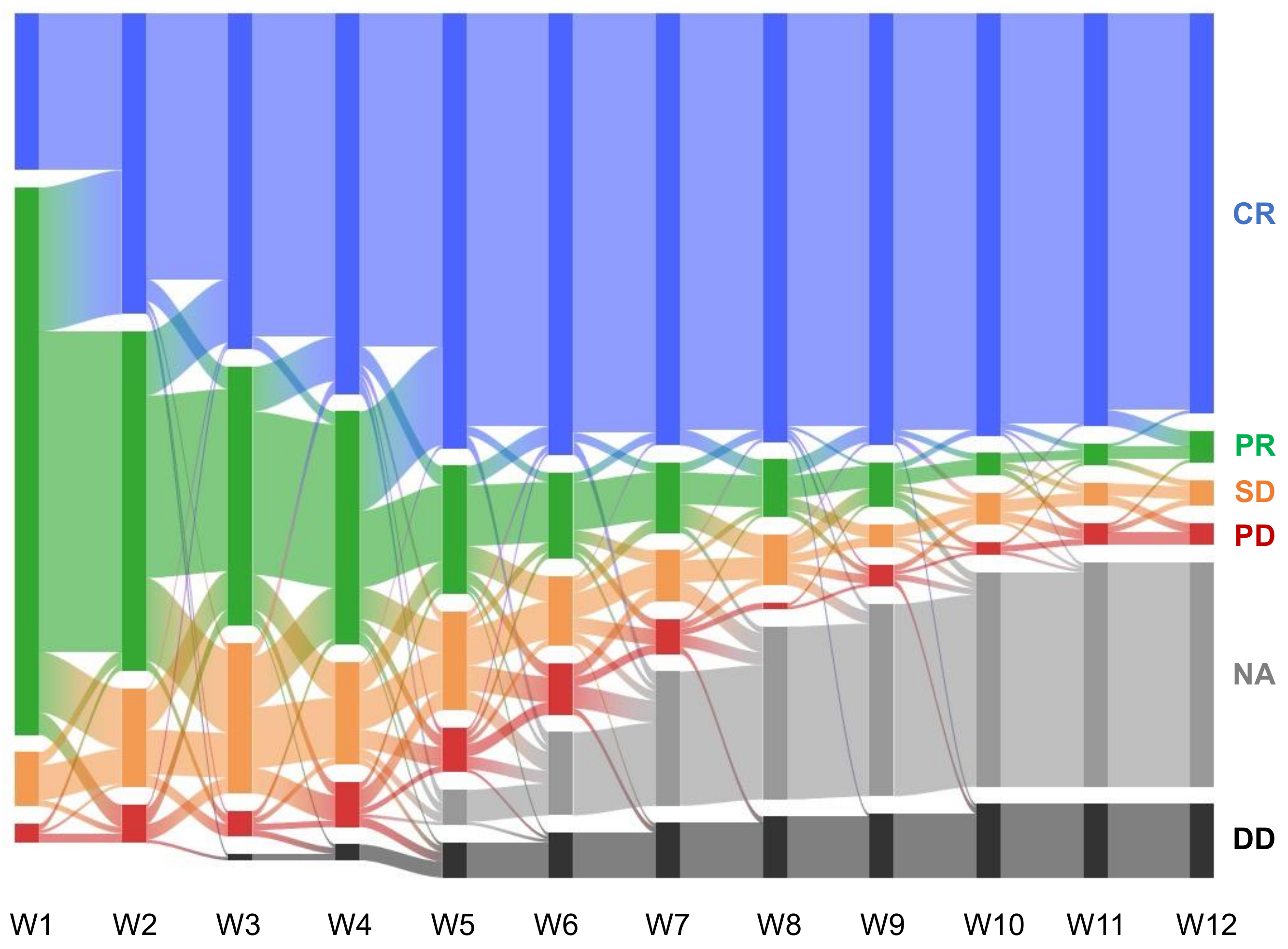
3.1. Data Structure, Acquisition, and Model Training
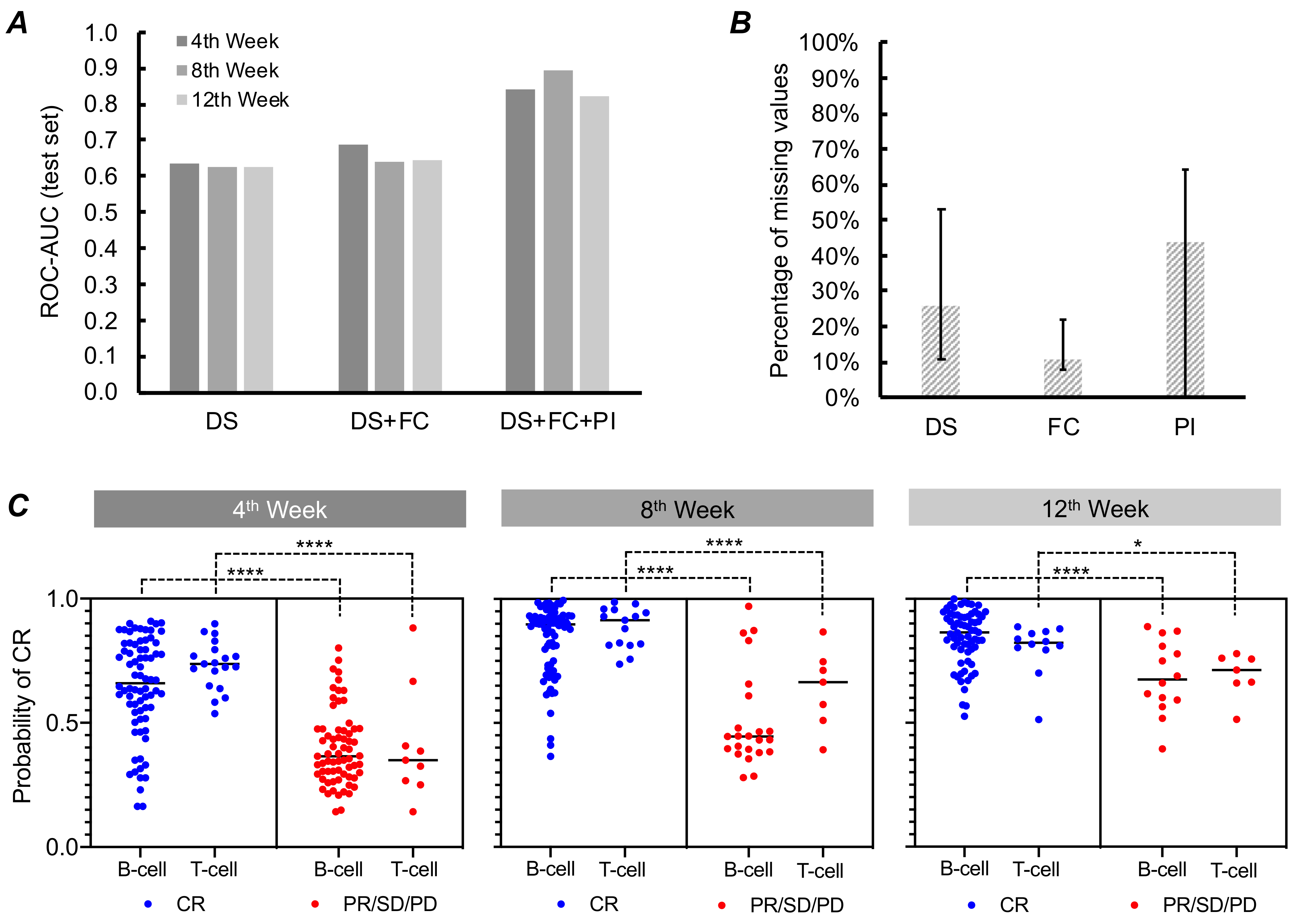
3.2. Model Performance Based on DS and FC Data
3.3. Model Performance Based on DS, FC, and PI Data
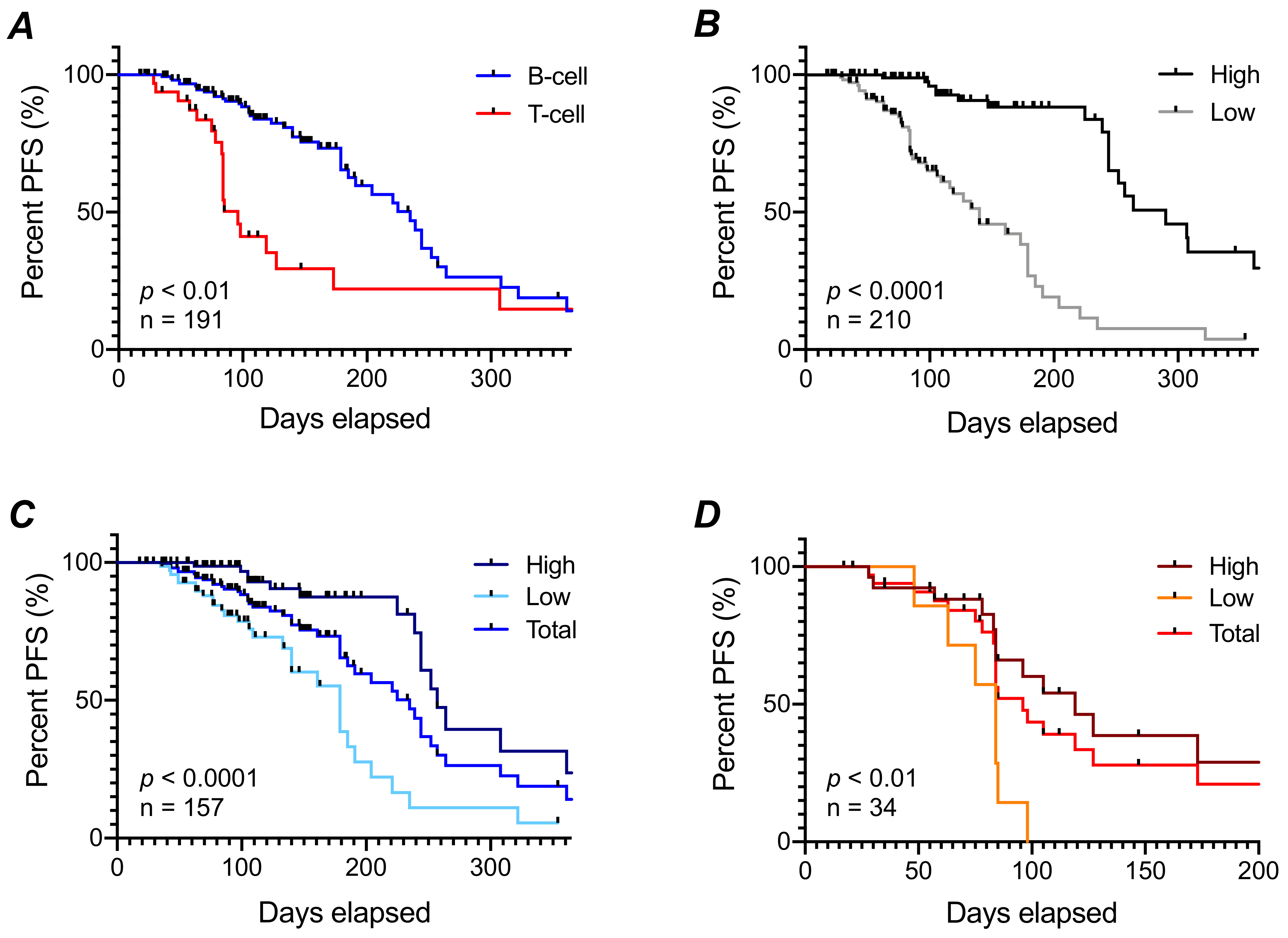
3.4. Progression-Free Survival Prediction
4. Discussion
5. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Carr, K.M.; Rosenblatt, K.; Petricoin, E.F.; Liotta, L.A. Genomic and Proteomic Approaches for Studying Human Cancer: Prospects for True Patient-Tailored Therapy. Hum. Genom. 2004, 1, 134–140. [Google Scholar] [CrossRef][Green Version]
- Coates, A.S.; Winer, E.P.; Goldhirsch, A.; Gelber, R.D.; Gnant, M.; Piccart-Gebhart, M.J.; Thürlimann, B.; Senn, H.J.; André, F.; Baselga, J.; et al. Tailoring Therapies-Improving the Management of Early Breast Cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann. Oncol. 2015, 26, 1533–1546. [Google Scholar] [CrossRef]
- Yersal, O.; Barutca, S. Biological Subtypes of Breast Cancer: Prognostic and Therapeutic Implications. World J. Clin. Oncol. 2014, 5, 412–424. [Google Scholar] [CrossRef]
- Slamon, D.; Eiermann, W.; Robert, N.; Pienkowski, T.; Martin, M.; Press, M.; Mackey, J.; Glaspy, J.; Chan, A.; Pawlicki, M.; et al. Adjuvant Trastuzumab in HER2 Positive Breast Cancer. N. Engl. J. Med. 2011, 365, 1273–1283. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, J.F.; Marx, K.A.; Hoffman, P.E.; Gee, A.G.; O’Neil, P.; Ujwal, M.L.; Hotchikiss, J. Applications of Machine Learning and High-Dimensional Visualization in Cancer Detection, Diagnosis, and Management. Ann. N. Y. Acad. Sci. 2006, 1020, 239–262. [Google Scholar] [CrossRef]
