The Performance of Three Immune Assays to Assess the Serological Status of Cattle Experimentally Exposed to Mycoplasma bovis
Abstract
1. Introduction
2. Materials and Methods
2.1. Mycoplasma Culture and DNA Isolation
2.2. Amplification and Cloning of the VspA RA1 Motif
2.3. Expression of Recombinant Polypeptides
2.4. SDS-PAGE and Western Blotting
2.5. Antibodies and Bovine Sera Used in This Study
2.5.1. Monoclonal Antibodies:
2.5.2. Canadian Cattle Sera
2.5.3. English Cattle Sera
2.5.4. Australian Cattle Sera
2.5.5. Control Sera from Commercial Serological ELISA Assays
2.6. Commercial ELISA Assays
2.7. Western Blot Analyses with the S-vspA_RA1 Polypeptide
2.8. Detection of Seroconversion to M. bovis
2.9. Onset of Detectable Antibodies to M. bovis Post-Exposure
2.10. Statistical Methods
3. Results
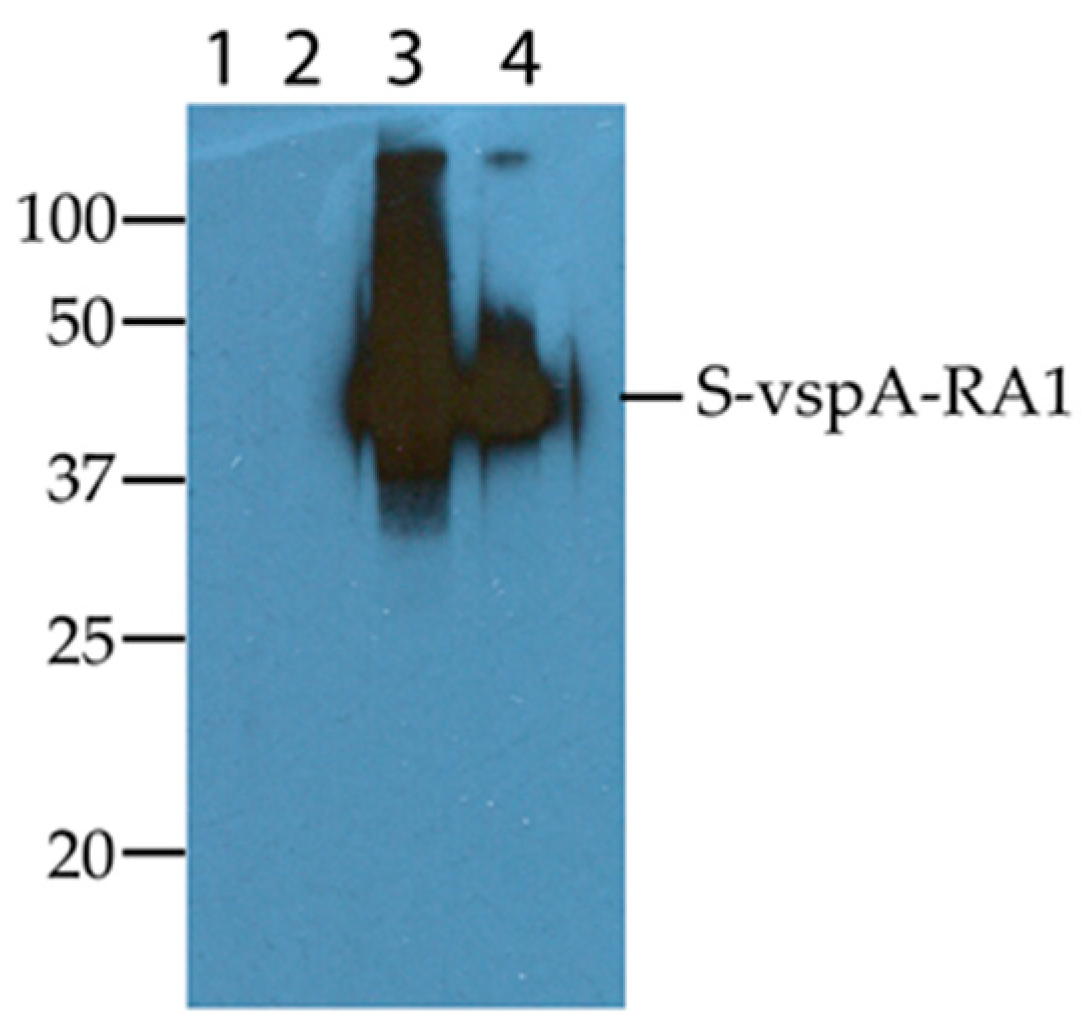
3.1. Expression of the vspA-RA1 Motif
3.2. Immunoreactivity of the S-vspA_RA1 Recombinant Polypeptide
3.3. Reactivity of Canadian Cattle Sera
- Western blot: The Day 0 serum samples from Canadian cattle (n = 5) had varying reactivity with the S-vspA_RA1 recombinant polypeptide (Table 1). Three of the five Day 0 samples tested negative by Western blot, while the other two samples tested positive (Table 1). The Day 68 samples (n = 9) were reactive with the S-vspA_RA1 recombinant polypeptide and were deemed to be positive (Table 1).
