Detection and Characterisation of Anaplasma marginale and A. centrale in South Africa
Abstract
1. Introduction
2. Classification of Anaplasma Species
| Superkingdom | Bacteria |
| Phylum | Proteobacteria |
| Class | Alpha-proteobacteria |
| Order | Rickettsiales |
| Family | Anaplasmataceae |
| Genus | Anaplasma |
| Species | A. marginale (type species) |
| A. bovis | |
| A. caudatum | |
| A. centrale | |
| A. ovis | |
| A. phagocytophilum | |
| A. platys | |
| Not formally described: | |
| A. capra | |
| Anaplasma sp. (Omatjenne) |
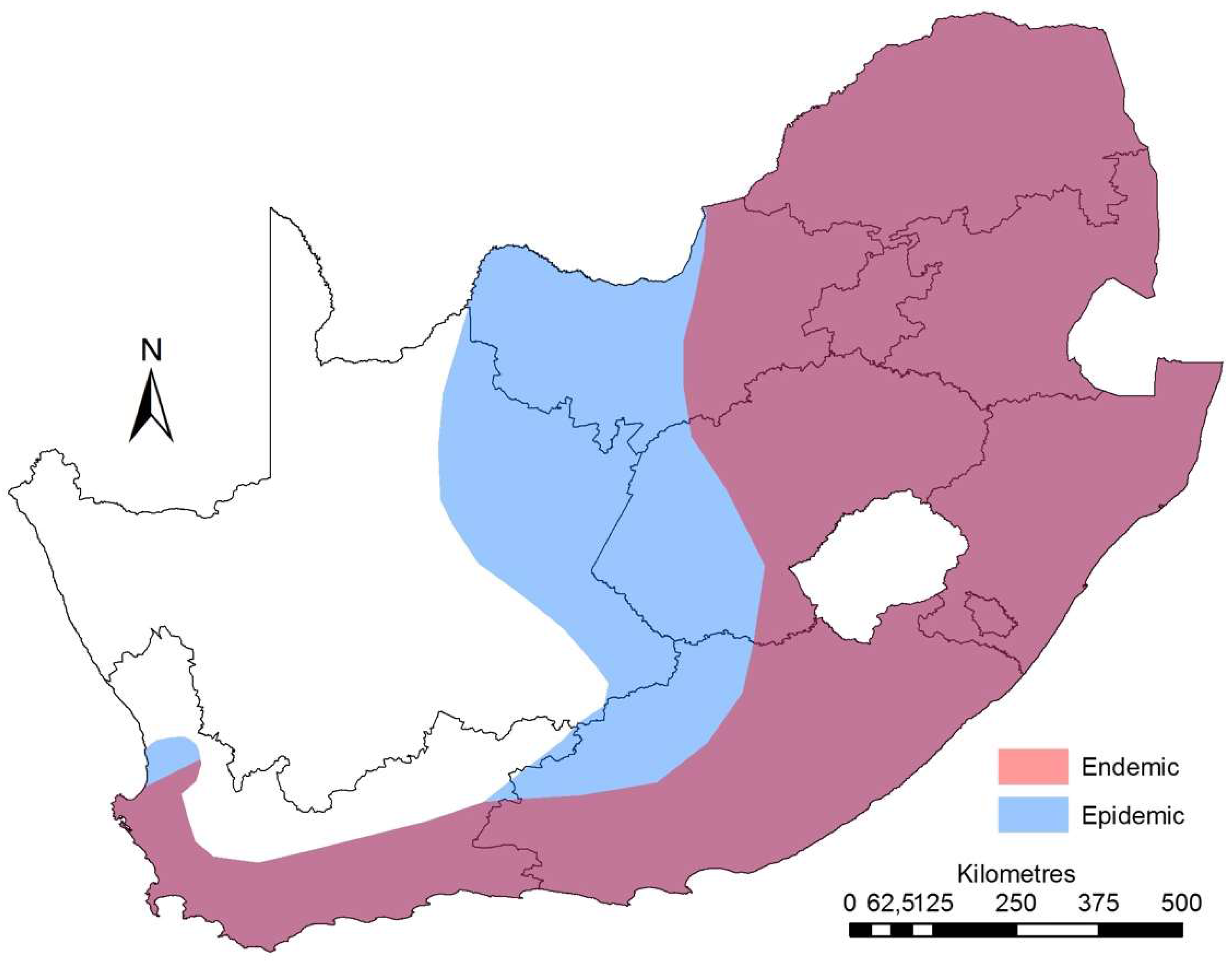
3. Epidemiology
4. Detection of A. marginale and A. centrale in South Africa
5. Genetic Diversity of A. marginale and A. centrale in South Africa
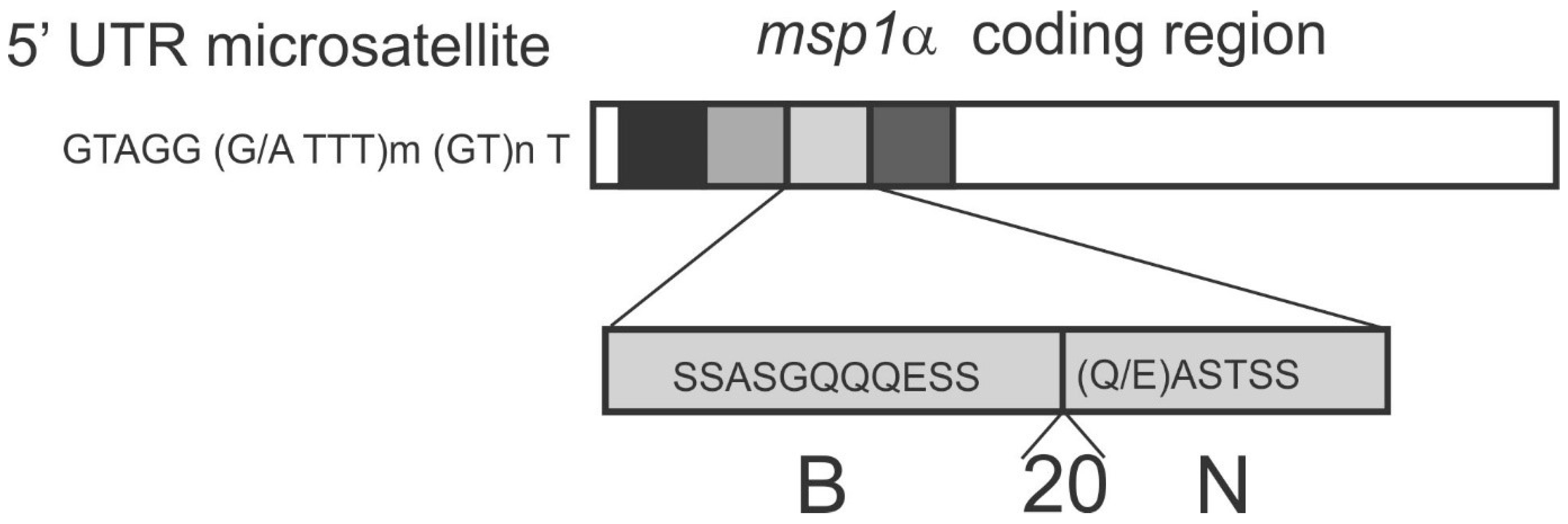
5.1. msp1α Genotyping of A. marginale
5.2. msp1aS Genotyping of A. centrale
6. Conclusions
Acknowledgments
- Hove P., Brayton K.A., Liebenberg J., Pretorius A., Oosthiuzen M.C., Mtshali M.S., Mutshembele A., Noh S.M., Collins N.E. 2017. Molecular characterization of vaccine candidates from Anaplasma marginale strains in South Africa. 9th Tick and Tick-borne Pathogen Conference & 1st Asia Pacific Rickettsia Conference, Cairns, Australia, 27 August–1 September 2017.
- Chaisi M.E., Hove P., Oosthuizen M.C., Brayton K.A., Collins N.E. 2017. Anaplasma marginale and A. centrale are widespread in cattle in South Africa. 9th Tick and Tick-borne Pathogen Conference & 1st Asia Pacific Rickettsia Conference, Cairns, Australia, 27 August–1 September 2017.