- Zhou, M.; Scott, J.; Chaudhury, B.; Hall, L.; Goldgof, D.; Yeom, K.W.; Iv, M.; Ou, Y.; Kalpathy-Cramer, J.; Napel, S.; et al. Radiomics in Brain Tumor: Image Assessment, Quantitative Feature Descriptors, and Machine-Learning Approaches. Am. J. Neuroradiol. 2018, 39, 208–216. [Google Scholar] [CrossRef]
- Mobadersany, P.; Yousefi, S.; Amgad, M.; Gutman, D.A.; Barnholtz-Sloan, J.S.; Velázquez Vega, J.E.; Brat, D.J.; Cooper, L.A.D. Predicting Cancer Outcomes from Histology and Genomics Using Convolutional Networks. Proc. Natl. Acad. Sci. USA 2018, 115, E2970–E2979. [Google Scholar] [CrossRef]
- Malta, T.M.; Sokolov, A.; Gentles, A.J.; Burzykowski, T.; Poisson, L.; Weinstein, J.N.; Kamińska, B.; Huelsken, J.; Omberg, L.; Gevaert, O.; et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 2018, 173, 338–354.e15. [Google Scholar] [CrossRef]
- Goldenberg, S.L.; Nir, G.; Salcudean, S.E. A New Era: Artificial Intelligence and Machine Learning in Prostate Cancer. Nat. Rev. Urol. 2019, 16, 391–403. [Google Scholar] [CrossRef]
- Bohannan, Z.; Pudupakam, R.S.; Koo, J.; Horwitz, H.; Tsang, J.; Polley, A.; Han, E.J.; Fernandez, E.; Park, S.; Swartzfager, D.; et al. Predicting Likelihood of in Vivo Chemotherapy Response in Canine Lymphoma Using Ex Vivo Drug Sensitivity and Immunophenotyping Data in a Machine Learning Model. Vet. Comp. Oncol. 2021, 19, 160–171. [Google Scholar] [CrossRef]
- Shipp, M.A.; Ross, K.N.; Tamayo, P.; Weng, A.P.; Kutok, J.L.; Aguiar, R.C.T.; Gaasenbeek, M.; Angelo, M.; Reich, M.; Pinkus, G.S.; et al. Diffuse Large B-Cell Lymphoma Outcome Prediction by Gene-Expression Profiling and Supervised Machine Learning. Nat. Med. 2002, 8, 68–74. [Google Scholar] [CrossRef]
- Mucaki, E.J.; Zhao, J.Z.L.; Lizotte, D.J.; Rogan, P.K. Predicting Responses to Platin Chemotherapy Agents with Biochemically-Inspired Machine Learning. Signal Transduct. Target. Ther. 2019, 4, 1. [Google Scholar] [CrossRef]