3.4. Reactivity of English Cattle Sera
- BIO K302 ELISA: The Day 0 and Day 28 samples from the unexposed animal and the Day 0 samples from the two exposed animals tested negative in the BIO K302 ELISA (Table 2). Only one of the Day 28 samples from the two exposed animals tested positive (Table 2). The “cut-off control” and the “positive control” tested negative and positive, respectively (Table 2).
- BIO K260 ELISA: The Day 0 and Day 28 samples from the unexposed animal and the Day 0 samples from the two exposed animals tested negative in the BIO K260 ELISA (Table 2). Only one of the Day 28 samples from the two exposed animals tested positive (Table 2). The “cut-off control” and the “positive control” tested negative and positive, respectively (Table 2).
- Western blot: The immunoreactivities of the English cattle sera with the S-vspA_RA1 polypeptide from the two animals before and after exposure to M. bovis were consistent with the results of an in-house ELISA, with the Day 0 samples testing negative and the Day 28 samples testing positive (Table 2). The two control samples used in the in-house ELISA were both reactive in the Western blot assay and were deemed positive. Neither the Day 0 nor the Day 28 sample from the unexposed animal reacted with the S-vspA_RA1 polypeptide (Table 2).
3.5. Reactivity of Australian Cattle Sera
- BIO K302 ELISA: All samples (Day 0 and Day 24) from the group (n = 5) that were not exposed to M. bovis tested negative in the BIO K302 ELISA (Table 3). Two Day 0 samples from the M. bovis exposed group tested positive (APCAH32 and APCAH48) in this ELISA (Table 3). Eleven of the Day 24 samples from the exposed group (n = 30) tested positive in the BIO K302 ELISA (Table 3).
- Western blot: The S-vspA_RA1 Western blotting results were compared to the results of the MilA M. bovis ELISA (Table 3) [25]. None of the Day 0 or Day 24 serum samples from Australian calves (n = 5) that were not exposed to the M. bovis reacted with the S-vspA_RA1 (Table 3). Three of the Day 0 samples from the Australian calves that were subsequently exposed to M. bovis (n = 30) reacted with the S-vspA_RA1 polypeptide, while the remaining Day 0 samples did not react. The Day 24 samples from these animals also reacted in the Western blot. In total, 18 of the Day 24 samples from this group reacted with S-vspA_RA1 (Table 3).
3.6. Reactivity of Bio-X Diagnostics Control Sera
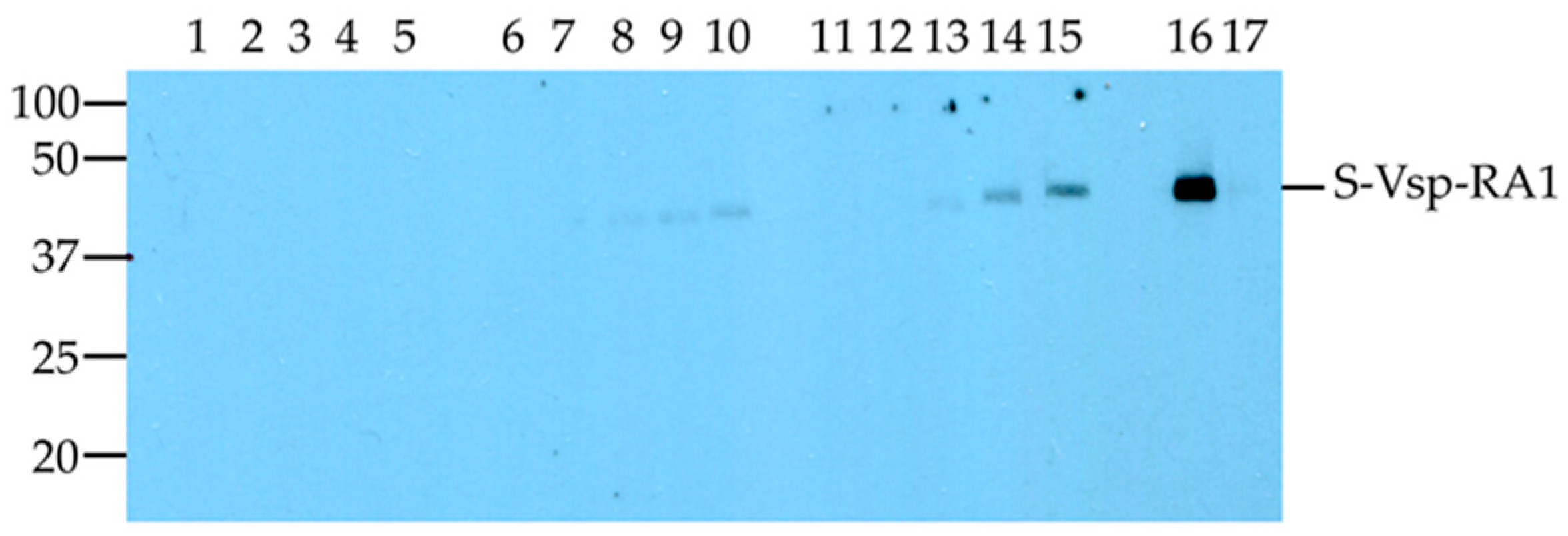
- Western blot: The reactivities of the ELISA control sera obtained from the three batches of the BIO K302 kit, the four batches of the BIO K260 kit, and the four batches of the pentavalent BIO K284 ELISA kit with the S-vspA_RA1 polypeptide are summarised in Table 4 (representative examples supplemental file Figure S1). The reactivity of all Bio-X Diagnostics sera with the SUMO-CAT polypeptide was also assessed. All positive control sera showed reactivity with S-vspA_RA1 and no reactivity was evident with any of the negative control sera tested (Table 4, representative examples supplemental file Figure S1). None of positive or negative sera from the BIO K302 or BIO K260 kits reacted with the SUMO-CAT polypeptide (Table 4). The positive control sera from the pentavalent BIO K284 reacted with the S-vspA_RA1 polypeptide and with the SUMO-CAT polypeptide (Table 4).
