MicroRNAs as Biomarkers in Canine Osteosarcoma: A New Future?
Abstract
1. Introduction
2. Canine Osteosarcoma
3. What Are MicroRNAs?
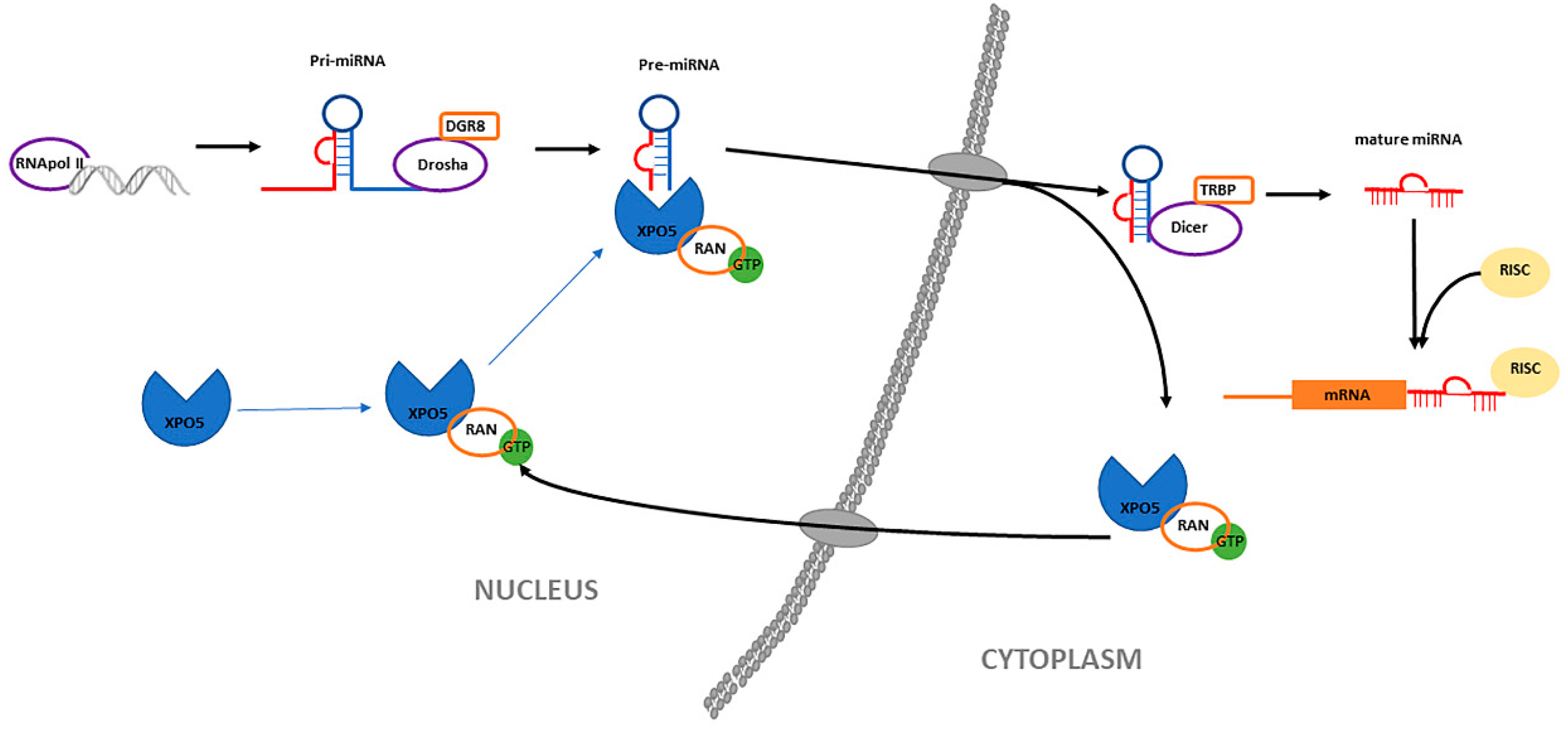
3.1. Biogenesis of MiRNAs
3.2. MiRNAs as Biomarkers
4. MiRNAs in Canine Osteosarcoma Cells
4.1. miR-1 and miR-133b
4.2. miR-9
4.3. miR-196a
4.4. MicroRNAs in the 14q32 Locus
4.5. miR-34a
4.6. Cluster of miR-106b-25 (miR-106b, miR-25 and miR-93-5p)
5. Circulating MicroRNAs and Their Potential as Non-Invasive Biomarkers
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chang, L.; Asatrian, G.; Dry, S.M.; James, A.W. Circulating tumor cells in sarcomas: A brief review. Med. Oncol. 2015, 32, 430. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, D.L.; Duval, D.L.; Regan, D.P.; Thamm, D.H. Canine sarcomas as a surrogate for the human disease. Pharmacol. Ther. 2018, 188, 80–96. [Google Scholar] [CrossRef] [PubMed]
- Sarver, A.L.; Thayanithy, V.; Scott, M.C.; Cleton-Jansen, A.-M.; Hogendoorn, P.C.; Modiano, J.F.; Subramanian, S. MicroRNAs at the human 14q32 locus have prognostic significance in osteosarcoma. Orphanet J. Rare Dis. 2013, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Schiffman, J.D.; Breen, M. Comparative oncology: What dogs and other species can teach us about humans with cancer. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370. [Google Scholar] [CrossRef] [PubMed]
- Séguin, B. Canine Soft Tissue Sarcomas: Can Being a Dog’s Best Friend Help a Child? Front. Oncol. 2017, 7, 285. [Google Scholar] [CrossRef]
- Simpson, S.; Dunning, M.D.; de Brot, S.; Grau-Roma, L.; Mongan, N.P.; Rutland, C.S. Comparative review of human and canine osteosarcoma: Morphology, epidemiology, prognosis, treatment and genetics. Acta Vet. Scand. 2017, 59, 71. [Google Scholar] [CrossRef]
- Fang, Y.; Guo, J.; Wu, S.; Li, X.; Zhao, J.; Li, Y.; Guo, S.; Mu, Y.; Kong, Q.; Liu, Z. Cellular reprogramming by single-cell fusion with mouse embryonic stem cells in pig. J. Cell. Physiol. 2019. [Google Scholar] [CrossRef]
- Longhi, A.; Pasini, A.; Cicognani, A.; Baronio, F.; Pellacani, A.; Baldini, N.; Bacci, G. Height as a risk factor for osteosarcoma. J. Pediatr. Hematol. Oncol. 2005, 27, 314–318. [Google Scholar] [CrossRef]
- Ottaviani, G.; Jaffe, N. The etiology of osteosarcoma. Cancer Treat. Res. 2009, 152, 15–32. [Google Scholar] [CrossRef]
- Varshney, J.; Scott, M.C.; Largaespada, D.A.; Subramanian, S. Understanding the Osteosarcoma Pathobiology: A Comparative Oncology Approach. Vet. Sci. 2016, 3, 3. [Google Scholar] [CrossRef]
- Fenger, J.M.; London, C.A.; Kisseberth, W.C. Canine osteosarcoma: A naturally occurring disease to inform pediatric oncology. ILAR J. 2014, 55, 69–85. [Google Scholar] [CrossRef] [PubMed]
- Mirabello, L.; Troisi, R.J.; Savage, S.A. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the Surveillance, Epidemiology, and End Results Program. Cancer 2009, 115, 1531–1543. [Google Scholar] [CrossRef] [PubMed]
