A Century of Swine Influenza: Is It Really Just about the Pigs?
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Publications Related to Host
3.1.1. Annual Scientific Production
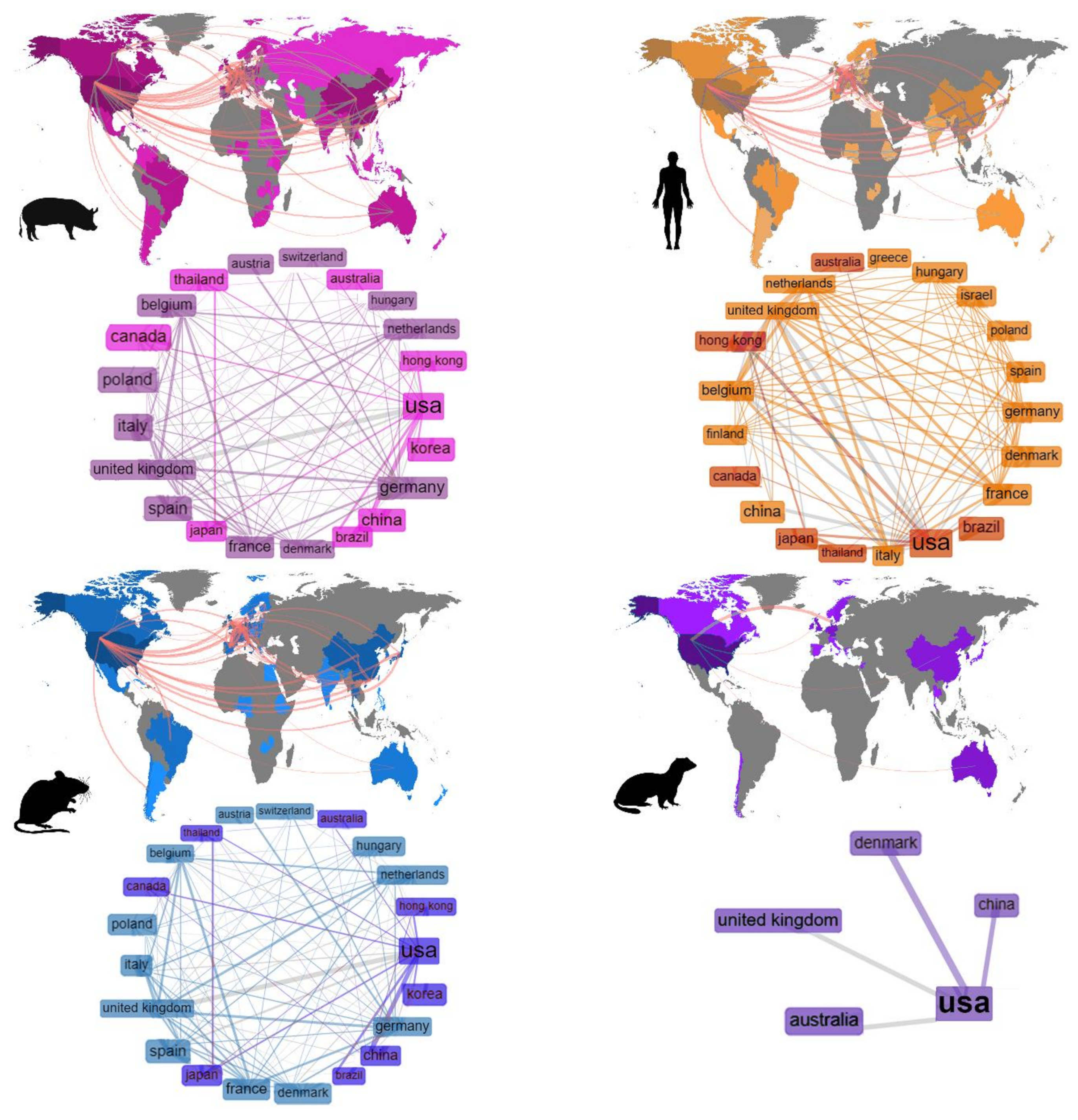
3.1.2. Collaboration Networks
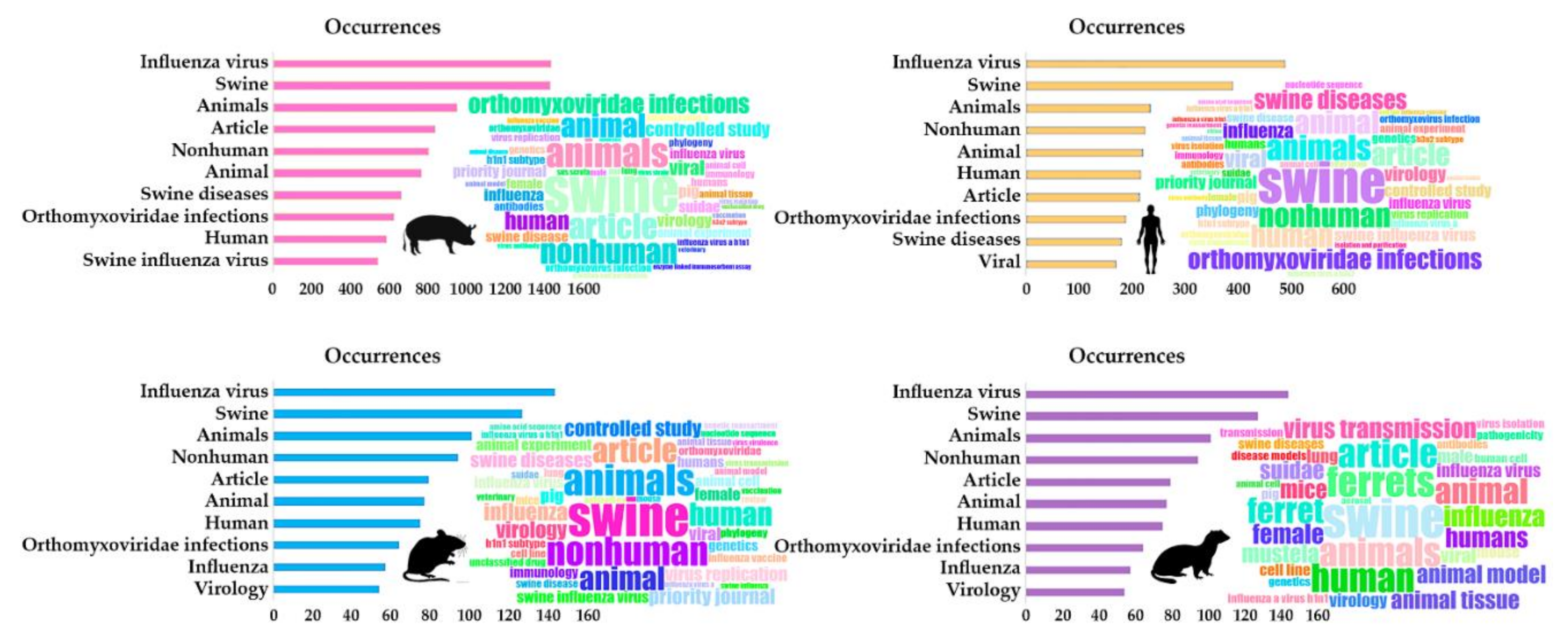
3.1.3. Keywords
3.2. Publications Related to SwIV Subtypes
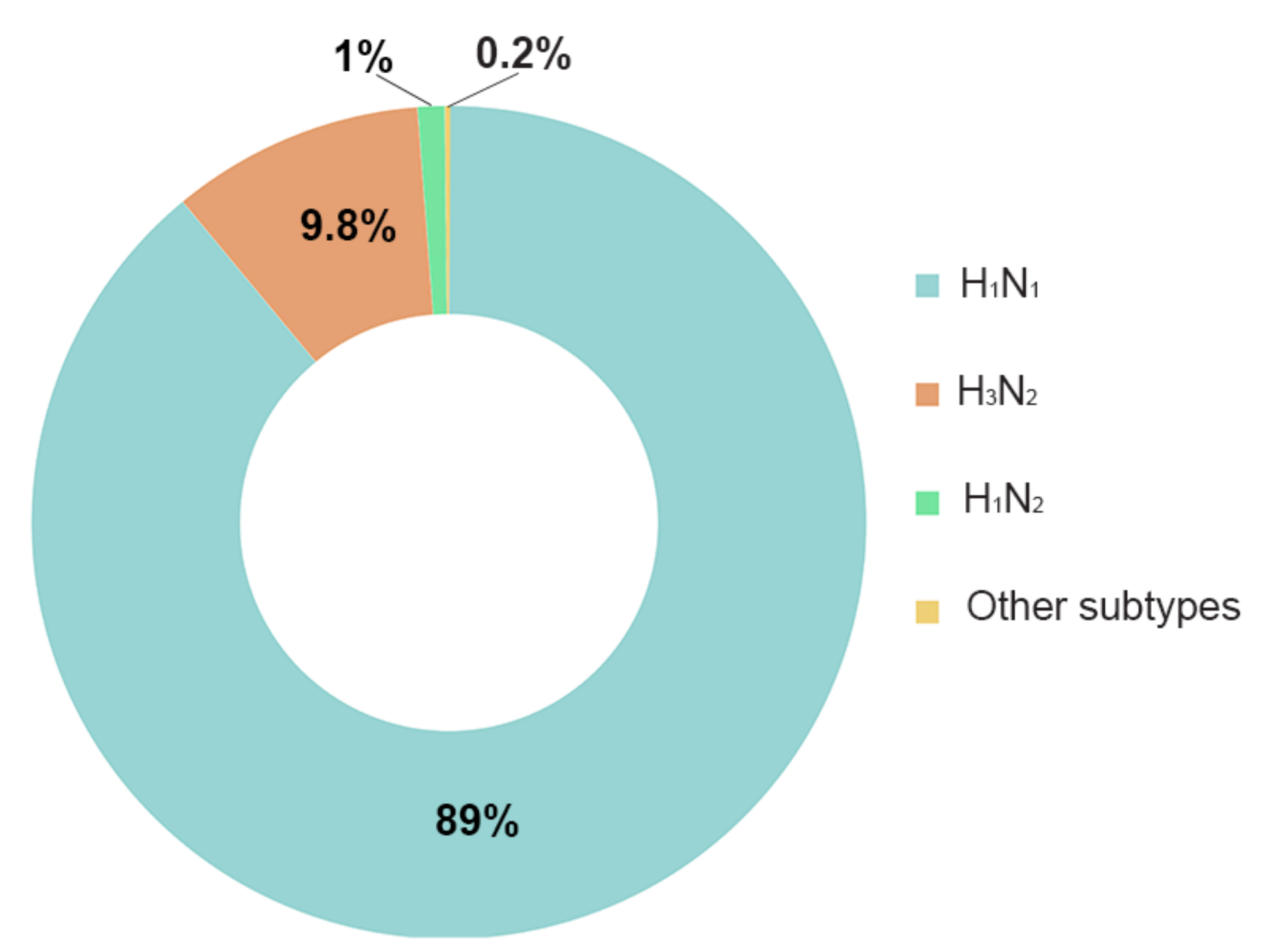
3.2.1. Most Common Subtypes
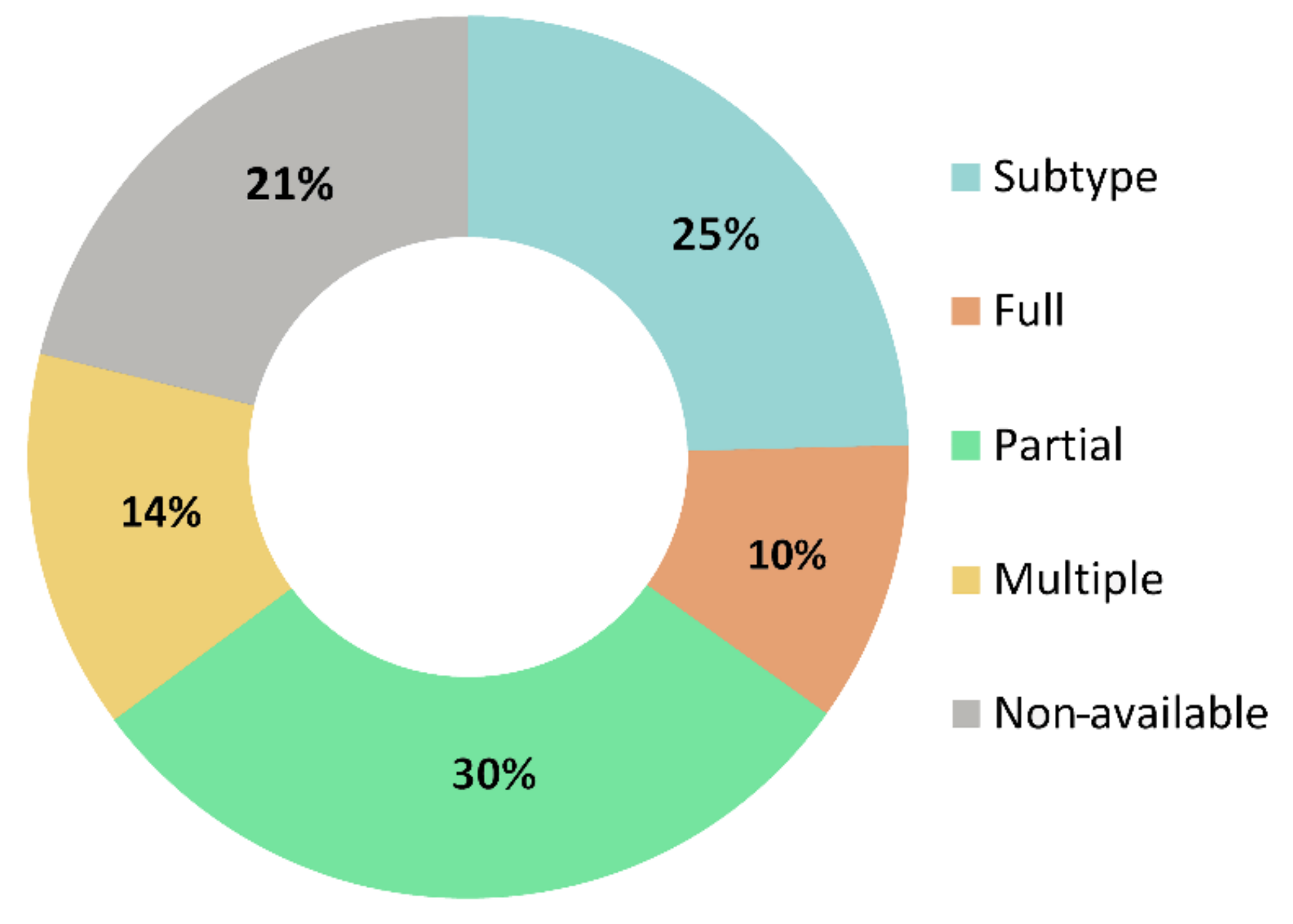
3.2.2. Available Genetic Information
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Crisci, E.; Mussá, T.; Fraile, L.; Montoya, M. Influenza virus in pigs. Mol. Immunol. 2013, 55, 200–211. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Key Facts about Swine Influenza (Swine Flu) in Pigs; CDC: Atlanta, GA, USA, 2014. [Google Scholar]
- Castrucci, M.R.; Donatelli, I.; Sidoli, L.; Barigazzi, G.; Kawaoka, Y.; Webster, R.G. Genetic reassortment between avian and human influenza A viruses in Italian pigs. Virology 1993, 193, 503–506. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Origin of 2009 H1N1 Flu (Swine Flu): Questions and Answers; CDC: Atlanta, GA, USA, 2009. [Google Scholar]
- Chastagner, A.; Enouf, V.; Peroz, D.; Hervé, S.; Lucas, P.; Quéguiner, S.; Gorin, S.; Beven, V.; Behillil, S.; Leneveu, P.; et al. Bidirectional Human–Swine Transmission of Seasonal Influenza A (H1N1) pdm09 Virus in Pig Herd, France, 2018. Emerg. Infect. Dis. 2019, 25, 1940. [Google Scholar] [CrossRef] [PubMed]