- Tahmassebi, A.; Wengert, G.J.; Helbich, T.H.; Bago-Horvath, Z.; Alaei, S.; Bartsch, R.; Dubsky, P.; Baltzer, P.; Clauser, P.; Kapetas, P.; et al. Impact of Machine Learning with Multiparametric Magnetic Resonance Imaging of the Breast for Early Prediction of Response to Neoadjuvant Chemotherapy and Survival Outcomes in Breast Cancer Patients. Investig. Radiol. 2019, 54, 110–117. [Google Scholar] [CrossRef]
- Chan, L.W.C.; Chan, T.; Cheng, L.F.; Mak, W.S. Machine Learning of Patient Similarity: A Case Study on Predicting Survival in Cancer Patient after Locoregional Chemotherapy. In Proceedings of the 2010 IEEE International Conference Bioinformatics and Biomedicine Workshops, BIBMW 2010, Hong Kong, China, 18 December 2010; pp. 467–470. [Google Scholar] [CrossRef]
- Ferroni, P.; Zanzotto, F.M.; Scarpato, N.; Riondino, S.; Nanni, U.; Roselli, M.; Guadagni, F. Risk Assessment for Venous Thromboembolism in Chemotherapy-Treated Ambulatory Cancer Patients. Med. Decis. Mak. 2017, 37, 234–242. [Google Scholar] [CrossRef]
- DeAndrés-Galiana, E.J.; Fernández-Martínez, J.L.; Luaces, O.; del Coz, J.J.; Fernández, R.; Solano, J.; Nogués, E.A.; Zanabilli, Y.; Alonso, J.M.; Payer, A.R.; et al. On the Prediction of Hodgkin Lymphoma Treatment Response. Clin. Transl. Oncol. 2015, 17, 612–619. [Google Scholar] [CrossRef]
- Bisong, E. Building Machine Learning and Deep Learning Models on Google Cloud Platform: A Comprehensive Guide for Beginners; Bisong, E., Ed.; Springer: Ottawa, ON, Canada, 2019; ISBN 978-1-4842-4470-8. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Pretttenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Beretta, L.; Santaniello, A. Nearest Neighbor Imputation Algorithms: A Critical Evaluation. BMC Med. Inform. Decis. Mak. 2016, 16, 74. [Google Scholar] [CrossRef]
