Using Manual and Computer-Based Text-Mining to Uncover Research Trends for Apis mellifera
Abstract
1. Introduction
2. Methods
2.1. Dataset
2.2. Author-Assigned Keywords’ Analysis
2.2.1. Pre-Processing the Dataset
2.2.2. Temporal Trends
2.2.3. Cluster Analysis
2.2.4. Network Analysis
2.3. Computer-Based Keywords’ Analysis
2.3.1. Preparing and Pre-Processing the Corpora
2.3.2. Applying Topic Modeling
3. Results
3.1. Dataset
3.2. Author-Assigned Keywords’ Analysis
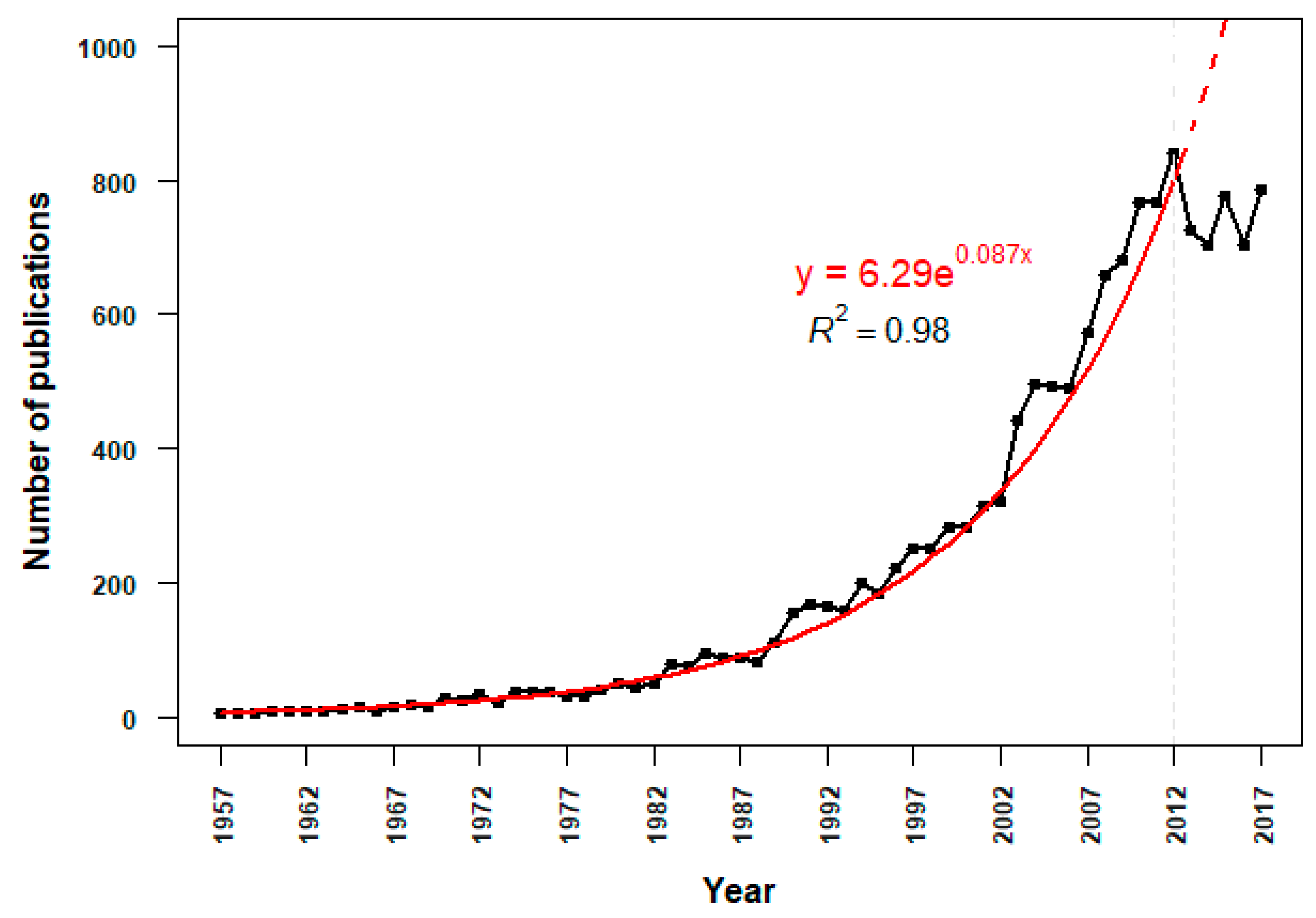
3.2.1. Temporal Trends
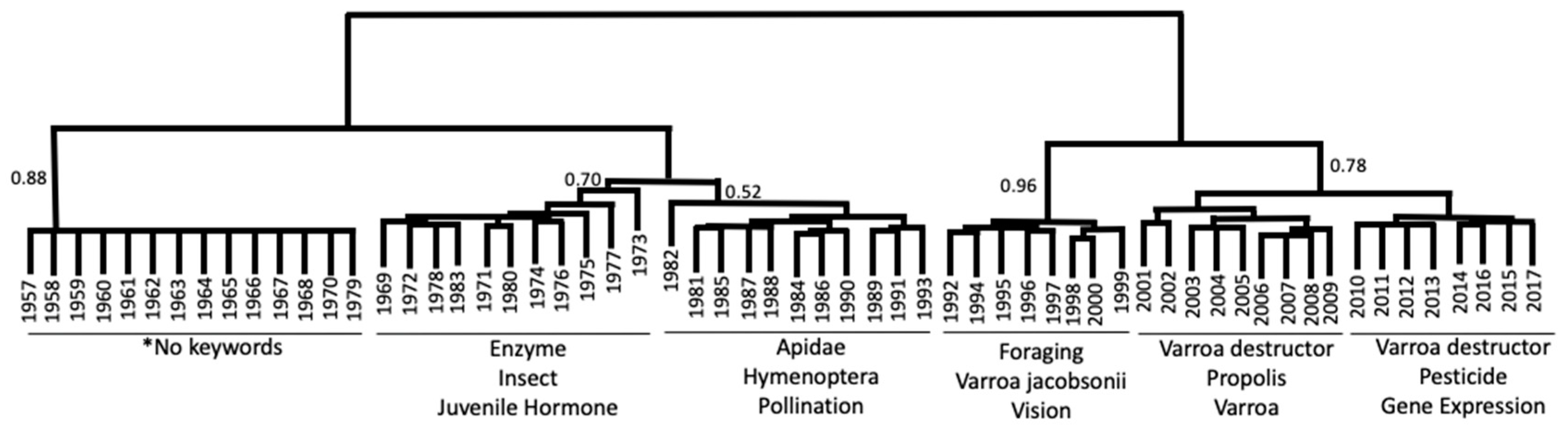
3.2.2. Cluster Analysis
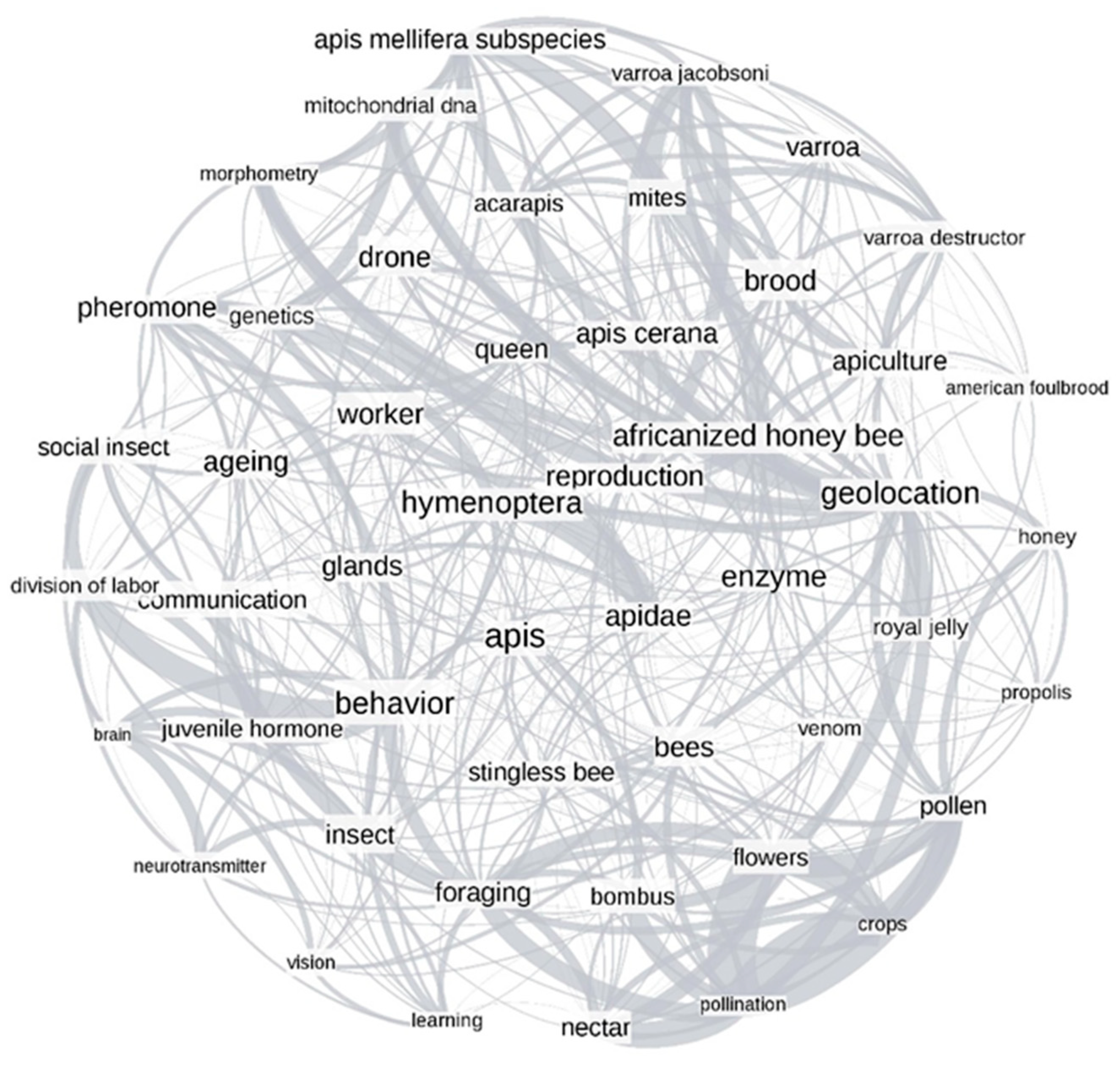
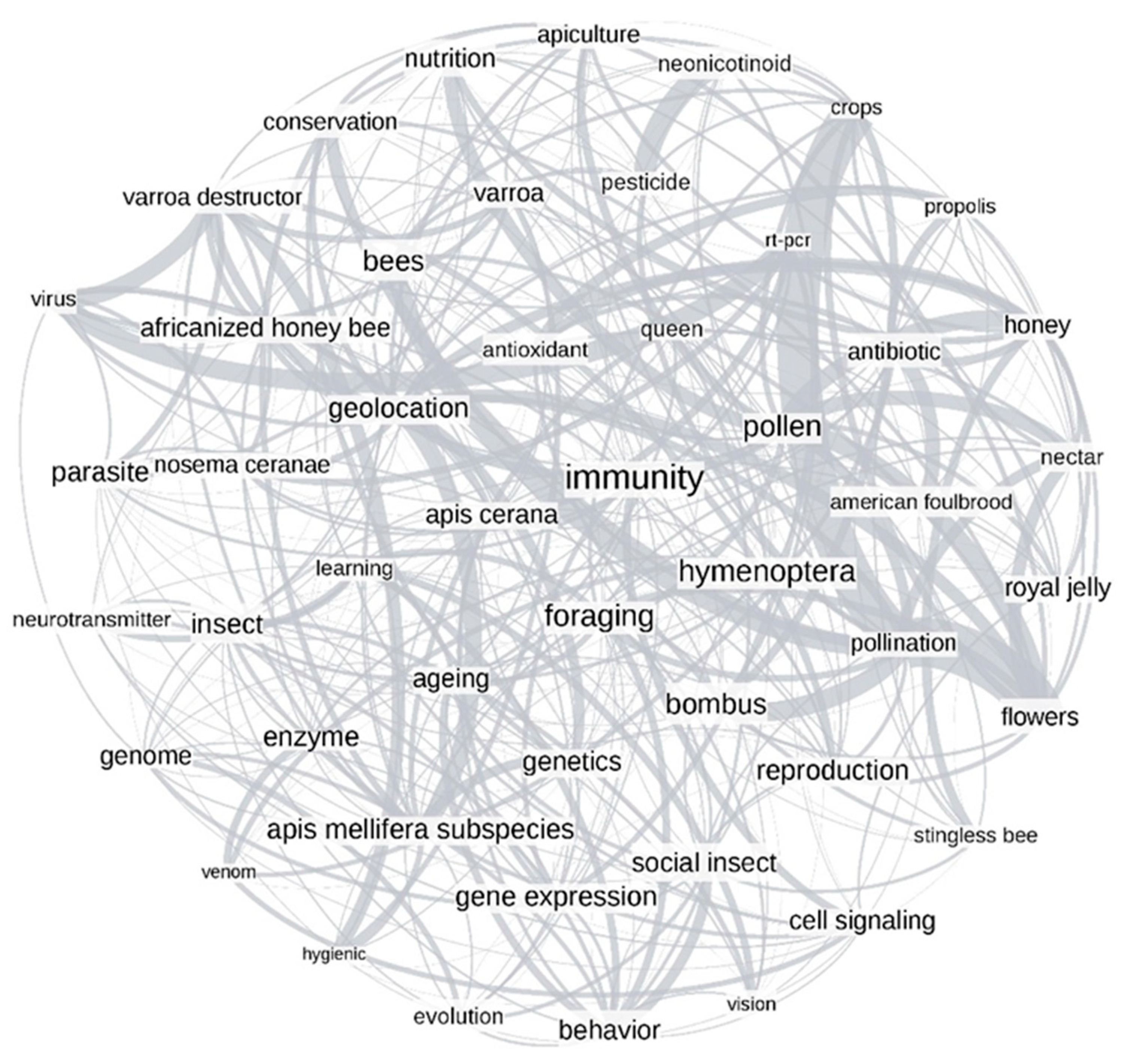
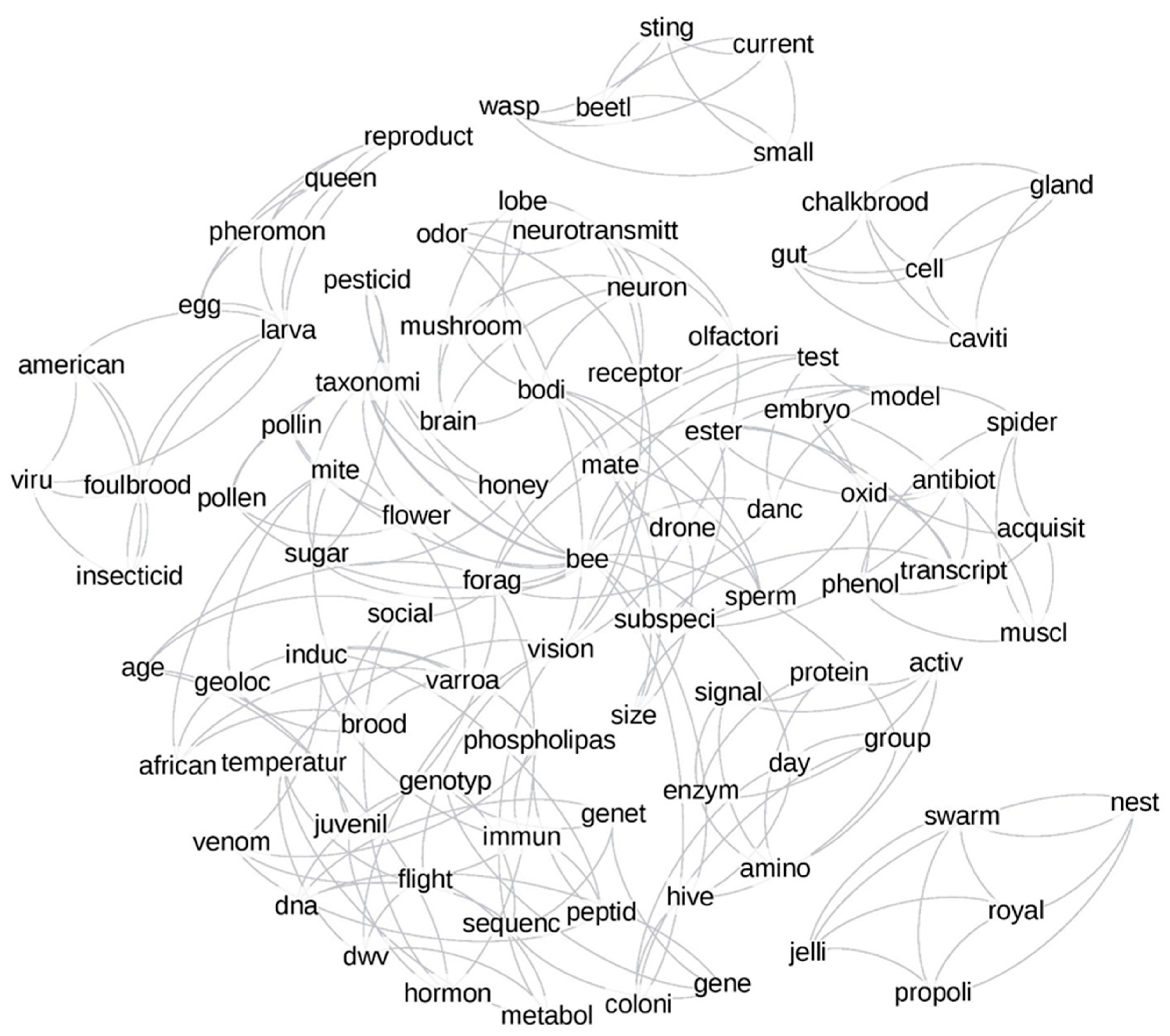
3.2.3. Network Analysis
3.3. Computer-Based Topic Modeling
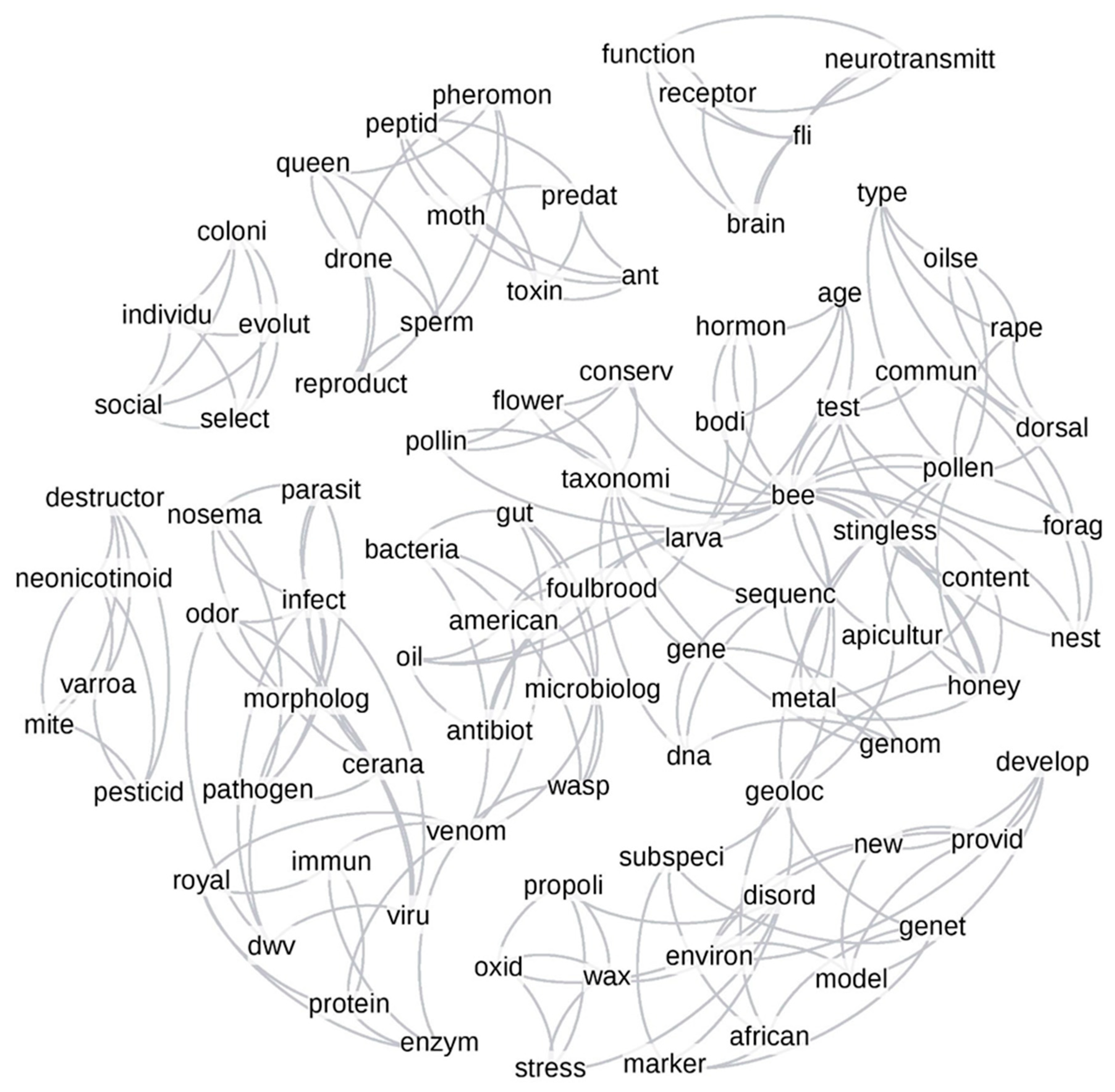
3.4. Author-Assigned vs. Computer-Based Networks
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Data Availability
Abbreviations
| CCD | Colony Collapse Disorder |
