Infectious Bronchitis Virus in Egypt: Genetic Diversity and Vaccination Strategies
Abstract
1. Introduction
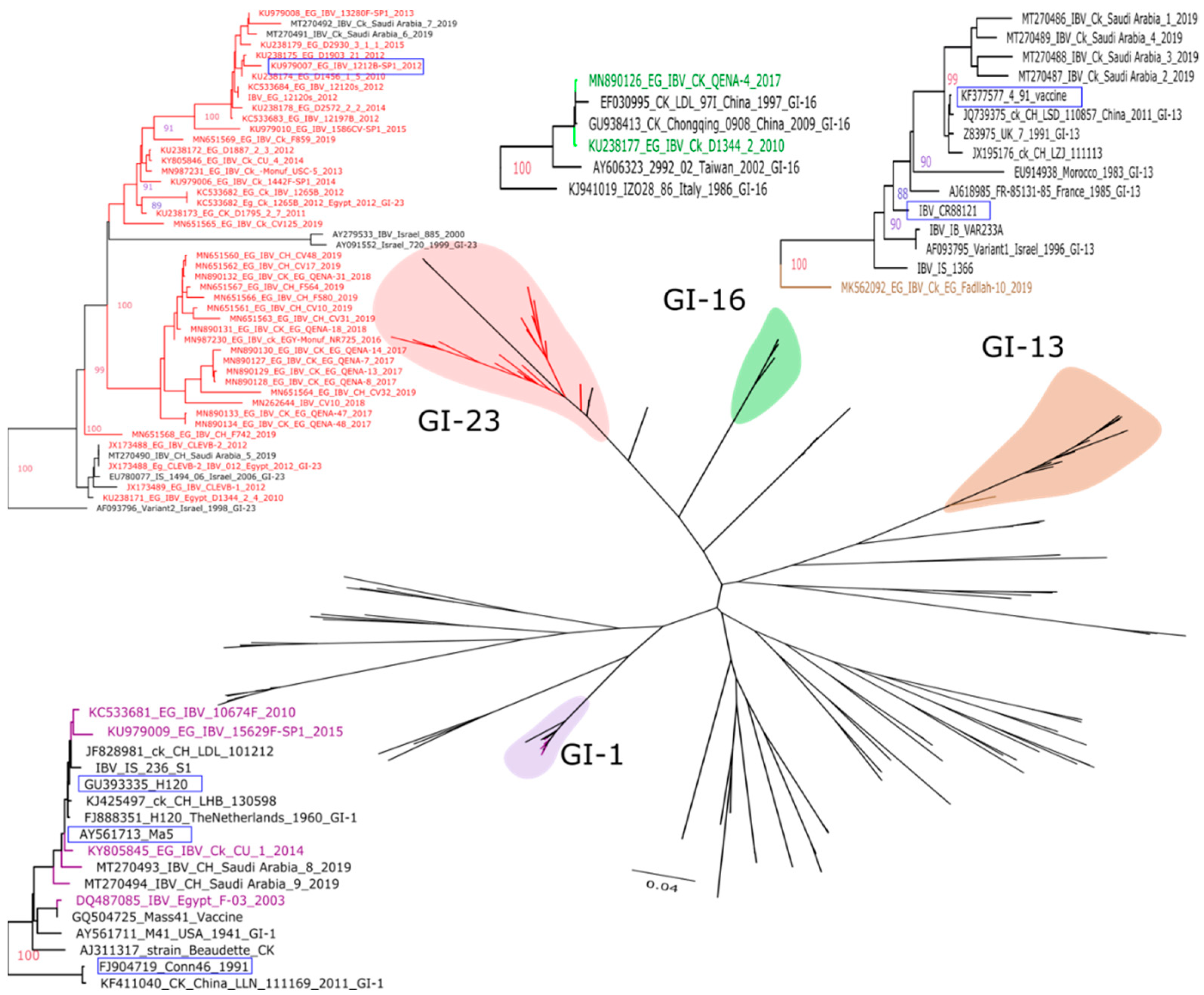
2. Multiple IBV Genotypes Circulating in Egypt
3. Genetic Drift and Recombination of the Egyptian IBV Strains
4. IBV Vaccines in Egypt
5. Perspectives and Recommendations for a Better Control Strategy
Author Contributions
Funding
Conflicts of Interest
References
- De Wit, J.J.S.; Cook, J.K.A. Spotlight on avian pathology: Infectious bronchitis virus. Avian Pathol. 2019, 48, 393–395. [Google Scholar] [CrossRef] [PubMed]
- Miłek, J.; Blicharz-Domańska, K. Coronaviruses in avian species—Review with focus on epidemiology and diagnosis in wild birds. J. Vet. Res. 2018, 62, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Rohaim, M.A.; El Naggar, R.F.; Helal, A.M.; Bayoumi, M.M.; El-Saied, M.A.; Ahmed, K.A.; Shabbir, M.Z.; Munir, M. Genetic diversity and phylodynamics of avian coronaviruses in Egyptian wild birds. Viruses 2019, 11, 57. [Google Scholar] [CrossRef]
- Payne, S. Chapter 17—Family coronaviridae. In Viruses; Payne, S., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 149–158. [Google Scholar] [CrossRef]
- Ennaji, Y.; Khataby, K.; Ennaji, M.M. Infectious bronchitis virus in poultry: Molecular epidemiology and factors leading to the emergence and reemergence of novel strains of infectious bronchitis virus. Emerg. Reemerg. Viral Pathog. 2020, 31–44. [Google Scholar] [CrossRef]
- De Wit, S.J.J.; Cook, J.K.A.; van der Heijden, H.M.J.F. Infectious bronchitis virus variants: A review of the history, current situation and control measures. Avian Pathol. 2011, 40, 223–235. [Google Scholar] [CrossRef]
- Valastro, V.; Holmes, E.C.; Britton, P.; Fusaro, A.; Jackwood, M.W.; Cattoli, G.; Monne, I. S1 gene-based phylogeny of infectious bronchitis virus: An attempt to harmonize virus classification. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2016, 39, 349–364. [Google Scholar] [CrossRef]
- Shan, D.; Fang, S.; Han, Z.; Ai, H.; Zhao, W.; Chen, Y.; Jiang, L.; Liu, S. Effects of hypervariable regions in spike protein on pathogenicity, tropism, and serotypes of infectious bronchitis virus. Virus Res. 2018, 250, 104–113. [Google Scholar] [CrossRef]
- Huang, Y.-P.; Lee, H.-C.; Cheng, M.-C.; Wang, C.-H. S1 and N gene analysis of avian infectious bronchitis viruses in Taiwan. Avian Dis. 2004, 48, 581–589. [Google Scholar] [CrossRef]
- Mase, M.; Tsukamoto, K.; Imai, K.; Yamaguchi, S. Phylogenetic analysis of avian infectious bronchitis virus strains isolated in Japan. Arch. Virol. 2004, 149, 2069–2078. [Google Scholar] [CrossRef]
- Abro, S.H.; Renström, L.H.; Ullman, K.; Isaksson, M.; Zohari, S.; Jansson, D.S.; Belák, S.; Baule, C. Emergence of novel strains of avian infectious bronchitis virus in Sweden. Vet. Microbiol. 2012, 155, 237–246. [Google Scholar] [CrossRef]