- White, I.R.; Royston, P.; Wood, A.M. Multiple Imputation Using Chained Equations: Issues and Guidance for Practice. Stat. Med. 2011, 30, 377–399. [Google Scholar] [CrossRef]
- Peng, H.; Long, F.; Ding, C. Feature Selection Based on Mutual Information: Criteria of Max-Dependency, Max-Relevance, and Min-Redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Burton, J.H.; Garrett-Mayer, E.; Thamm, D.H. Evaluation of a 15-Week CHOP Protocol for the Treatment of Canine Multicentric Lymphoma. Vet. Comp. Oncol. 2013, 11, 306–315. [Google Scholar] [CrossRef]
- Ross, B.C. Mutual Information between Discrete and Continuous Data Sets. PLoS ONE 2014, 9, e87357. [Google Scholar] [CrossRef]
- Zandvliet, M. Canine Lymphoma: A Review. Vet. Q. 2016, 36, 76–104. [Google Scholar] [CrossRef] [PubMed]
- Curran, K.; Thamm, D.H. Retrospective Analysis for Treatment of Naïve Canine Multicentric Lymphoma with a 15-Week, Maintenance-Free CHOP Protocol. Vet. Comp. Oncol. 2016, 14, 147–155. [Google Scholar] [CrossRef]
- Rebhun, R.B.; Kent, M.S.; Borrofka, S.A.E.B.; Frazier, S.; Skorupski, K.; Rodriguez, C.O. CHOP Chemotherapy for the Treatment of Canine Multicentric T-Cell Lymphoma. Vet. Comp. Oncol. 2011, 9, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Daters, A.T.; Mauldin, G.E.; Mauldin, G.N.; Brodsky, E.M.; Post, G.S. Evaluation of a Multidrug Chemotherapy Protocol with Mitoxantrone Based Maintenance (CHOP-MA) for the Treatment of Canine Lymphoma. Vet. Comp. Oncol. 2010, 8, 11–22. [Google Scholar] [CrossRef]
- Henriques, J.; Felisberto, R.; Constantino-Casas, F.; Cabeçadas, J.; Dobson, J. Peripheral Blood Cell Ratios as Prognostic Factors in Canine Diffuse Large B-Cell Lymphoma Treated with CHOP Protocol. Vet. Comp. Oncol. 2020, 19, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, H.F.; Weaver, V.M.; Tlsty, T.D.; Bergers, G. Tumor Microenvironment and Progression. J. Surg. Oncol. 2011, 103, 468–474. [Google Scholar] [CrossRef]
