1. Introduction
Bovine respiratory disease (BRD) is a significant cause of morbidity and mortality among beef cattle globally.
Pasteurella multocida (
P. multocida) and
Mannheimia haemolytica (
M. haemolytica), which are commensal Gram-negative bacteria in the upper respiratory tract of animals, represent the major bacterial causative agents for BRD [
1]. Under stresses including environmental, managemental, and/or infectious factors, those agents often produce mild to severe clinical signs. The incubation period varies from 3 to 5 days. In peracute cases, sudden death within 24–36 h with clear clinical signs may be observed. In chronic cases, they may cause permanent lung damage such as fibrosis, adhesions and/or abscesses that are affecting the performance [
1]. The clinical manifestations include a rise in temperature, respiratory distress with nasal discharge, and frothing from the mouth, and then recumbency and death may be the result [
2]. Disease incidence is most often in 6–24 month-old animals and groups of less than 10 animals. The disease was seasonal, occurring only in rainy seasons of the year, and victims were only cattle and buffaloes [
2]. Thus, early recognition and treatment of BRD are so important [
3]. Respiratory disorders in animal production sectors in Egypt were reported to cause a considerable economic loss due to lower productivity and death [
4].
Pasteurella multocida is a zoonotic bacterium causing hemorrhagic septicaemia (HS), which is a major disease of cattle and buffaloes occurring as catastrophic epizootics in many African and Asian countries characterized by an acute, highly fatal septicemia with high morbidity and mortality [
3]. In humans,
P. multocida has been tightly associated with animal exposure, usually involving soft-tissue sites within 24 h after animal bites, especially dog or cat, or scratch wounds. Serious respiratory tract infections including pneumonia, empyema, and lung abscesses are typically found in patients with underlying pulmonary disease. In more serious cases, a bacteremia can occur by spreading from a localized bite wound or from another localized source of infection, such as pneumonia, meningitis, or arthritis. Varieties of other serious invasive infections such as meningitis, endocarditis, and peritonitis, have also been reported, but are rare [
5].
HS is caused by certain serotypes of
P. multocida including B:2 in Asia and E:2 in Africa based on the Carter and Heddleston system, which is corresponding to the newer B:6 and E:6 serotypes in Namioka–Carter classification [
6]. OIE added that A:1 and A:3 serotypes have been incriminated in an HS-like syndrome in bovines in India with mainly characterized by pneumonia ended with death. On the other hand,
M. haemolytica (formerly
Pasteurella haemolytica) is considered one of the most important pathogens in ruminants of all ages and it is the principal cause of bovine and ovine pneumonic pasteurellosis or shipping fever. It is responsible for considerable economic losses to the livestock industries all over the world [
3,
7]. Two biotypes have been recognized for the taxon
Pasteurella haemolytica: biotype A, an isolate that ferments L-arabinose, and biotype T, an isolate that ferments trehalose [
8]. Based on capsular polysaccharide antigen typing using the indirect haemagglutination test,
P. haemolytica complex has been identified as 17 serotypes; including 13 A serotypes (A:1, A:2, A:5, A:6, A:7, A:8, A:9, A:11, A:12, A:13, A:14, A:16, and A:17) and 4 T serotypes (serotypes 3, 4, 10 and 15) [
8].
P. haemolytica biotype A was later allocated to a new genus
Mannheimia and renamed as
M. haemolytica, while the four T serotypes were named
Bibersteinia trehalosi. Later, serotype A:11 was classified into a new taxon as
M. glucosida, due to its different biochemical profile, leaving 12 serotypes of
M. haemolytica [
9].
Many virulence genes are important in the pathogenesis of
P. multocida, such as fimbriae and adhesins (
nanB and
nanH), a variety of outer membrane proteins (OMPs) such as protectins (
ompA,
ompH,
omp87 and
plpB), and toxins such as dermonecrotoxin (
toxA) [
1,
10,
11]. These virulence factors facilitate the colonization and invasion of
P. multocida through impairing the host defense mechanisms, destruction of host tissues, and/or stimulation of a noxious host inflammatory response [
12]. The
toxA and the OMPs-encoding genes have been suggested as epidemiological markers as they are found in high prevalence in pneumonic
P. multocida isolates [
13].