- Kida, H.; Ito, T.; Yasuda, J.; Shimizu, Y.; Itakura, C.; Shortridge, K.F.; Kawaoka, Y.; Webster, R.G. Potential for transmission of avian influenza viruses to pigs. J. Gen. Virol. 1994, 75, 2183–2188. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Kahn, R.E.; Richt, J.A. The pig as a mixing vessel for influenza viruses: Human and veterinary implications. J. Mol. Genet. Med. Int. J. Biomed. Res. 2009, 3, 158. [Google Scholar] [CrossRef]
- Parrish, C.R.; Voorhees, I.E.H. H3N8 and H3N2 canine influenza viruses: Understanding these new viruses in dogs. Vet. Clin. Small Anim. Pract. 2019, 49, 643–649. [Google Scholar] [CrossRef]
- Scholtissek, C. Pigs as ‘mixing vessels’ for the creation of new pandemic influenza A viruses. Med. Princ. Pract. 1990, 2, 65–71. [Google Scholar] [CrossRef]
- Swayne, D.E.; Pantin-Jackwood, M.J.; Kapczynski, D.; Spackman, E.; Suarez, D.L. Susceptibility of Poultry to Pandemic (H1N1) 2009 Virus. Emerg. Infect. Dis. 2009, 15, 2061–2063. [Google Scholar] [CrossRef]
- King, A.M.; Lefkowitz, E.; Adams, M.J.; Carstens, E.B. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier: Amsterdam, The Netherlands, 2011; Volume 9. [Google Scholar]
- Memorandums, M.-L. A revision of the system of nomenclature for influenza viruses: A WHO memorandum. Bull. World Health Organ. 1980, 58, 585–591. [Google Scholar]
- Van Der Schot, A.A.; Phillips, C.J. Publication bias in animal welfare scientific literature. J. Agric. Environ. Ethics 2013, 26, 945–958. [Google Scholar] [CrossRef]
- Colquhoun, H.L.; Levac, D.; O’Brien, K.K.; Straus, S.; Tricco, A.C.; Perrier, L.; Kastner, M.; Moher, D. Scoping reviews: Time for clarity in definition, methods, and reporting. J. Clin. Epidemiol. 2014, 67, 1291–1294. [Google Scholar] [CrossRef] [PubMed]