| CI | Confidence Interval |
| LDA | Latent Dirichlet Allocation |
| PCR | Polymerase Chain Reaction |
| RT-PCR | Reverse Transcription Polymerase Chain Reaction |
References
- Morse, R.A.; Calderone, N.W. The value of honey bees as pollinators of US crops in 2000. Bee Cult. 2000, 128, 1–15. [Google Scholar]
- Partap, U. The pollination role of honey bees. In Honey Bees of Asia; Hepburn, H., Radloff, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 227–255. [Google Scholar] [CrossRef]
- Aizen, M.A.; Garibaldi, L.A.; Cunningham, S.A.; Klein, A.M. Long-term global trends in crop yield and production reveal no current pollination shortage but increasing pollinator dependency. Curr. Biol. 2008, 18, 1572–1575. [Google Scholar] [CrossRef]
- Menzel, R. The honey bee as a model for understanding the basis of cognition. Nat. Rev. Neurosci. 2012, 13, 758–768. [Google Scholar] [CrossRef]
- Cridge, A.G.; Lovegrove, M.R.; Skelly, J.G.; Taylor, S.E.; Petersen, G.E.L.; Cameron, R.C.; Dearden, P.K. The honey bee as a model insect for developmental genetics. Genesis 2017, 55, e23019. [Google Scholar] [CrossRef] [PubMed]
- van der Zee, R.; Pisa, L.; Andonov, S.; Brodschneider, R.; Charrière, J.-D.; Chlebo, R.; Coffey, M.F.; Crailsheim, K.; Dahle, B.; Gajda, A.; et al. Managed honey bee colony losses in Canada, China, Europe, Israel and Turkey, for the winters of 2008–9 and 2009–10. J. Apic. Res. 2012, 51, 100–114. [Google Scholar] [CrossRef]
- Liu, Z.; Chen, C.; Niu, Q.; Qi, W.; Yuan, C.; Su, S.; Liu, S.; Zhang, Y.; Zhang, X.; Ji, T.; et al. Survey results of honey bee (Apis mellifera) colony losses in China (2010–2013). J. Apic. Res. 2016, 55, 29–37. [Google Scholar] [CrossRef]
- vanEngelsdorp, D.; Meixner, M.D. A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J. Invertebr. Pathol. 2010, 103, S80–S95. [Google Scholar] [CrossRef]