- De Wit, J.J.; Nieuwenhuisen-van Wilgen, J.; Hoogkamer, A.; van de Sande, H.; Zuidam, G.J.; Fabri, T.H. Induction of cystic oviducts and protection against early challenge with infectious bronchitis virus serotype D388 (genotype QX) by maternally derived antibodies and by early vaccination. Avian Pathol. 2011, 40, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Toffan, A.; Bonci, M.; Bano, L.; Bano, L.; Valastro, V.; Vascellari, M.; Capua, I.; Terregino, C. Diagnostic and clinical observation on the infectious bronchitis virus strain Q1 in Italy. Vet. Ital. 2013, 49, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Toffan, A.; Monne, I.; Terregino, C.; Cattoli, G.; Hodobo, C.T.; Gadaga, B.; Makaya, P.V.; Mdlongwa, E.; Swiswa, S. QX-like infectious bronchitis virus in Africa. Vet. Rec. 2011, 169, 589. [Google Scholar] [CrossRef] [PubMed]
- Ababneh, M.; Dalab, A.E.; Alsaad, S.; Al-Zghoul, M. Presence of infectious bronchitis virus strain CK/CH/LDL/97I in the Middle East. ISRN Vet. Sci. 2012, 2012, 201721. [Google Scholar] [CrossRef] [PubMed]
- Amin, O.G.M.; Valastro, V.; Salviato, A.; Drago, A.; Cattoli, G.; Monne, I. Circulation of QX-like infectious bronchitis virus in the Middle East. Vet. Rec. 2012, 171, 530. [Google Scholar] [CrossRef]
- Ahmed, H.N. Incidence and Treatment of Some Infectious Viral Respiratory Diseases of Poultry in Egypt. Ph.D. Thesis, Cairo University, Giza, Egypt, 1954. [Google Scholar]
- Davelaar, F.G.; Kouwenhoven, B.; Burger, A.G. Occurrence and significance of infectious bronchitis virus variant strains in egg and broiler production in the Netherlands. Vet. Q. 1984, 6, 114–120. [Google Scholar] [CrossRef]
- Taha, M.; Moustafa, M.M.; Mohamed, S.; Attia, S.A.; Hatem, M.E. Effect of both mycoplasma galliseplicum and infectious bronchitis virus classical and variant strains on the egg production of broiler breeder flocks. Vet. Med. 1991, 39, 169–179. [Google Scholar]
- Rohaim, M.A.; El Naggar, R.F.; Hamoud, M.M.; Bazid, A.I.; Gamal, A.M.; Laban, S.E.; Abdel-Sabour, M.A.; Nasr, S.A.E.; Zaki, M.M.; Shabbir, M.Z.; et al. Emergence and genetic analysis of variant pathogenic 4/91 (serotype 793/B) infectious bronchitis virus in Egypt during 2019. Virus Genes 2019, 55, 720–725. [Google Scholar] [CrossRef]
- Madbouly, H.M.; Abdel-Moneim, A.S.; Gelb, J.J.; Landman, B.S. Molecular characterization of three Egyptian isolates of infectious bronchitis virus. Vet. Med. J. Giza 2002, 50, 1053–1064. [Google Scholar]
- Abdel-Moneim, A.S.; Madbouly, H.M.; Gelb, J., Jr.; Ladman, B.S. Isolation and identification of Egypt/Beni-Suef/01 a novel genotype of infectious bronchitis virus. Vet. Med. J. Giza 2002, 50, 1065–1078. [Google Scholar]