M. haemolytica can colonize and establish infection in the lungs due to various virulence factors, including capsule, adhesins, lipopolysaccharide (LPS), OMPs, and various proteases [
14]. The virulence of
M. haemolytica is linked to different virulence genes, such as leukotoxin (
lkt), leukotoxin C (
lktC), putative adhesin (
ahs), O-sialoglycoprotease (
gcp), outer membrane lipoprotein (
gs60), transferring-binding protein B (
tbpB) and UDP-N-acetyl-D-glucosamine-2-epimerase (
nmaA). Characterization of these genes provides important information about the pathogenicity of
M. haemolytica [
14,
15]
The present study was designed to perform serotyping and genotyping of P. multocida and M. haemolytica isolates obtained from pneumonic calves in North Upper Egypt Governorates, as well as characterization of virulence-associated genes in both bacteria.
4. Discussion
Bacterial infections causing pneumonia in calves can be fatal. The pathogens
P. multocida and
M. haemolytica are the two most common bacterial agents causing calves’ pneumonia in Egypt and worldwide [
7]. Respiratory disorders in animal production units in Egypt were reported to cause a considerable loss due to lower productivity and death [
4]. One of the challenges of bovine respiratory medicine is the early recognition and treatment of clinical cases of BRD. As
P. multocida and
M. haemolytica are commensals in the upper respiratory tract of animals, they represent the main bacterial etiology for BRD. Successful treatment occurs if antibiotics were given at the initial stages of the disease [
27].
Pasteurella multocida-polymerase chain reaction (PM-PCR) and capsular PCR assays are highly specific rapid efficient tools for the diagnosis of
Pasteurella species, especially in epidemiological studies [
28,
29]. Capsular PCR assay was found to be a very convenient and reliable method for serogrouping of
P. multocida, in contrast to the conventional serogrouping method, which is very slow and requires the production and maintenance of a battery of hyperimmune sera, which is difficult to produce [
29,
30]. Primers for
P. multocida were designed to detect a fragment of the
kmt1 gene encoding the outer membrane protein, producing an amplification product unique to all strains of
P. multocida [
31]. The present PCR method was designed to identify
P. multocida serogroup B strains by amplification of the
kmt1 gene with genotyping of it depending on the cap loci at
bcbD that is highly specific for serogroup B.
The present study focused on molecular identification as well as serotyping and genotyping of
P. multocida and M. haemolytica isolates from pneumonic calves in North Upper Egypt governorates. The PCR assay was applied as a confirmatory identification of
P. multocida and
M. haemolytica isolates using the
kmt1 and
rpt2 universal genes, respectively. The results of PCR reveal that 87.9% and 100% of
P. multocida isolates and
M. haemolytica isolates, respectively, were positive for the corresponding universal gene. The PCR results of
P. multocida using the
kmt1 gene are in agreement with the results obtained by Balakrishnan and Roy [
32], who identified
P. multocida strains recovered from sheep using the primers
kmt1. In addition, Abbas et al. [
31] identified the
P. multocida isolates from different hosts using the primers
kmt1 and they also considered these primers unique to all strains of
P. multocida. Meanwhile, the obtained results for the detection of the
rpt2 gene in
M. haemolytica isolates are similar to those previously studied by Ryan and Lo [
33]. Serotyping of
P. multocida and
M. haemolytica was conducted only on the PCR-positive isolates for universal genes. The results of the serotyping of
P. multocida isolates (
n = 29) using capsular type B antisera indicate that 86.2% isolates were serotype B:2, while 13.8% of isolates were untyped. These results run hand in hand with reports by Elshemey and Abd-Elrahman [
2], who serotyped fifty
P. multocida isolates from HS outbreak in cattle and buffalo in Alexandria province and found
P. multocida type B:2 in 100% of strains. In addition, Abbas et al. [
31] reported that 100% of
P. multocida isolates from HS cases of cattle and buffalo in different governorates of Egypt belonged to serotype B:2. Regarding the untyped
P. multocida isolates, after capsular and somatic antigen detection, some of the isolates cannot be differentiated because they may react similarly in both the antigens [
34]. Furthermore, the agglutination of homologous antiserum may fail [
31]. Passive hemagglutination has a substantial concern, as that test can be rendered ineffective by the loss of
P. multocida capsule after repeated subcultures in vitro [
35]. Moreover, the agglutination failure of serogroups A, D, and F with homologous antisera is one of the main causes of reduced sensitivity in this phenotypic test [
36]. We speculated that the untyped isolates might be related to serogroups other than B, especially capsular group E, which is the most common in Africa. This point is supported by what was reported by OIE [