- Khumalo, Z.T.H., Collins, N.E., Chaisi, M.E., Brayton, K.A., Quan, M., Chaisi, M.E., Oosthuizen, M.C. 2017. Confirmation of Anaplasma marginale variety centrale (Theiler 1911) as a separate species, Anaplasma centrale (non Theiler 1911) sp. nov., comb. nov (Ristic & Kreier 1984). 9th Tick and Tick-borne Pathogen Conference & 1st Asia Pacific Rickettsia Conference, Cairns, Australia, 27 August–1 September 2017.
Author Contributions
Conflicts of Interest
References
- De Waal, D.T. Anaplasmosis control and diagnosis in South Africa. Ann. N. Y. Acad. Sci. 2000, 916, 474–483. [Google Scholar] [CrossRef]
- Theiler, A. Anaplasma marginale (Gen. and spec. nov.): The marginal points in the blood of cattle suffering from a specific disease. In Report of the government veterinary bacteriologist, 1908–1909; Government Printing and Stationery Office: Pretoria, South Africa, 1910; pp. 1–65. [Google Scholar]
- Theiler, A. Anaplasma marginale (Gen. and spec. nova.): A protozoon of cattle; a cause of the so-called Gall-sickness. Transvaal Med. J. 1910, 110–111. [Google Scholar]
- Dumler, J.S.; Barbet, A.F.; Bekker, C.P.; Dasch, G.A.; Palmer, G.H.; Ray, S.C.; Rikihisa, Y.; Rurangirwa, F.R. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: Unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 2001, 51, 2145–2165. [Google Scholar] [CrossRef] [PubMed]
- Potgieter, F.T.; Stoltsz, W.H. Bovine Anaplasmosis. In Infectious Diseases of Livestock; Coetzer, J.A.W., Tustin, R.C., Eds.; Oxford University Press: Cape Town, South Africa, 2004; pp. 594–616. [Google Scholar]
- Merck. Anaplasmosis. Available online: http://www.merckvetmanual.com/circulatory-system/blood-parasites/anaplasmosis (accessed 20 December 2017).
- Mtshali, M.S.; de la Fuente, J.; Ruybal, P.; Kocan, K.M.; Vicente, J.; Mbati, P.A.; Shkap, V.; Blouin, E.F.; Mohale, N.E.; Moloi, T.P.; et al. Prevalence and genetic diversity of Anaplasma marginale strains in cattle in South Africa. Zoonoses Public Health 2007, 54, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Theiler, A. Gallsickness of imported cattle and the protective inoculation against this disease. Agric. J. Union South Africa. 1912, 3, 1–11. [Google Scholar]
- Aubry, P.; Geale, D.W. A review of bovine anaplasmosis. Transbound. Emerg. Dis. 2011, 58, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Kocan, K.M.; de la Fuente, J.; Guglielmone, A.A.; Melendez, R.D. Antigens and alternatives for control of Anaplasma marginale infection in cattle. Clin. Microbiol. Rev. 2003, 16, 698–712. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.G. Dynamics and impact of tick-borne diseases of cattle. Trop. Anim. Health Prod. 1997, 29, 1S–3S. [Google Scholar] [CrossRef] [PubMed]
- Bock, R.E.; de Vos, A.J. Immunity following use of Australian tick fever vaccine: A review of the evidence. Aust. Vet. J. 2001, 79, 832–839. [Google Scholar] [CrossRef] [PubMed]
- Brayton, K.A.; Dark, M.J.; Palmer, G.H. Anaplasma. In Genome Mapping and Genomics in Animal-Associated Microbes; Nene, V., Kole, C., Eds.; Springer-Verlag: Berlin, Germany, 2009; pp. 85–116. [Google Scholar]
- Allsopp, M.; Visser, E.S.; du Plessis, J.L.; Vogel, S.W.; Allsopp, B.A. Different organisms associated with heartwater as shown by analysis of 16S ribosomal RNA gene sequences. Vet. Parasitol. 1997, 71, 283–300. [Google Scholar] [CrossRef]