- Meir, R.; Rosenblut, E.; Perl, S.; Kass, N.; Ayali, G.; Perk, S.; Hemsani, E. Identification of a novel nephropathogenic infectious bronchitis virus in Israel. Avian Dis. 2004, 48, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Abdel-Moneim, A.S.; El-Kady, M.F.; Ladman, B.S.; Gelb, J. S1 gene sequence analysis of a nephropathogenic strain of avian infectious bronchitis virus in Egypt. Virol. J. 2006, 3, 78. [Google Scholar] [CrossRef] [PubMed]
- Zanaty, A.; Arafa, A.S.; Hagag, N.; El-Kady, M. Genotyping and pathotyping of diversified strains of infectious bronchitis viruses circulating in Egypt. World J. Virol. 2016, 5, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Abozeid, H.H.; Paldurai, A.; Khattar, S.K.; Afifi, M.A.; El-Kady, M.F.; El-Deeb, A.H.; Samal, S.K. Complete genome sequences of two avian infectious bronchitis viruses isolated in Egypt: Evidence for genetic drift and genetic recombination in the circulating viruses. Infect. Genet. Evol. 2017, 53, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Selim, K.; Arafa, A.S.; Hussein, H.A.; El-Sanousi, A.A. Molecular characterization of infectious bronchitis viruses isolated from broiler and layer chicken farms in Egypt during 2012. Int. J. Vet. Sci. Med. 2013, 1, 102–108. [Google Scholar] [CrossRef]
- Abdel-Moneim, A.S.; Afifi, M.A.; El-Kady, M.F. Emergence of a novel genotype of avian infectious bronchitis virus in Egypt. Arch. Virol. 2012, 157, 2453–2457. [Google Scholar] [CrossRef]
- Zanaty, A.; Naguib, M.M.; El-Husseiny, M.H.; Mady, W.; Hagag, N.; Arafa, A.S. The sequence of the full spike S1 glycoprotein of infectious bronchitis virus circulating in Egypt reveals evidence of intra-genotypic recombination. Arch. Virol. 2016, 161, 3583–3587. [Google Scholar] [CrossRef]
- Moharam, I.; Sultan, H.; Hassan, K.; Ibrahim, M.; Shany, S.; Shehata, A.A.; Abo-ElKhair, M.; Pfaff, F.; Höper, D.; El Kady, M.; et al. Emerging infectious bronchitis virus (IBV) in Egypt: Evidence for an evolutionary advantage of a new S1 variant with a unique gene 3ab constellation. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2020, 85, 104433. [Google Scholar] [CrossRef]
- Sabra, M.; Abdellatif, W.; Ahmed, A.; Osman, N. Molecular characterization and phylogenetic analysis of full-length S1 gene of GI-16 and GI-23 infectious bronchitis virus in Qena, Egypt. J. World’s Poult. Res. 2020, 10, 71–80. [Google Scholar] [CrossRef]
- Abdel-Sabour, M.A.; Al-Ebshahy, E.M.; Khaliel, S.A.; Abdel-Wanis, N.A.; Yanai, T. Isolation and molecular characterization of novel infectious bronchitis virus variants from vaccinated broiler flocks in Egypt. Avian Dis. 2017, 61, 307–310. [Google Scholar] [CrossRef]