- Ru, G.; Terracini, B.; Glickman, L.T. Host related risk factors for canine osteosarcoma. Vet. J. 1998, 156, 31–39. [Google Scholar] [CrossRef]
- Lopez, C.M.; Yu, P.Y.; Zhang, X.; Yilmaz, A.S.; London, C.A.; Fenger, J.M. MiR-34a regulates the invasive capacity of canine osteosarcoma cell lines. PLoS ONE 2018, 13, e0190086. [Google Scholar] [CrossRef]
- Klein, M.J.; Siegal, G.P. Osteosarcoma: Anatomic and histologic variants. Am. J. Clin. Pathol. 2006, 125, 555–581. [Google Scholar] [CrossRef]
- Leonardo, L.; Laura, P.; Serena, B.M. miR-1 and miR-133b expression in canine osteosarcoma. Res. Vet. Sci. 2018, 117, 133–137. [Google Scholar] [CrossRef]
- Maeda, J.; Yurkon, C.R.; Fujisawa, H.; Kaneko, M.; Genet, S.C.; Roybal, E.J.; Rota, G.W.; Saffer, E.R.; Rose, B.J.; Hanneman, W.H.; et al. Genomic instability and telomere fusion of canine osteosarcoma cells. PLoS ONE 2012, 7, e43355. [Google Scholar] [CrossRef]
- Selvarajah, S.; Yoshimoto, M.; Ludkovski, O.; Park, P.C.; Bayani, J.; Thorner, P.; Maire, G.; Squire, J.A.; Zielenska, M. Genomic signatures of chromosomal instability and osteosarcoma progression detected by high resolution array CGH and interphase FISH. Cytogenet. Genome Res. 2008, 122, 5–15. [Google Scholar] [CrossRef]
- Thomas, R.; Wang, H.J.; Tsai, P.-C.; Langford, C.F.; Fosmire, S.P.; Jubala, C.M.; Getzy, D.M.; Cutter, G.R.; Modiano, J.F.; Breen, M. Influence of genetic background on tumor karyotypes: Evidence for breed-associated cytogenetic aberrations in canine appendicular osteosarcoma. Chromosome Res. 2009, 17, 365–377. [Google Scholar] [CrossRef]
- Heishima, K.; Meuten, T.; Yoshida, K.; Mori, T.; Thamm, D.H. Prognostic significance of circulating microRNA-214 and -126 in dogs with appendicular osteosarcoma receiving amputation and chemotherapy. BMC Vet. Res. 2019, 15, 39. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Mitani, S.; Du, H.; Hall, D.H.; Driscoll, M.; Chalfie, M. Combinatorial control of touch receptor neuron expression in Caenorhabditis elegans. Development 1993, 119, 773–783. [Google Scholar] [PubMed]
- Chen, X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef] [PubMed]
- Doench, J.G.; Sharp, P.A. Specificity of microRNA target selection in translational repression. Genes Dev. 2004, 18, 504–511. [Google Scholar] [CrossRef] [PubMed]
- Heneghan, H.M.; Miller, N.; Kerin, M.J. MiRNAs as biomarkers and therapeutic targets in cancer. Curr. Opin. Pharmacol. 2010, 10, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- Ballarino, M.; Pagano, F.; Girardi, E.; Morlando, M.; Cacchiarelli, D.; Marchioni, M.; Proudfoot, N.J.; Bozzoni, I. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol. Cell. Biol. 2009, 29, 5632–5638. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Morlando, M.; Ballarino, M.; Gromak, N.; Pagano, F.; Bozzoni, I.; Proudfoot, N.J. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 2008, 15, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, M.T.; Czaplinski, K.; Gorlich, D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 2004, 10, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear export of microRNA precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef]