- Li, H.; Zheng, Y.-C.; Ma, L.; Jia, N.; Jiang, B.-G.; Jiang, R.-R.; Huo, Q.-B.; Wang, Y.-W.; Liu, H.-B.; Chu, Y.-L.; et al. Human infection with a novel tick-borne Anaplasma species in China: A surveillance study. Lancet Infect. Dis. 2015, 15, 663–670. [Google Scholar] [CrossRef]
- Ristic, M. Anaplasmosis. In Infectious diseases of man and animals; Weinman, D., Ristic, D., Eds.; Academic Press: New York, NY, USA, 1968; pp. 473–542. [Google Scholar]
- Tindall, B.J.; Ross, H.N.M.; Grant, W.D. Validation of the publication of new names and new combinations previously effectively published outside the IJSB. List Number 15. Int. J. Syst. Bacteriol. 1984, 34, 355–357. [Google Scholar]
- Ristic, M.; Kreier, J.P. Genus Anaplasma. In Bergey’s Manual of Systematic Bacteriology; Kreig, N.R., Holt, J.G., Eds.; The Williams & Wilkins Co.: Baltomore, MD, USA, 1984; pp. 720–722. [Google Scholar]
- Khumalo, Z.T.H.; Brayton, K.A.; Collins, N.E.; Chaisi, M.E.; Quan, M.; Oosthuizen, M.C. Evidence confirming the phylogenetic position of Anaplasma centrale (ex Theiler 1911) Ristic & Kreier 1984. Int. J. Syst. Evol. Microbiol. (under review).
- Khumalo, Z.T.H.; Catanese, H.N.; Liesching, N.; Hove, P.; Collins, N.E.; Chaisi, M.E.; Gebremedhin, A.H.; Oosthuizen, M.C.; Brayton, K.A. Characterization of Anaplasma marginale subsp. centrale strains by use of msp1aS genotyping reveals a wildlife reservoir. J. Clin. Microbiol. 2016, 54, 2503–2512. [Google Scholar] [CrossRef] [PubMed]
- Brayton, K.A.; Kappmeyer, L.S.; Herndon, D.R.; Dark, M.J.; Tibbals, D.L.; Palmer, G.H.; McGuire, T.C.; Knowles, D.P. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 844–849. [Google Scholar] [CrossRef] [PubMed]
- Herndon, D.R.; Palmer, G.H.; Shkap, V.; Knowles, D.P.; Brayton, K.A. Complete genome sequence of Anaplasma marginale subsp. centrale. J. Bacteriol. 2010, 192, 379–380. [Google Scholar] [CrossRef] [PubMed]
- Mutshembele, A.M.; Cabezas-Cruz, A.; Mtshali, M.S.; Thekisoe, O.M.M.; Galindo, R.C.; de la Fuente, J. Epidemiology and evolution of the genetic variability of Anaplasma marginale in South Africa. Ticks Tick. Borne. Dis. 2014, 5, 624–631. [Google Scholar] [CrossRef] [PubMed]
- Nyangiwe, N.; Horak, I.G.; van der Mescht, L.; Matthee, S. Range expansion of the economically important Asiatic blue tick, Rhipicephalus microplus, in South Africa. J. S. Afr. Vet. Assoc. 2017, 88, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Potgieter, F.T.; van Rensburg, L. Tick transmission of Anaplasma centrale. Onderstepoort J. Vet. Res. 1987, 54, 5–7. [Google Scholar] [PubMed]
- Khumalo, Z.T.H. Anaplasma Centrale in South Africa; Occurrence, Phylogeny and gGenetic Diversity. Ph.D. Thesis, University of Pretoria, Pretoria, South Africa, 2017. [Google Scholar]
- Neitz, W.O. Bovine anaplasmosis: the transmission of Anaplasma marginale to a black wildebeest (Connochaetes gnou). Onderstepoort J. Vet. Sci. Anim. Ind. 1935, 5, 9–11. [Google Scholar]
- Potgieter, F.T. Epizootiology and control of anaplasmosis in South Africa. J. S. Afr. Vet. Assoc. 1979, 50, 367–372. [Google Scholar] [PubMed]
- Rajput, Z.I.; Hu, S.; Chen, W.; Arijo, A.G.; Xiao, C. Importance of ticks and their chemical and immunological control in livestock. J. Zhejiang Univ. Sci. B 2006, 7, 912–921. [Google Scholar] [CrossRef] [PubMed]
- Potgieter, F.T.; van Rensburg, L. Infectivity virulence and immunogenicity of Anaplasma centrale live blood vaccine. Onderstepoort J Vet Res 1983, 50, 29–31. [Google Scholar] [PubMed]