6] and Farooq et al. [
27], in that serotypes B:2 and E:2 were the most common serotypes of
P. multocida associated with HS in animals in Asia and Africa, respectively.
The serotyping of
M. haemolytica isolates (
n = 15) showed that 60% and 40% of isolates belonged to serotype 2 and serotype 1, respectively. These results were supported by other studies [
15,
37,
38], in which
M. haemolytica serotypes (A:1, A:2, and A:6) were the most prevalent isolates recovered from cattle with BRD. Singh et al. [
14] and Kabeta et al. [
39] reported that HS is mainly caused by
M. haemolytica serotype 1, and the disease is most commonly found in calves. Apart from outer membrane proteins and capsular antigens, the virulence-associated genes (
toxA,
nanB,
oma87, and others) are playing important roles in the pathogenesis of
P. multocida [
40]. These virulence factors facilitate the colonization and invasion of
P. multocida through impairing the host defense mechanisms, destruction of host tissues, and/or stimulation of a noxious host inflammatory response [
12]. The
toxA and the OMPs-encoding genes have been suggested as epidemiological markers and are found in high prevalence in pneumonic
P. multocida isolates [
13]. PCR-based methods have been used to ascertain their distribution in strains recovered from a wide range of sources and disease conditions [
40]. The virulence of
M. haemolytica is linked to different virulence genes including
lkt, especially
lktC,
gcp, and other genes, and characterization of these genes provides important information about the pathogenicity of
M. haemolytica [
14,
15].
In the current study, PCR assay was applied on five MDR
P. multocida isolates to determine
nanB,
omp87, and
toxA virulence genes and on five
M. haemolytica isolates to determine
ssa,
gcp, and
lktC virulence genes. The results indicate that all the tested
P. multocida isolates harbored all the tested genes (100%); meanwhile, four of the tested
M. haemolytica isolates (80%) harbored both
gcp and
lktC, of which three isolates only (60%) harbored
ssa gene. The obtained PCR results of
P. multocida using
the toxA gene agreed with data obtained by Devi et al. [
41], who concluded that the
toxA gene is an important marker gene for defining the pathogenic potential of
P. multocida strains. Meanwhile, Ewers et al. [
40] recorded that the
toxA was found only in 12.5% of all isolates from small ruminants in Germany. In addition, Vougidou et al. [
13] reported that some of the genes including the
toxA and the OMPs-encoding genes have been suggested as epidemiological markers. On the contrary, Sarangi et al. [
19] reported that all the virulence-associated genes, except the
toxA gene, were found to be regularly distributed among
P. multocida isolates. The results of PCR using fimbriae and adhesins encoding gene (
nanB) and outer membrane proteins encoding gene (
omp87) are supported by those reported by Katsuda et al. [
11], who analyzed 378
P. multocida isolates using PCR and detected the presence of
nanB and
omp87 in most of the isolates. In addition, Jamali et al. [
10] examined 141