- Potts, S.G.; Biesmeijer, J.C.; Kremen, C.; Neumann, P.; Schweiger, O.; Kunin, W.E. Global pollinator declines: Trends, impacts and drivers. Trends Ecol. Evol. 2010, 25, 345–353. [Google Scholar] [CrossRef]
- Moritz, R.F.A.; Erler, S. Lost colonies found in a data mine: Global honey trade but not pests or pesticides as a major cause of regional honey bee colony declines. Agric. Ecosyst. Environ. 2016, 216, 44–50. [Google Scholar] [CrossRef]
- Holden, C. Report warns of looming pollination crisis in North America. Science 2006, 314, 397. [Google Scholar] [CrossRef]
- vanEngelsdorp, D.; Underwood, R.; Caron, D.; Hayes Jr, J. An estimate of managed colony losses in the winter of 2006-2007: A report commissioned by the apiary inspectors of America. Am. Bee J. 2007, 147, 599–603. [Google Scholar]
- Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honey bee Apis mellifera. Nature 2006, 443, 931–949. [Google Scholar] [CrossRef] [PubMed]
- Grozinger, C.M.; Robinson, G.E. The power and promise of applying genomics to honey bee health. Curr. Opin. Insect Sci. 2015, 10, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Bornmann, L.; Mutz, R. Growth rates of modern science: A bibliometric analysis based on the number of publications and cited references. J. Assoc. Inf. Sci. Technol. 2015, 66, 2215–2222. [Google Scholar] [CrossRef]
- Comeau, D.C.; Wei, C.H.; Islamaj Dogan, R.; Lu, Z. PMC text mining subset in BioC: About three million full-text articles and growing. Bioinformatics 2019, 35, 3533–3535. [Google Scholar] [CrossRef]
- McAlister, F.A.; Clark, H.D.; van Walraven, C.; Straus, S.E.; Lawson, F.M.E.; Moher, D.; Mulrow, C.D. The medical review article revisited: Has the science improved? Ann. Intern. Med. 1999, 131, 947–951. [Google Scholar] [CrossRef]
- Strube, M.J.; Gardner, W.; Hartmann, D.P. Limitations, liabilities, and obstacles in reviews of the literature: The current status of meta-analysis. Clin. Psychol. Rev. 1985, 5, 63–78. [Google Scholar] [CrossRef]
- Pautasso, M. Publication growth in biological sub-fields: Patterns, predictability and sustainability. Sustainability 2012, 4, 3234–3247. [Google Scholar] [CrossRef]
- Mooney, R.J.; Bunescu, R. Mining knowledge from text using information extraction. SIGKDD Explor. Newsl. 2005, 7, 3–10. [Google Scholar] [CrossRef]
- Blei, D.M.; Ng, A.Y.; Jordan, M.I.; Lafferty, J. Latent dirichlet allocation. J. Mach. Learn. Res. 2003, 3, 993–1022. [Google Scholar]
- Khalili, M.; Rahimi-Movaghar, A.; Shadloo, B.; Mojtabai, R.; Mann, K.; Amin-Esmaeili, M. Global scientific production on illicit drug addiction: A two-decade analysis. Eur. Addict. Res. 2018, 24, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Meyer, D.; Hornik, K.; Feinerer, I. Text mining infrastructure in R. J. Stat. Softw. 2008, 25, 1–54. [Google Scholar] [CrossRef]
- Porter, M.F. Snowball: A language for Stemming Algorithms. Available online: http://snowball.tartarus.org/texts/introduction (accessed on 18 February 2019).