- Bigalke, R.D. Laboratory and field observations on the use of Anaplasma centrale as a vaccine against anaplasmosis. Zimbabwe Vet. J. 1980, 11, 21–22. [Google Scholar]
- Palmer, G.H.; McElwain, T.F. Molecular basis for vaccine development against anaplasmosis and babesiosis. Vet. Parasitol. 1995, 57, 233–253. [Google Scholar] [CrossRef]
- Palmer, G.H.; Rurangirwa, F.R.; Kocan, K.M.; Brown, W.C. Molecular basis for vaccine development against the ehrlichial pathogen Anaplasma marginale. Parasitol. Today 1999, 15, 281–286. [Google Scholar] [CrossRef]
- Agnes, J.T.; Brayton, K.A.; LaFollett, M.; Norimine, J.; Brown, W.C.; Palmer, G.H. Identification of Anaplasma marginale outer membrane protein antigens conserved between A. marginale sensu stricto strains and the live A. marginale subsp. centrale vaccine. Infect. Immun. 2011, 79, 1311–1318. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.E.; Siems, W.F.; Palmer, G.H.; Brayton, K.A.; Mcguire, T.C.; Norimine, J.; Brown, W.C. Identification of novel antigenic proteins in a complex Anaplasma marginale outer membrane immunogen by mass spectrometry and genomic mapping. Infect. Immun. 2005, 73, 8109–8118. [Google Scholar] [CrossRef] [PubMed]
- Noh, S.M.; Brayton, K.A.; Brown, W.C.; Norimine, J.; Munske, G.R.; Davitt, C.M.; Palmer, G.H. Composition of the surface proteome of Anaplasma marginale and its role in protective immunity induced by outer membrane immunization. Infect. Immun. 2008, 76, 2219–2226. [Google Scholar] [CrossRef] [PubMed]
- Noh, S.M.; Zhuang, Y.; Futse, J.E.; Brown, W.C.; Brayton, K.A.; Palmer, G.H. The immunization-induced antibody response to the Anaplasma marginale major surface protein 2 and its association with protective immunity. Vaccine 2010, 28, 3741–3747. [Google Scholar] [CrossRef] [PubMed]
- Palmer, G.H.; Brown, W.C.; Noh, S.M.; Brayton, K.A. Genome-wide screening and identification of antigens for rickettsial vaccine development. FEMS Immunol. Med. Microbiol. 2012, 64, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Ducken, D.R.; Brown, W.C.; Alperin, D.C.; Brayton, K.A.; Reif, K.E.; Turse, J.E.; Palmer, G.H.; Noh, S.M. Subdominant outer membrane antigens in Anaplasma marginale: Conservation, antigenicity, and protective capacity using recombinant protein. PLoS ONE 2015, 10, e0129309. [Google Scholar] [CrossRef] [PubMed]
- Allred, D.R.; McGuire, T.C.; Palmer, G.H.; Leib, S.R.; Harkins, T.M.; McElwain, T.F.; Barbet, A.F. Molecular basis for surface antigen size polymorphisms and conservation of a neutralization-sensitive epitope in Anaplasma marginale. Proc. Natl. Acad. Sci. USA 1990, 87, 3220–3224. [Google Scholar] [CrossRef] [PubMed]
- Brown, W.C.; McGuire, T.C.; Mwangi, W.; Kegerreis, K.A.; Macmillan, H.; Lewin, H.A.; Palmer, G.H. Major histocompatibility complex class II DR-restricted memory CD4+ T-lymphocytes recognize conserved immunodominant epitopes of Anaplasma marginale major surface protein 1a. Infect. Immun. 2002, 70, 5521–5532. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Garcia-Garcia, J.C.; de la Fuente, J.; Kocan, K.M.; Blouin, E.F.; Halbur, T.; Onet, V.C.; Saliki, J.T. Mapping of B-cell epitopes in the N-terminal repeated peptides of Anaplasma marginale major surface protein 1a and characterization of the humoral immune response of cattle immunized with recombinant and whole organism antigens. Vet. Immunol. Immunopathol. 