- Pham, M.T.; Rajić, A.; Greig, J.D.; Sargeant, J.M.; Papadopoulos, A.; McEwen, S.A. A scoping review of scoping reviews: Advancing the approach and enhancing the consistency. Res. Synth. Methods 2014, 5, 371–385. [Google Scholar] [CrossRef] [PubMed]
- Peters, M.D.; Godfrey, C.M.; Khalil, H.; McInerney, P.; Parker, D.; Soares, C.B. Guidance for conducting systematic scoping reviews. Int. J. Evid. Based Healthc. 2015, 13, 141–146. [Google Scholar] [CrossRef]
- Almeida, F.; De Paula, L.G. The Place of Uncertainty in Heterodox Economics Journals: A Bibliometric Study. J. Econ. Issues 2019, 53, 553–562. [Google Scholar] [CrossRef]
- Alonso, J.M.; Castiello, C.; Mencar, C. A bibliometric analysis of the explainable artificial intelligence research field. In Proceedings of the International Conference on Information Processing and Management of Uncertainty in Knowledge-Based Systems, Cádiz, Spain, 11–15 June 2018; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Anglada-Tort, M.; Sanfilippo, K.R.M. Visualizing Music Psychology: A Bibliometric Analysis of Psychology of Music, Music Perception, and Musicae Scientiae from 1973 to 2017. Music Sci. 2019, 2, 2059204318811786. [Google Scholar] [CrossRef]
- Arfaoui, A.; Ibrahimi, K.; Trabelsi, F. Biochar application to soil under arid conditions: A bibliometric study of research status and trends. Arab. J. Geosci. 2019, 12, 45. [Google Scholar] [CrossRef]