- Ansiaux, R.; Baudelet, C.; Jordan, B.F.; Beghein, N.; Sonveaux, P.; De Wever, J.; Martinive, P.; Grégoire, V.; Feron, O.; Gallez, B. Thalidomide Radiosensitizes Tumors through Early Changes in the Tumor Microenvironment. Clin. Cancer Res. 2005, 11, 743–750. [Google Scholar]
- Polyak, K.; Haviv, I.; Campbell, I.G. Co-Evolution of Tumor Cells and Their Microenvironment. Trends Genet. 2009, 25, 30–38. [Google Scholar] [CrossRef]
- Romano, F.R.; Heinze, C.R.; Barber, L.G.; Mason, J.B.; Freeman, L.M. Association between Body Condition Score and Cancer Prognosis in Dogs with Lymphoma and Osteosarcoma. J. Vet. Intern. Med. 2016, 30, 1179–1186. [Google Scholar] [CrossRef]
- García-Laencina, P.J.; Sancho-Gómez, J.L.; Figueiras-Vidal, A.R. Pattern Classification with Missing Data: A Review. Neural Comput. Appl. 2010, 19, 263–282. [Google Scholar] [CrossRef]
- Jerez, J.M.; Molina, I.; García-Laencina, P.J.; Alba, E.; Ribelles, N.; Martín, M.; Franco, L. Missing Data Imputation Using Statistical and Machine Learning Methods in a Real Breast Cancer Problem. Artif. Intell. Med. 2010, 50, 105–115. [Google Scholar] [CrossRef]
- Bertsimas, D.; Pawlowski, C.; Zhuo, Y.D. From Predictive Methods to Missing Data Imputation: An Optimization Approach. J. Mach. Learn. Res. 2018, 18, 1–39. [Google Scholar]
| Parameter | CR Prediction Study Population (N = 242) | PFS Prediction Study Population (N = 210) |
|---|---|---|
| Age | ||
| Median ± SD | 9 ± 3.2 | 8 ± 3.2 |
| Range | 1 to 17 years | 2 to 16 years |
| Sex | ||
| Male | 56% | 57% |
| Female | 41% | 42% |
| Unknown | 3% | 1% |
| Relapse vs. Naïve | ||
| Naïve | 90% | 93% |
| Relapse | 6% | 5% |
| Unknown | 4% | 2% |
| Immunophenotype | ||
| B | 71% | 75% |
| T | 15% | 16% |
| Others | 14% | 9% |