2004, 98, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Deringer, J.R.; Forero-Becerra, E.G.; Ueti, M.W.; Turse, J.E.; Futse, J.E.; Noh, S.M.; Palmer, G.H.; Brown, W.C. Identification of a T-cell epitope that is globally conserved among outer membrane proteins (OMPs) OMP7, OMP8, and OMP9 of Anaplasma marginale strains and with OMP7 from the A. marginale subsp. centrale vaccine strain. Clin. Vaccine Immunol. 2017, 24, e00406-16. [Google Scholar] [CrossRef] [PubMed]
- Gale, K. Anaplasma marginale: Detection of carrier cattle by PCR-ELISA. Int. J. Parasitol. 1996, 26, 1103–1109. [Google Scholar] [CrossRef]
- Visser, E.S.; McGuire, T.C.; Palmer, G.H.; Davis, W.C.; Shkap, V.; Pipano, E.; Knowles, D.P. The Anaplasma marginale msp5 gene encodes a 19-kilodalton protein conserved in all recognized Anaplasma species. Infect. Immun. 1992, 60, 5139–5144. [Google Scholar] [PubMed]
- Bekker, C.P.J.; de Vos, S.; Taoufik, A.; Sparagano, O.A.E.; Jongejan, F. Simultaneous detection of Anaplasma and Ehrlichia species in ruminants and detection of Ehrlichia ruminantium in Amblyomma variegatum ticks by reverse line blot hybridization. Vet. Microbiol. 2002, 89, 223–238. [Google Scholar] [CrossRef]
- Chaisi, M.E.; Baxter, J.R.; Hove, P.; Choopa, C.N.; Oosthuizen, M.C.; Brayton, K.A.; Khumalo, Z.T.H.; Mutshembele, A.M.; Mtshali, M.S.; Collins, N.E. Comparison of three nucleic acid-based tests for detecting Anaplasma marginale and Anaplasma centrale in cattle. Onderstepoort J. Vet. Res. 2017, 84, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Machado, R.Z.; Teixeira, M.M.G.; Rodrigues, A.C.; André, M.R.; Gonçalves, L.R.; Barbosa da Silva, J.; Pereira, C.L. Molecular diagnosis and genetic diversity of tick-borne Anaplasmataceae agents infecting the African buffalo Syncerus caffer from Marromeu Reserve in Mozambique. Parasit. Vectors 2016, 9, 454. [Google Scholar] [CrossRef] [PubMed]
- Sisson, D.; Hufschmid, J.; Jolles, A.; Beechler, B.; Jabbar, A. Molecular characterisation of Anaplasma species from African buffalo (Syncerus caffer) in Kruger National Park, South Africa. Ticks Tick. Borne. Dis. 2017, 8, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Molad, T.; Mazuz, M.L.; Fleiderovitz, L.; Fish, L.; Savitsky, I.; Krigel, Y.; Leibovitz, B.; Molloy, J.; Jongejan, F.; Shkap, V. Molecular and serological detection of A. centrale- and A. marginale-infected cattle grazing within an endemic area. Vet. Microbiol. 2006, 113, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Carelli, G.; Decaro, N.; Lorusso, A.; Elia, G.; Lorusso, E.; Mari, V.; Ceci, L.; Buonavoglia, C. Detection and quantification of Anaplasma marginale DNA in blood samples of cattle by real-time PCR. Vet. Microbiol. 2007, 124, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Decaro, N.; Carelli, G.; Lorusso, E.; Lucente, M.S.; Greco, G.; Lorusso, A.; Radogna, A.; Ceci, L.; Buonavoglia, C. Duplex real-time polymerase chain reaction for simultaneous detection and quantification of Anaplasma marginale and Anaplasma centrale. J. Vet. Diagnostic Investig. 2008, 20, 606–611. [Google Scholar] [CrossRef] [PubMed]
- Lew, A.E.; Bock, R.E.; Minchin, C.M.; Masaka, S. A msp1-alpha polymerase chain reaction assay for specific detection and differentiation of Anaplasma marginale isolates. Vet. Microbiol. 2002, 86, 325–335. [Google Scholar] [CrossRef]