- Ballandonne, M. The historical roots (1880–1950) of recent contributions (2000–2017) to ecological economics: Insights from reference publication year spectroscopy. J. Econ. Methodol. 2019, 26, 307–326. [Google Scholar] [CrossRef]
- Kimberly, V.W.; Deen, J. Global trends in infectious diseases of swine. Proc. Natl. Acad. Sci. USA 2018, 115, 11495–11500. [Google Scholar]
- Nafade, V.; Nash, M.; Huddart, S.; Pande, T.; Gebreselassie, N.; Lienhardt, C.; Pai, M. A bibliometric analysis of tuberculosis research, 2007–2016. PLoS ONE 2018, 13, e0199706. [Google Scholar] [CrossRef]
- Wiethoelter, A.K.; Beltrán-Alcrudo, D.; Kock, R.; Mor, S.M. Global trends in infectious diseases at the wildlife–livestock interface. Proc. Natl. Acad. Sci. USA 2015, 112, 9662–9667. [Google Scholar] [CrossRef]
- Moral-Munoz, J.A.; Herrera-Viedma, E.; Espejo, A.S.; Cobo, M.-J. Software tools for conducting bibliometric analysis in science: An up-to-date review. El Prof. Inf. 2020, 29, e290103. [Google Scholar] [CrossRef]
- Baskaran, C.; Sivakami, N. Swine influenza research output: A bibliometric analysis. SRELS J. Inf. Manag. 2014, 51, 13–20. [Google Scholar]
- Ramakrishnan, J.; Sankar, G.R.; Thavamani, K. Literature on Swine Flu: A Bibliometric Analysis. Int. J. Inf. Dissem. Technol. 2016, 6, 193–198. [Google Scholar]
- Aria, M.; Cuccurullo, C. bibliometrix: An R-tool for comprehensive science mapping analysis. J. Informetr. 2017, 11, 959–975. [Google Scholar] [CrossRef]
- WHO. Standardization of Terminology for the Influenza Virus Variants Infecting Humans: Update; WHO: Geneva, Switzerland, 2014. [Google Scholar]