- Wu, J.; Qi, X.; Liu, L.; Hu, X.; Liu, J.; Yang, J.; Yang, J.; Lu, L.; Zhang, Z.; Ma, S.; et al. Emerging Epigenetic Regulation of Circular RNAs in Human Cancer. Mol. Ther. Nucleic Acids 2019, 16, 589–596. [Google Scholar] [CrossRef]
- Bennasser, Y.; Chable-Bessia, C.; Triboulet, R.; Gibbings, D.; Gwizdek, C.; Dargemont, C.; Kremer, E.J.; Voinnet, O.; Benkirane, M. Competition for XPO5 binding between Dicer mRNA, pre-miRNA and viral RNA regulates human Dicer levels. Nat. Struct. Mol. Biol. 2011, 18, 323–327. [Google Scholar] [CrossRef]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Grishok, A.; Pasquinelli, A.E.; Conte, D.; Li, N.; Parrish, S.; Ha, I.; Baillie, D.L.; Fire, A.; Ruvkun, G.; Mello, C.C. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 2001, 106, 23–34. [Google Scholar] [CrossRef]
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.E.; Bálint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef]
- Ketting, R.F.; Fischer, S.E.; Bernstein, E.; Sijen, T.; Hannon, G.J.; Plasterk, R.H. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001, 15, 2654–2659. [Google Scholar] [CrossRef]
- Shivdasani, R.A. MicroRNAs: Regulators of gene expression and cell differentiation. Blood 2006, 108, 3646–3653. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ding, M.; Duan, X.; Feng, X.; Wang, P.; Jiang, Q.; Cheng, Z.; Zhang, W.; Yu, S.; Yao, W.; et al. Diagnostic Value of Plasma MicroRNAs for Lung Cancer Using Support Vector Machine Model. J. Cancer 2019, 10, 5090–5098. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed]
- Ruby, J.G.; Stark, A.; Johnston, W.K.; Kellis, M.; Bartel, D.P.; Lai, E.C. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007, 17, 1850–1864. [Google Scholar] [CrossRef] [PubMed]
- Fromm, B.; Billipp, T.; Peck, L.E.; Johansen, M.; Tarver, J.E.; King, B.L.; Newcomb, J.M.; Sempere, L.F.; Flatmark, K.; Hovig, E.; et al. A Uniform System for the Annotation of Vertebrate microRNA Genes and the Evolution of the Human microRNAome. Annu. Rev. Genet. 2015, 49, 213–242. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Pang, M.; Agarwal, V.; Chen, Z.J. Interspecies regulation of microRNAs and their targets. Biochim. Biophys. Acta 2008, 1779, 735–742. [Google Scholar] [CrossRef]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef]
- Alegre, F.; Ormonde, A.R.; Snider, K.M.; Woolard, K.; Yu, A.-M.; Wittenburg, L.A. A genetically engineered microRNA-34a prodrug demonstrates anti-tumor activity in a canine model of osteosarcoma. PLoS ONE 2018, 13, e0209941. [Google Scholar] [CrossRef]
- Heishima, K.; Mori, T.; Ichikawa, Y.; Sakai, H.; Kuranaga, Y.; Nakagawa, T.; Tanaka, Y.; Okamura, Y.; Masuzawa, M.; Sugito, N.; et al. MicroRNA-214 and MicroRNA-126 Are Potential Biomarkers for Malignant Endothelial Proliferative Diseases. Int. J. Mol. Sci. 2015, 16, 25377–25391. [Google Scholar] [CrossRef]