- Hove, P.; Chaisi, M.E.; Brayton, K.A.; Ganesan, H.; Catanese, H.N.; Mtshali, M.S.; Mutshembele, A.M.; Oosthuizen, M.C.; Collins, N.E. Co-infections with multiple genotypes of Anaplasma marginale in cattle indicate pathogen diversity. Parasit. Vectors 2018, 11, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Byaruhanga, C.; Collins, N.E.; Knobel, D.; Chaisi, M.E.; Vorster, I.; Steyn, H.C.; Oosthuizen, M.C. Molecular investigation of tick-borne haemoparasite infections among transhumant zebu cattle in Karamoja Region, Uganda. Vet. Parasitol. Reg. Stud. Reports 2016, 3–4, 27–35. [Google Scholar] [CrossRef]
- Pfitzer, S.; Oosthuizen, M.C.; Bosman, A.-M.; Vorster, I.; Penzhorn, B.L. Tick-borne blood parasites in nyala (Tragelaphus angasii, Gray 1849) from KwaZulu-Natal, South Africa. Vet. Parasitol. 2011, 176, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Gubbels, J.M.; de Vos, A.P.; van der Weide, M.; Viseras, J.; Schouls, L.M.; de Vries, E.; Jongejan, F. Simultaneous detection of bovine Theileria and Babesia species by reverse line blot hybridization. J. Clin. Microbiol. 1999, 37, 1782–1789. [Google Scholar] [PubMed]
- Rodríguez, J.-L.; Palmer, G.H.; Knowles, D.P.; Brayton, K.A. Distinctly different msp2 pseudogene repertoires in Anaplasma marginale strains that are capable of superinfection. Gene 2005, 361, 127–132. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente, J.; Ruybal, P.; Mtshali, M.S.; Naranjo, V.; Shuqing, L.; Mangold, A.J.; Rodríguez, S.D.; Jiménez, R.; Vicente, J.; Moretta, R.; et al. Analysis of world strains of Anaplasma marginale using major surface protein 1a repeat sequences. Vet. Microbiol. 2007, 119, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Palmer, G.H.; Rurangirwa, F.R.; McElwain, T.F. Strain composition of the ehrlichia Anaplasma marginale within persistently infected cattle, a mammalian reservoir for tick transmission. J. Clin. Microbiol. 2001, 39, 631–635. [Google Scholar] [CrossRef] [PubMed]
- De la Fuente, J.; Van Den Bussche, R.A.; Garcia-Garcia, J.C.; Rodríguez, S.D.; García, M.A.; Guglielmone, A.A.; Mangold, A.J.; Friche Passos, L.M.; Barbosa Ribeiro, M.F.; Blouin, E.F.; et al. Phylogeography of New World isolates of Anaplasma marginale based on major surface protein sequences. Vet. Microbiol. 2002, 88, 275–285. [Google Scholar] [CrossRef]
- Ybañez, A.P.; Ybañez, R.H.D.; Claveria, F.G.; Cruz-Flores, M.J.; Xuenan, X.; Yokoyama, N.; Inokuma, H. High genetic diversity of Anaplasma marginale detected from Philippine cattle. J. Vet. Med. Sci. 2014, 76, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Peña, A.; Naranjo, V.; Acevedo-Whitehouse, K.; Mangold, A.J.; Kocan, K.M.; de la Fuente, J. Phylogeographic analysis reveals association of tick-borne pathogen, Anaplasma marginale, MSP1a sequences with ecological traits affecting tick vector performance. BMC Biol. 2009, 7, 57. [Google Scholar] [CrossRef] [PubMed]