- Murtagh, F.; Legendre, P. Ward”s hierarchical agglomerative clustering method: Which algorithms implement ward”s criterion? J. Classif. 2014, 31, 274–295. [Google Scholar] [CrossRef]
- Hennig, C. Cluster-wise assessment of cluster stability. Comput. Stat. Data Anal. 2007, 52, 258–271. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2015. [Google Scholar]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the International AAAI Conference on Web and Social Media, San Jose, CA, USA, 17–20 March 2009; pp. 361–362. [Google Scholar]
- Wasserman, S.; Faust, K. Social Network Analysis: Methods and Applications; Cambridge University Press: Cambridge, UK, 1994; Volume 8. [Google Scholar]
- Opsahl, T.; Panzarasa, P. Clustering in weighted networks. Soc. Netw. 2009, 31, 155–163. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The igraph software package for complex network research. InterJournal Complex Systems 2006, 1695, 1–9. Available online: https://igraph.org (accessed on 5 March 2019).
- Blei, D.M.; Lafferty, J.D. Correlated topic models. In Proceedings of the 18th International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, December 2006; Weiss, Y., Schölkopf, B., Platt, J., Eds.; MIT Press: Cambridge, MA, USA, 2006; pp. 147–154. [Google Scholar]
- Newman, D.; Lau, J.H.; Grieser, K.; Baldwin, T. Automatic evaluation of topic coherence. In Human Language Technologies: The 2010 Annual Conference of the North American Chapter of the Association for Computational Linguistics; Association for Computational Linguistics: Los Angeles, CA, CA, 2010; pp. 100–108. [Google Scholar]
- Chang, J.; Boyd-Graber, J.; Gerrish, S.; Wang, C.; Blei, D.M. Reading tea leaves: How humans interpret topic models. In Proceedings of the 22nd International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, December 2009; Bengio, Y., Schuumans, D., Lafferty, J.D., Williams, C.K.I., Culotta, A., Eds.; MIT Press: Cambridge, MA, USA, 2009; pp. 288–296. [Google Scholar]
- Chang, J.; Blei, D. Relational topic models for document networks. In Proceedings of the Twelfth International Conference on Artificial Intelligence and Statistics, Clearwater Beach, FL, USA, 16–19 April 2009; pp. 81–88. [Google Scholar]
- Roder, M.; Both, A.; Hinneburg, A. Exploring the space of topic coherence measures. In Proceedings of the Eighth ACM International Conference on Web Search and Data Mining, Shanghai, China, 31 January–2 February 2015; pp. 399–408. [Google Scholar]
- Sievert, C.; Shirley, K.E. LDAvis: A method for visualizing and interpreting topics. In Proceedings of the Workshop on Interactive Language Learning, Visualization, and Interfaces, Baltimore, MD, USA, 27 June 2014; pp. 63–70. [Google Scholar]
- Jinha, A.E. Article 50 million: An estimate of the number of scholarly articles in existence. Learn. Publ. 2010, 23, 258–263. [Google Scholar] [CrossRef]
- Burnham, J.F. Scopus database: A review. Biomed. Digit. Libr. 2006, 3, e1. [Google Scholar] [CrossRef]
- Lamb, C. Open access publishing models: Opportunity or threat to scholarly and academic publishers? Learn. Publ. 2004, 17, 143–150. [Google Scholar] [CrossRef]
- Fields, S. The interplay of biology and technology. Proc. Natl. Acad. Sci. USA 2001, 98, 10051–10054. [Google Scholar] [CrossRef]
- Lawrence, P.A. The politics of publication. Nature 2003, 422, 259–261. [Google Scholar] [CrossRef]
- Robinson, G.E.; Evans, J.D.; Maleszka, R.; Robertson, H.M.; Weaver, D.B.; Worley, K.; Gibbs, R.A.; Weinstock, G.M. Sweetness and light: Illuminating the honey bee genome. Insect Mol. Biol. 2006, 15, 535–539. [Google Scholar] [CrossRef] [PubMed]
- Pettis, J.S.; Delaplane, K.S. Coordinated responses to honey bee decline in the USA. Apidologie 2010, 41, 256–263. [Google Scholar] [CrossRef]