P. multocida isolates for the detection of different virulence genes and found that
omp87 and
nanB genes were present in all isolates. On the other hand, the obtained PCR results of
M. haemolytica are similar to those recorded by Singh et al. [
14], who recorded
lkt as species-specific for ruminants. In addition, Klima et al. [
15] detected
lktC and
gcp in all
M. haemolytica isolates. Meanwhile, Klima et al. [
42] recorded OMPs serine protease encoding (
ssa) gene as one of the top ten antigens detected among 240
M. haemolytica. Moreover, Ayalew et al. [
43] previously identified
M. haemolytica OMPs that may be an important immunogen, including serotype 1-specific antigen (
ssa1) by using immunoproteomic analyses.
In the present study, the
toxA gene of
P. multocida serotype B:2 and
lktC gene of
M. haemolytica serotype 2 (the most prevalent serotype) were sequenced. Regarding
P. multocida toxA gene, amino acid and nucleotide sequence analysis showed great homology between the Egyptian strain and the fourteen
P. multocida strains uploaded in gene bank representing different clinical lesions, hosts, and localities worldwide. Generally, the sequence identities between the isolated Egyptian strain and different
P. multocida strains uploaded from gene bank revealed 99.4–100% homology. This could suggest the high pathogenicity of the isolated strain and its high affinity to cause respiratory problems in infected animals [
44], where
toxA plays an important in destruction of lung tissues, and/or stimulation of a noxious host inflammatory response. Therefore, the
toxA gene is considered an epidemiological marker found mostly in pneumonic
P. multocida isolates [
12]. Pullinger et al. [
45] reported that the
P. multocida toxin (PMT) acts as a potent mitogen. Sequence analysis of the structural gene for PMT,
toxA, suggested that it was horizontally acquired because it had a low G+C content relative to the
P. multocida genome [
45]. Concerning
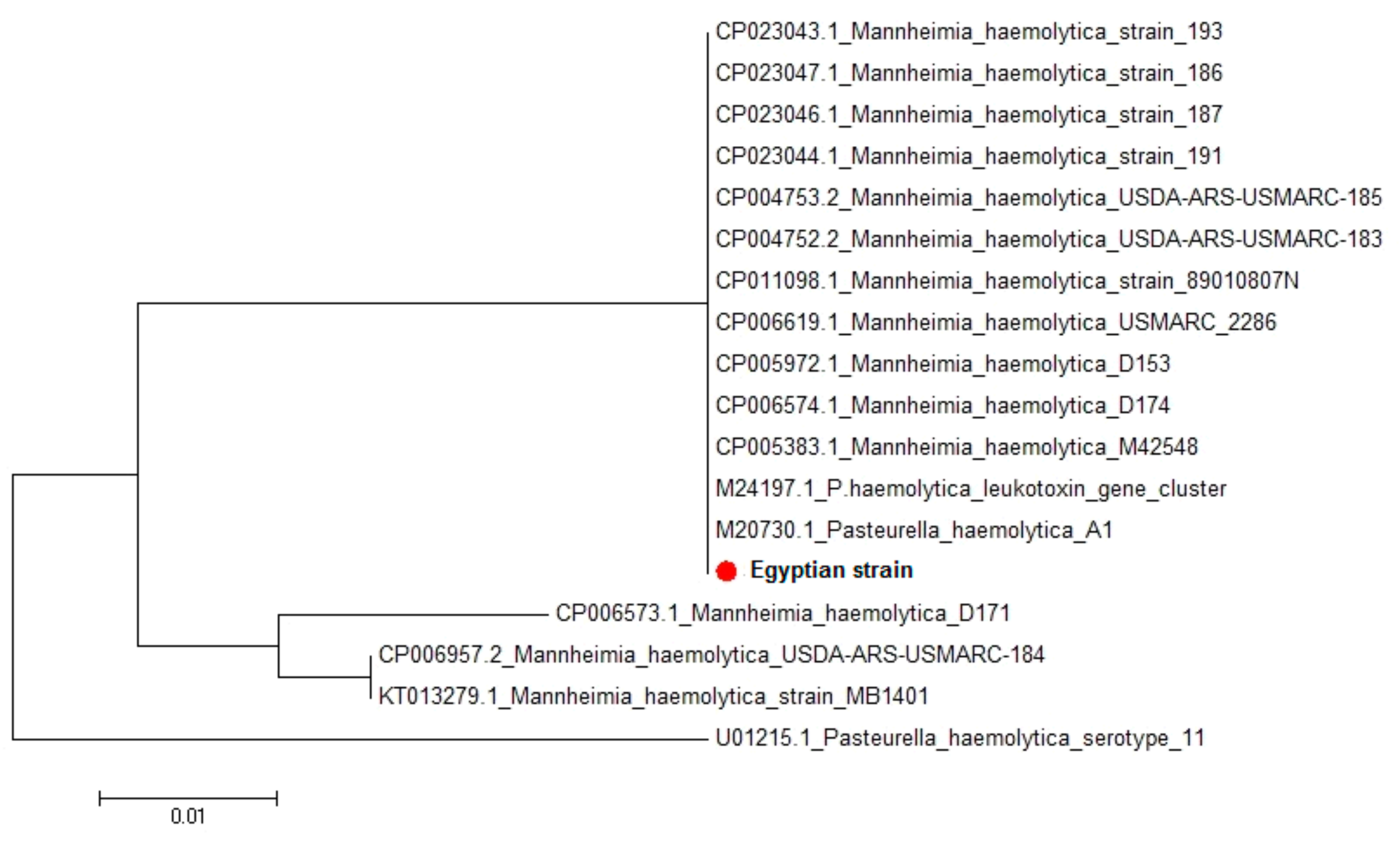
M. haemolytica lktC gene, amino acid and nucleotide sequence analysis also indicated great homology between the Egyptian strain and the seventeen
M. haemolytica strains uploaded in genebank representing different clinical lesions, hosts, and localities worldwide. Sequence identities between the isolated Egyptian strain and different
M. haemolytica strains uploaded in genebank revealed 92.6–100% homology. This leukotoxin has been implicated as a major virulence factor in the pathogenesis of
M. haemolytica, helping in the colonization and invasion of the lung tissues by impairing the primary lung defense mechanism and subsequent immune response or by the induction of inflammation as a consequence of leukocyte lysis [
46]. Therefore, characterization of such a gene provides important information about the pathogenicity of
M. haemolytica [
14]. Moreover, Highlander et al. [
47] reported that
M. haemolytica secreted a 102-kilodalton leukotoxin that was believed to be involved in the pathogenesis of severe bovine pneumonia and considered
lktC as the activator for leukotoxin (
lktA).