- Heishima, K.; Mori, T.; Sakai, H.; Sugito, N.; Murakami, M.; Yamada, N.; Akao, Y.; Maruo, K. MicroRNA-214 Promotes Apoptosis in Canine Hemangiosarcoma by Targeting the COP1-p53 Axis. PLoS ONE 2015, 10, e0137361. [Google Scholar] [CrossRef] [PubMed]
- Nohata, N.; Hanazawa, T.; Enokida, H.; Seki, N. microRNA-1/133a and microRNA-206/133b clusters: Dysregulation and functional roles in human cancers. Oncotarget 2012, 3, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Novello, C.; Pazzaglia, L.; Cingolani, C.; Conti, A.; Quattrini, I.; Manara, M.C.; Tognon, M.; Picci, P.; Benassi, M.S. miRNA expression profile in human osteosarcoma: Role of miR-1 and miR-133b in proliferation and cell cycle control. Int. J. Oncol. 2013, 42, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Arora, H.; Qureshi, R.; Jin, S.; Park, A.K.; Park, W.Y. miR-9 and let-7g enhance the sensitivity to ionizing radiation by suppression of NFκB1. Exp. Mol. Med. 2011, 43, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Zhang, Y.; Liu, Y.; Zhou, M.; Lu, Y.; Yuan, L.; Zhang, C.; Hong, M.; Wang, S.; Li, X. MiR-106b induces cell radioresistance via the PTEN/PI3K/AKT pathways and p21 in colorectal cancer. J. Transl. Med. 2015, 13. [Google Scholar] [CrossRef]
- Pazzaglia, L.; Leonardi, L.; Conti, A.; Novello, C.; Quattrini, I.; Montanini, L.; Roperto, F.; Del Piero, F.; Di Guardo, G.; Piro, F.; et al. miR-196a expression in human and canine osteosarcomas: A comparative study. Res. Vet. Sci. 2015, 99, 112–119. [Google Scholar] [CrossRef]
- Thayanithy, V.; Sarver, A.L.; Kartha, R.V.; Li, L.; Angstadt, A.Y.; Breen, M.; Steer, C.J.; Modiano, J.F.; Subramanian, S. Perturbation of 14q32 miRNAs-cMYC gene network in osteosarcoma. Bone 2012, 50, 171–181. [Google Scholar] [CrossRef]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—Micrornas with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef]
- Choi, N.; Park, J.; Lee, J.-S.; Yoe, J.; Park, G.Y.; Kim, E.; Jeon, H.; Cho, Y.M.; Roh, T.-Y.; Lee, Y. miR-93/miR-106b/miR-375-CIC-CRABP1: A novel regulatory axis in prostate cancer progression. Oncotarget 2015, 6, 23533–23547. [Google Scholar] [CrossRef]
- Hudson, R.S.; Yi, M.; Esposito, D.; Glynn, S.A.; Starks, A.M.; Yang, Y.; Schetter, A.J.; Watkins, S.K.; Hurwitz, A.A.; Dorsey, T.H.; et al. MicroRNA-106b-25 cluster expression is associated with early disease recurrence and targets caspase-7 and focal adhesion in human prostate cancer. Oncogene 2013, 32, 4139–4147. [Google Scholar] [CrossRef]
- Kan, T.; Sato, F.; Ito, T.; Matsumura, N.; David, S.; Cheng, Y.; Agarwal, R.; Paun, B.C.; Jin, Z.; Olaru, A.V.; et al. The miR-106b-25 polycistron, activated by genomic amplification, functions as an oncogene by suppressing p21 and Bim. Gastroenterology 2009, 136, 1689–1700. [Google Scholar] [CrossRef]