- Moritz, R.F.A.; de Miranda, J.; Fries, I.; Le Conte, Y.; Neumann, P.; Paxton, R.J. Research strategies to improve honey bee health in Europe. Apidologie 2010, 41, 227–242. [Google Scholar] [CrossRef]
- de Guzman, L.I.; Rinderer, T.E. Identification and comparison of Varroa species infesting honey bees. Apidologie 1999, 30, 85–95. [Google Scholar] [CrossRef]
- Anderson, D.L.; Trueman, J.W. Varroa jacobsoni (Acari: Varroidae) is more than one species. Exp. Appl. Acarol. 2000, 24, 165–189. [Google Scholar] [CrossRef]
- Ritter, W.; Leclercq, E.; Koch, W. Observations on bee and Varroa and mite populations in infested honey bee colonies. Apidologie 1984, 15, 389–399. [Google Scholar] [CrossRef]
- Nazzi, F.; Le Conte, Y. Ecology of Varroa destructor, the major ectoparasite of the Western honey bee, Apis mellifera. Annu. Rev. Entomol. 2016, 61, 417–432. [Google Scholar] [CrossRef]
- Decourtye, A.; Alaux, C.; Le Conte, Y.; Henry, M. Toward the protection of bees and pollination under global change: Present and future perspectives in a challenging applied science. Curr. Opin. Insect Sci. 2019, 35, 123–131. [Google Scholar] [CrossRef]
- Paton, D.C. Honey bees in the Australian environment: Does Apis mellifera disrupt or benefit the native biota? Bioscience 1993, 43, 95–103. [Google Scholar] [CrossRef]
- Hung, K.-L.J.; Kingston, J.M.; Albrecht, M.; Holway, D.A.; Kohn, J.R. The worldwide importance of honey bees as pollinators in natural habitats. Proc. R. Soc. B 2018, 285, e20172140. [Google Scholar] [CrossRef]
- Grozinger, C.M.; Flenniken, M.L. Bee viruses: Ecology, pathogenicity, and impacts. Annu. Rev. Entomol. 2019, 64, 205–226. [Google Scholar] [CrossRef] [PubMed]
- Trapp, J.; McAfee, A.; Foster, L.J. Genomics, transcriptomics and proteomics: Enabling insights into social evolution and disease challenges for managed and wild bees. Mol. Ecol. 2017, 26, 718–739. [Google Scholar] [CrossRef] [PubMed]
- Boutin, S.; Alburaki, M.; Mercier, P.-L.; Giovenazzo, P.; Derome, N. Differential gene expression between hygienic and non-hygienic honey bee (Apis mellifera L.) hives. BMC Genom. 2015, 16, e500. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Kryger, P.; Le Conte, Y.; Moritz, R.F.A. Survival and immune response of drones of a Nosemosis tolerant honey bee strain towards N. ceranae infections. J. Invertebr. Pathol. 2012, 109, 297–302. [Google Scholar] [CrossRef]
- Cox-Foster, D.L.; Conlan, S.; Holmes, E.C.; Palacios, G.; Evans, J.D.; Moran, N.A.; Quan, P.-L.; Briese, T.; Hornig, M.; Geiser, D.M.; et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 2007, 318, 283–287. [Google Scholar] [CrossRef]
- Qin, X.; Evans, J.D.; Aronstein, K.A.; Murray, K.D.; Weinstock, G.M. Genome sequences of the honey bee pathogens Paenibacillus larvae and Ascosphaera apis. Insect Mol. Biol. 2006, 15, 715–718. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amiri, E.; Waiker, P.; Rueppell, O.; Manda, P. Using Manual and Computer-Based Text-Mining to Uncover Research Trends for Apis mellifera. Vet. Sci. 2020, 7, 61. https://doi.org/10.3390/vetsci7020061
Amiri E, Waiker P, Rueppell O, Manda P. Using Manual and Computer-Based Text-Mining to Uncover Research Trends for Apis mellifera. Veterinary Sciences. 2020; 7(2):61. https://doi.org/10.3390/vetsci7020061
Chicago/Turabian StyleAmiri, Esmaeil, Prashant Waiker, Olav Rueppell, and Prashanti Manda. 2020. "Using Manual and Computer-Based Text-Mining to Uncover Research Trends for Apis mellifera" Veterinary Sciences 7, no. 2: 61. https://doi.org/10.3390/vetsci7020061
APA StyleAmiri, E., Waiker, P., Rueppell, O., & Manda, P. (2020). Using Manual and Computer-Based Text-Mining to Uncover Research Trends for Apis mellifera. Veterinary Sciences, 7(2), 61. https://doi.org/10.3390/vetsci7020061