- Ghetas, A.M.; Kutkat, M.A.; Amer, M.M.; Awaad, M.H.H. Isolation and molecular identification of IBV isolates in different governorates in Egypt. J. Egypt. Soc. Parasitol. 2016, 46, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Lisowska, A.; Sajewicz-Krukowska, J.; Fusaro, A.; Pikula, A.; Domanska-Blicharz, K. First characterization of a Middle-East GI-23 lineage (Var2-like) of infectious bronchitis virus in Europe. Virus Res. 2017, 242, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Jiang, Y.; Low, S.; Wang, Z.; Nam, S.J.; Liu, W.; Kwang, J. Characterization of three infectious bronchitis virus isolates from China associated with proventriculus in vaccinated chickens. Avian Dis. 2001, 45, 416–424. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.W.; Jackwood, M.W. Evidence of genetic diversity generated by recombination among avian coronavirus IBV. Arch. Virol. 2000, 145, 2135–2148. [Google Scholar] [CrossRef]
- Jenkins, G.M.; Rambaut, A.; Pybus, O.G.; Holmes, E.C. Rates of molecular evolution in RNA viruses: A quantitative phylogenetic analysis. J. Mol. Evol. 2002, 54, 156–165. [Google Scholar] [CrossRef]
- Kusters, J.G.; Jager, E.J.; Lenstra, J.A.; Koch, G.; Posthumus, W.P.; Meloen, R.H.; van der Zeijst, B.A. Analysis of an immunodominant region of infectious bronchitis virus. J. Immunol. 1989, 143, 2692–2698. [Google Scholar]
- Kant, A.; Koch, G.; van Roozelaar, D.J.; Kusters, J.G.; Poelwijk, F.A.; van der Zeijst, B.A. Location of antigenic sites defined by neutralizing monoclonal antibodies on the S1 avian infectious bronchitis virus glycopolypeptide. J. Gen. Virol. 1992, 73 Pt 3, 591–596. [Google Scholar] [CrossRef]
- Cavanagh, D.; Davis, P.J.; Cook, J.K.; Li, D.; Kant, A.; Koch, G. Location of the amino acid differences in the S1 spike glycoprotein subunit of closely related serotypes of infectious bronchitis virus. Avian Pathol. 1992, 21, 33–43. [Google Scholar] [CrossRef]
- Pasternak, A.O.; Spaan, W.J.M.; Snijder, E.J. Nidovirus transcription: How to make sense…? J. Gen. Virol. 2006, 87, 1403–1421. [Google Scholar] [CrossRef]
- Simon-Loriere, E.; Holmes, E.C. Why do RNA viruses recombine? Nat. Rev. Microbiol. 2011, 9, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.M.; Jackwood, M.W. Molecular cloning and sequence comparison of the S1 glycoprotein of the Gray and JMK strains of avian infectious bronchitis virus. Virus Genes 1995, 9, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Thor, S.W.; Hilt, D.A.; Kissinger, J.C.; Paterson, A.H.; Jackwood, M.W. Recombination in avian gamma-coronavirus infectious bronchitis virus. Viruses 2011, 3, 1777–1799. [Google Scholar] [CrossRef]
- Naguib, M.M.; Höper, D.; Arafa, A.S.; Setta, A.M.; Abed, M.; Monne, I.; Beer, M.; Harder, T.C. Full genome sequence analysis of a newly emerged QX-like infectious bronchitis virus from Sudan reveals distinct spots of recombination. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2016, 46, 42–49. [Google Scholar] [CrossRef]
- Kiss, I.; Mató, T.; Homonnay, Z.; Tatár-Kis, T.; Palya, V. Successive occurrence of recombinant infectious bronchitis virus strains in restricted area of Middle East. Virus Evol. 2016, 2. [Google Scholar] [CrossRef]