- Viboud, C.; Grais, R.F.; Lafont, B.A.P.; Miller, M.A.; Simonsen, L. Multinational impact of the 1968 Hong Kong influenza pandemic: Evidence for a smoldering pandemic. J. Infect. Dis. 2005, 192, 233–248. [Google Scholar] [CrossRef] [PubMed]
- Brooke, Z.M.; Bielby, J.; Nambiar, K.; Carbone, C. Correlates of research effort in carnivores: Body size, range size and diet matter. PLoS ONE 2014, 9, e93195. [Google Scholar] [CrossRef]
- Mongeon, P.; Paul-Hus, A. The journal coverage of Web of Science and Scopus: A comparative analysis. Scientometrics 2016, 106, 213–228. [Google Scholar] [CrossRef]
- Frick, M. 2016 Report on Tuberculosis Research Funding Trends, 2005–2015: No Time to Lose; Treatment Action Group: New York, NY, USA, 2016. [Google Scholar]
- Bai, J.; Li, W.; Huang, Y.-M.; Guo, Y. Bibliometric study of research and development for neglected diseases in the BRICS. Infect. Dis. Poverty 2016, 5, 89. [Google Scholar] [CrossRef]
- Johnson, P.N.; Mueller, J. Updating the accounts: Global mortality of the 1918-1920 “Spanish” influenza pandemic. Bull. Hist. Med. 2002, 76, 105–115. [Google Scholar] [CrossRef]
- Taubenberger, K.J.; Moren, D.M. 1918 Influenza: The mother of all pandemics. Rev. Biomed. 2006, 17, 69–79. [Google Scholar] [CrossRef]
- Nelson, M.I.; Viboud, C.; Simonsen, L.; Bennett, R.T.; Griesemer, S.B.; George, K.S.; Taylor, J.; Spiro, D.J.; Sengamalay, N.A.; Ghedin, E.; et al. Multiple reassortment events in the evolutionary history of H1N1 influenza A virus since 1918. PLoS Pathog. 2008, 4, e1000012. [Google Scholar] [CrossRef]