- Li, Y.; Tan, W.; Neo, T.W.L.; Aung, M.O.; Wasser, S.; Lim, S.G.; Tan, T.M.C. Role of the miR-106b-25 microRNA cluster in hepatocellular carcinoma. Cancer Sci. 2009, 100, 1234–1242. [Google Scholar] [CrossRef]
- Petrocca, F.; Visone, R.; Onelli, M.R.; Shah, M.H.; Nicoloso, M.S.; de Martino, I.; Iliopoulos, D.; Pilozzi, E.; Liu, C.-G.; Negrini, M.; et al. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 2008, 13, 272–286. [Google Scholar] [CrossRef]
- Poliseno, L.; Salmena, L.; Riccardi, L.; Fornari, A.; Song, M.S.; Hobbs, R.M.; Sportoletti, P.; Varmeh, S.; Egia, A.; Fedele, G.; et al. Identification of the miR-106b~25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci. Signal. 2010, 3, ra29. [Google Scholar] [CrossRef]
- Zhang, R.; Li, F.; Wang, W.; Wang, X.; Li, S.; Liu, J. The effect of antisense inhibitor of miRNA 106b∼25 on the proliferation, invasion, migration, and apoptosis of gastric cancer cell. Tumor Biol. 2016, 37, 10507–10515. [Google Scholar] [CrossRef]
- He, Y.; Yu, B. MicroRNA-93 promotes cell proliferation by directly targeting P21 in osteosarcoma cells. Exp. Ther. Med. 2017, 13, 2003–2011. [Google Scholar] [CrossRef]
- Montanini, L.; Lasagna, L.; Barili, V.; Jonstrup, S.P.; Murgia, A.; Pazzaglia, L.; Conti, A.; Novello, C.; Kjems, J.; Perris, R.; et al. MicroRNA cloning and sequencing in osteosarcoma cell lines: Differential role of miR-93. Cell Oncol. 2012, 35, 29–41. [Google Scholar] [CrossRef]
- Leonardi, L.; Benassi, M.S.; Pollino, S.; Locaputo, C.; Pazzaglia, L. miR-106B-25 Cluster expression: A comparative human and canine osteosarcoma study. Vet. Rec. Open 2020, 7, e000379. [Google Scholar] [CrossRef]
- Fu, X.; Tian, J.; Zhang, L.; Chen, Y.; Hao, Q. Involvement of microRNA-93, a new regulator of PTEN/Akt signaling pathway, in regulation of chemotherapeutic drug cisplatin chemosensitivity in ovarian cancer cells. FEBS Lett. 2012, 586, 1279–1286. [Google Scholar] [CrossRef]
- Karimian, A.; Ahmadi, Y.; Yousefi, B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair 2016, 42, 63–71. [Google Scholar] [CrossRef]
- Zhang, T.M.; Zhang, J.; Zhou, D.J.; Wang, C.L. Expression of FATS in non-small cell lung cancer and its relationship with prognosis. Zhonghua Zhong Liu Za Zhi 2019, 41, 826–830. [Google Scholar] [CrossRef]
- Jiang, L.; Paone, S.; Caruso, S.; Atkin-Smith, G.K.; Phan, T.K.; Hulett, M.D.; Poon, I.K.H. Determining the contents and cell origins of apoptotic bodies by flow cytometry. Sci. Rep. 2017, 7, 14444. [Google Scholar] [CrossRef]
- Kawano, M.; Tanaka, K.; Itonaga, I.; Ikeda, S.; Iwasaki, T.; Tsumura, H. microRNA-93 promotes cell proliferation via targeting of PTEN in Osteosarcoma cells. J. Exp. Clin. Cancer Res. 2015, 34, 76. [Google Scholar] [CrossRef]
- Yu, D.; Wang, J.; Zou, H.; Feng, T.; Chen, L.; Li, J.; Qi, X.; Li, Z.; Duan, X.; Xu, C.; et al. Silencing of retrotransposon-derived imprinted gene RTL1 is the main cause for postimplantational failures in mammalian cloning. Proc. Natl. Acad. Sci. USA 2018, 115, 11071–11080. [Google Scholar] [CrossRef]