- Ruybal, P.; Moretta, R.; Perez, A.; Petrigh, R.; Zimmer, P.; Alcaraz, E.; Echaide, I.; Torioni de Echaide, S.; Kocan, K.M.; de la Fuente, J.; et al. Genetic diversity of Anaplasma marginale in Argentina. Vet. Parasitol. 2009, 162, 176–180. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, J.B.; André, M.R.; Machado, R.Z. Low genetic diversity of Anaplasma marginale in calves in an endemic area for bovine anaplasmosis in the state of São Paulo, Brazil. Ticks Tick. Borne. Dis. 2016, 7, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Ocampo Espinoza, V.; Vázquez, J.E.S.; Aguilar, M.D.; Ortiz, M.Á.G.; Alarcón, G.J.C.; Rodríguez, S.D. Anaplasma marginale: Lack of cross-protection between strains that share MSP1a variable region and MSP4. Vet. Microbiol. 2006, 114, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Palmer, G.H.; Knowles, D.P.; Rodriguez, J.L.; Gnad, D.P.; Hollis, L.C.; Marston, T.; Brayton, K.A. Stochastic transmission of multiple genotypically distinct Anaplasma marginale strains in a herd with high prevalence of Anaplasma infection. J. Clin. Microbiol. 2004, 42, 5381–5384. [Google Scholar] [CrossRef] [PubMed]
- Catanese, H.N.; Brayton, K.A.; Gebremedhin, A.H. RepeatAnalyzer: A tool for analysing and managing short-sequence repeat data. BMC Genomics 2016, 17, 422. [Google Scholar] [CrossRef] [PubMed]
- Brayton, K.A. Persistence and antigenic variation. In Intracellular Pathogens II: Rickettsiales; Palmer, G., Azad, F., Eds.; ASM Press: Washington DC, USA, 2012; pp. 366–390. [Google Scholar]
| Assay | Cost per sample (South African Rand - R) | Average throughput time | Comments on assay sensitivity | Technical skills & expensive equipment needed? |
|---|---|---|---|---|
| Light microscopic examination of Giemsa-stained smears [5,44] | R113 | 3 days | Low (106 A. marginale- infected erythrocytes per ml of blood) Best used during acute phase of infection | Low to Medium No |
| Msp5 competitive ELISA (cELISA) [5,45] | R140 | 4 days | Low to Medium Results in false negatives Detects Anaplasma to genus level only | Medium to High Yes |
| Reverse line blot (RLB) hybridisation [46,47] | R445 | 3 days | Medium to high Similar to PCR & higher than nPCR, but lower than qPCR | Medium to High Yes |
| Conventional PCR [48,49] | R250 | 2 days | Medium Similar to RLB | Medium to High Yes |
| Nested PCR [47,50] | R350 | 3 days | Medium Fails to detect genetic variant sequences leading to false negatives Less sensitive than RLB & qPCR | Medium to High Yes |
| Duplex quantitative real-time PCR (qPCR) [47,51,52] | R430 | 2 days | High (30 Anaplasma- infected erythrocytes per ml of blood) Detects parasites at very low levels Most sensitive test available in South Africa | Medium to High Yes |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hove, P.; Khumalo, Z.T.H.; Chaisi, M.E.; Oosthuizen, M.C.; Brayton, K.A.; Collins, N.E. Detection and Characterisation of Anaplasma marginale and A. centrale in South Africa. Vet. Sci. 2018, 5, 26. https://doi.org/10.3390/vetsci5010026
Hove P, Khumalo ZTH, Chaisi ME, Oosthuizen MC, Brayton KA, Collins NE. Detection and Characterisation of Anaplasma marginale and A. centrale in South Africa. Veterinary Sciences. 2018; 5(1):26. https://doi.org/10.3390/vetsci5010026
Chicago/Turabian StyleHove, Paidashe, Zamantungwa T. H. Khumalo, Mamohale E. Chaisi, Marinda C. Oosthuizen, Kelly A. Brayton, and Nicola E. Collins. 2018. "Detection and Characterisation of Anaplasma marginale and A. centrale in South Africa" Veterinary Sciences 5, no. 1: 26. https://doi.org/10.3390/vetsci5010026
APA StyleHove, P., Khumalo, Z. T. H., Chaisi, M. E., Oosthuizen, M. C., Brayton, K. A., & Collins, N. E. (2018). Detection and Characterisation of Anaplasma marginale and A. centrale in South Africa. Veterinary Sciences, 5(1), 26. https://doi.org/10.3390/vetsci5010026