| Clinical Stage | ||
| 2 | 1% | 2% |
| 3 | 40% | 43% |
| 4 | 21% | 19% |
| 5 | 6% | 3% |
| Not Available | 31% | 33% |
| Metrics | 4th Week | 8th Week | 12th Week |
|---|---|---|---|
| Accuracy | 0.804 | 0.891 | 0.827 |
| PPV | 0.824 | 0.894 | 0.879 |
| NPV | 0.791 | 0.875 | 0.500 |
| Sensitivity | 0.816 | 0.971 | 0.879 |
| Specificity | 0.800 | 0.636 | 0.500 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koo, J.; Choi, K.; Lee, P.; Polley, A.; Pudupakam, R.S.; Tsang, J.; Fernandez, E.; Han, E.J.; Park, S.; Swartzfager, D.; et al. Predicting Dynamic Clinical Outcomes of the Chemotherapy for Canine Lymphoma Patients Using a Machine Learning Model. Vet. Sci. 2021, 8, 301. https://doi.org/10.3390/vetsci8120301
Koo J, Choi K, Lee P, Polley A, Pudupakam RS, Tsang J, Fernandez E, Han EJ, Park S, Swartzfager D, et al. Predicting Dynamic Clinical Outcomes of the Chemotherapy for Canine Lymphoma Patients Using a Machine Learning Model. Veterinary Sciences. 2021; 8(12):301. https://doi.org/10.3390/vetsci8120301
Chicago/Turabian StyleKoo, Jamin, Kyucheol Choi, Peter Lee, Amanda Polley, Raghavendra Sumanth Pudupakam, Josephine Tsang, Elmer Fernandez, Enyang James Han, Stanley Park, Deanna Swartzfager, and et al. 2021. "Predicting Dynamic Clinical Outcomes of the Chemotherapy for Canine Lymphoma Patients Using a Machine Learning Model" Veterinary Sciences 8, no. 12: 301. https://doi.org/10.3390/vetsci8120301
APA StyleKoo, J., Choi, K., Lee, P., Polley, A., Pudupakam, R. S., Tsang, J., Fernandez, E., Han, E. J., Park, S., Swartzfager, D., Qi, N. S. X., Jung, M., Ocnean, M., Kim, H. U., & Lim, S. (2021). Predicting Dynamic Clinical Outcomes of the Chemotherapy for Canine Lymphoma Patients Using a Machine Learning Model. Veterinary Sciences, 8(12), 301. https://doi.org/10.3390/vetsci8120301