- Nelson, M.I.; Viboud, C.; Simonsen, L.; Bennett, R.T.; Griesemer, S.B.; George, K.S.; Taylor, J.; Spiro, D.J.; Sengamalay, N.A.; Ghedin, E.; et al. The emergence of pandemic influenza viruses. Protein Cell 2010, 1, 9–13. [Google Scholar]
- Anderson, T.K.; Macken, C.A.; Lewis, N.S.; Scheuermann, R.H.; Van Reeth, K.; Brown, I.H.; Swenson, S.L.; Simon, G.; Saito, T.; Berhane, Y.; et al. A phylogeny-based global nomenclature system and automated annotation tool for H1 hemagglutinin genes from swine influenza A viruses. MSphere 2016, 1, e00275-16. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frias-De-Diego, A.; Posey, R.; Pecoraro, B.M.; Fernandes Carnevale, R.; Beaty, A.; Crisci, E. A Century of Swine Influenza: Is It Really Just about the Pigs? Vet. Sci. 2020, 7, 189. https://doi.org/10.3390/vetsci7040189
Frias-De-Diego A, Posey R, Pecoraro BM, Fernandes Carnevale R, Beaty A, Crisci E. A Century of Swine Influenza: Is It Really Just about the Pigs? Veterinary Sciences. 2020; 7(4):189. https://doi.org/10.3390/vetsci7040189
Chicago/Turabian StyleFrias-De-Diego, Alba, Rachael Posey, Brittany M. Pecoraro, Rafaella Fernandes Carnevale, Alayna Beaty, and Elisa Crisci. 2020. "A Century of Swine Influenza: Is It Really Just about the Pigs?" Veterinary Sciences 7, no. 4: 189. https://doi.org/10.3390/vetsci7040189
APA StyleFrias-De-Diego, A., Posey, R., Pecoraro, B. M., Fernandes Carnevale, R., Beaty, A., & Crisci, E. (2020). A Century of Swine Influenza: Is It Really Just about the Pigs? Veterinary Sciences, 7(4), 189. https://doi.org/10.3390/vetsci7040189