- Ali, A.; Kilany, W.H.; Zain El-Abideen, M.A.; Sayed, M.E.; Elkady, M. Safety and efficacy of attenuated classic and variant 2 infectious bronchitis virus candidate vaccines. Poult. Sci. 2018, 97, 4238–4244. [Google Scholar] [CrossRef]
- Sultan, H.A.; Ali, A.; El Feil, W.K.; Bazid, A.H.I.; Zain El-Abideen, M.A.; Kilany, W.H. Protective efficacy of different live attenuated infectious bronchitis virus vaccination regimes against challenge with IBV variant-2 circulating in the Middle East. Front. Vet. Sci. 2019, 6, 341. [Google Scholar] [CrossRef]
- Cook, J.K.A.; Orbell, S.J.; Woods, M.A.; Huggins, M.B. Breadth of protection of the respiratory tract provided by different live-attenuated infectious bronchitis vaccines against challenge with infectious bronchitis viruses of heterologous serotypes. Avian Pathol. 1999, 28. [Google Scholar] [CrossRef]
- Cavanagh, D.; Naqi, S.A. Infectious Bronchitis. Dis. Poult. 2003, 11, 101–119. [Google Scholar] [CrossRef]
- Hassan, K.E.; Shany, S.A.S.; Ali, A.; Dahshan, A.-H.M.; El-Sawah, A.A.; El-Kady, M.F. Prevalence of avian respiratory viruses in broiler flocks in Egypt. Poult. Sci. 2016, 95, 1271–1280. [Google Scholar] [CrossRef]
- Abozeid, H.H.; Paldurai, A.; Varghese, B.P.; Khattar, S.K.; Afifi, M.A.; Zouelfakkar, S.; El-Deeb, A.H.; El-Kady, M.F.; Samal, S.K. Development of a recombinant Newcastle disease virus-vectored vaccine for infectious bronchitis virus variant strains circulating in Egypt. Vet. Res. 2019, 50, 12. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.T.; Wu, H.Y.; Wang, C.H. Genetic sequence changes related to the attenuation of avian infectious bronchitis virus strain TW2575/98. Virus Genes 2020, 56, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Jackwood, M.W.; Hall, D.; Handel, A. Molecular evolution and emergence of avian gammacoronaviruses. Infect. Genet. Evol. 2012, 12, 1305. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abozeid, H.H.; Naguib, M.M. Infectious Bronchitis Virus in Egypt: Genetic Diversity and Vaccination Strategies. Vet. Sci. 2020, 7, 204. https://doi.org/10.3390/vetsci7040204
Abozeid HH, Naguib MM. Infectious Bronchitis Virus in Egypt: Genetic Diversity and Vaccination Strategies. Veterinary Sciences. 2020; 7(4):204. https://doi.org/10.3390/vetsci7040204
Chicago/Turabian StyleAbozeid, Hassanein H., and Mahmoud M. Naguib. 2020. "Infectious Bronchitis Virus in Egypt: Genetic Diversity and Vaccination Strategies" Veterinary Sciences 7, no. 4: 204. https://doi.org/10.3390/vetsci7040204
APA StyleAbozeid, H. H., & Naguib, M. M. (2020). Infectious Bronchitis Virus in Egypt: Genetic Diversity and Vaccination Strategies. Veterinary Sciences, 7(4), 204. https://doi.org/10.3390/vetsci7040204