- Heishima, K.; Ichikawa, Y.; Yoshida, K.; Iwasaki, R.; Sakai, H.; Nakagawa, T.; Tanaka, Y.; Hoshino, Y.; Okamura, Y.; Murakami, M.; et al. Circulating microRNA-214 and -126 as potential biomarkers for canine neoplastic disease. Sci. Rep. 2017, 7, 2301. [Google Scholar] [CrossRef]
- Tavallaie, R.; McCarroll, J.; Le Grand, M.; Ariotti, N.; Schuhmann, W.; Bakker, E.; Tilley, R.D.; Hibbert, D.B.; Kavallaris, M.; Gooding, J.J. Nucleic acid hybridization on an electrically reconfigurable network of gold-coated magnetic nanoparticles enables microRNA detection in blood. Nat. Nanotechnol. 2018, 13, 1066–1071. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
| miRNAs | Expression Profiles in OSA | Target Genes or Proteins | Result of Aberrant miRNA Expression | Possible Uses |
|---|---|---|---|---|
| miR-1 and miR-133b | Reduced expression | MET and MCL1 | Cell proliferation and growth Cancer cells Invasion Cancer cells Survival Inhibition of apoptosis | Diagnostic biomarker for canine OSA |
| miR-9 | Overexpression | Gelsolin | Cell motility and invasive capacity increase Promotion of metastasis | Possible prognostic biomarker |
| miR-196a | Reduced expression | Annexin 1 | Facilitates migration and invasion Cancer cells adhesion and migration Mediates VEGF1 induced cell migration | Potential prognostic biomarker |
| 14q32 locus (miR-544, miR-369-3p, miR-134 and miR-382) | Reduced levels | cMYC | Promotion of metastasis Poorer survival rates Inhibition of apoptosis Tumorigenesis maintenance | Potential prognostic biomarker Potential therapeutic target |
| miR-34a | Reduced levels | MET, SIRT1, CDK6, VEGF1 | Mediates VEGF1 induced cell migration Shorter disease-free survival times | Potential prognostic biomarker Potential therapeutic target |
| miR-106b-25 cluster (miR-106b, miR-25 and miR-93-5p) | Overexpression | p21 | Cell proliferation and growth Cancer cells Invasion Cell cycle promotors Activation of tumorigenesis | Potential prognostic biomarker |
| Circulating miR-214 | Increased levels in circulation | Shorter survival times | Potential non-invasive predictive biomarker | |
| Circulating miR-126 | Increased levels in circulation | Longer survival times | Potential non-invasive predictive biomarker |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gourbault, O.; Llobat, L. MicroRNAs as Biomarkers in Canine Osteosarcoma: A New Future? Vet. Sci. 2020, 7, 146. https://doi.org/10.3390/vetsci7040146
Gourbault O, Llobat L. MicroRNAs as Biomarkers in Canine Osteosarcoma: A New Future? Veterinary Sciences. 2020; 7(4):146. https://doi.org/10.3390/vetsci7040146
Chicago/Turabian StyleGourbault, Olivia, and Lola Llobat. 2020. "MicroRNAs as Biomarkers in Canine Osteosarcoma: A New Future?" Veterinary Sciences 7, no. 4: 146. https://doi.org/10.3390/vetsci7040146
APA StyleGourbault, O., & Llobat, L. (2020). MicroRNAs as Biomarkers in Canine Osteosarcoma: A New Future? Veterinary Sciences, 7(4), 146. https://doi.org/10.3390/vetsci7040146