3.7. Immunoassay Se and Sp Estimates
3.8. Detection of Seroconversion
3.8.1. Detection of a Seroconversion Using the BIO K302 ELISA
3.8.2. Detection of Seroconversion Using the BIO K260 ELISA
3.8.3. Detection of Seroconversion Using Western Blotting
3.8.4. Time to Serodetection
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Arcangioli, M.A.; Duet, A.; Meyer, G.; Dernburg, A.; Bezille, P.; Poumarat, F.; Le Grand, D. The role of Mycoplasma bovis in bovine respiratory disease outbreaks in veal calf feedlots. Vet. J. 2008, 177, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Maunsell, F.P.; Woolums, A.R.; Francoz, D.; Rosenbusch, R.F.; Step, D.L.; Wilson, D.J.; Janzen, E.D. Mycoplasma bovis infections in cattle. J. Vet. Intern. Med. 2011, 25, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Radaelli, E.; Castiglioni, V.; Losa, M.; Benedetti, V.; Piccinini, R.; Nicholas, R.A.; Scanziani, E.; Luini, M. Outbreak of bovine clinical mastitis caused by Mycoplasma bovis in a north Italian herd. Res. Vet. Sci. 2011, 91, 251–253. [Google Scholar] [CrossRef] [PubMed]
- Bras, A.L.; Barkema, H.W.; Woodbury, M.R.; Ribble, C.S.; Perez-Casal, J.; Windeyer, M.C. Clinical presentation, prevalence, and risk factors associated with Mycoplasma bovis-associated disease in farmed bison (Bison bison) herds in western Canada. J. Am. Vet. Med. Assoc. 2017, 250, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Horwood, P.F.; Schibrowski, M.I.; Fowler, E.V.; Gibson, J.S.; Barnes, T.S.; Mahony, T.J. Is Mycoplasma bovis a missing component of the bovine respiratory disease complex in Australia? Aust. Vet. J. 2014, 92, 185–191. [Google Scholar] [CrossRef] [PubMed]
- Behrens, A.; Heller, M.; Kirchhoff, H.; Yogev, D.; Rosengarten, R. A family of phase- and size-variant membrane surface lipoprotein antigens (vsps) of Mycoplasma bovis. Infect. Immun. 1994, 62, 5075–5084. [Google Scholar] [PubMed]
- Beier, T.; Hotzel, H.; Lysnyansky, I.; Grajetzki, C.; Heller, M.; Rabeling, B.; Yogev, D.; Sachse, K. Intraspecies polymorphism of vsp genes and expression profiles of variable surface protein antigens (vsps) in field isolates of Mycoplasma bovis. Vet. Microbiol. 1998, 63, 189–203. [Google Scholar] [CrossRef]
- Brank, M.; Le Grand, D.; Poumarat, F.; Bezille, P.; Rosengarten, R.; Citti, C. Development of a recombinant antigen for antibody-based diagnosis of Mycoplasma bovis infection in cattle. Clin. Diagn. Lab. Immunol. 1999, 6, 861–867. [Google Scholar] [PubMed]
- Lysnyansky, I.; Sachse, K.; Rosenbusch, R.; Levisohn, S.; Yogev, D. The vsp locus of Mycoplasma bovis: Gene organization and structural features. J. Bacteriol. 1999, 181, 5734–5741. [Google Scholar] [PubMed]
- Rosengarten, R.; Yogev, D. Variant colony surface antigenic phenotypes within mycoplasma strain populations: Implications for species identification and strain standardization. J. Clin. Microbiol. 1996, 34, 149–158. [Google Scholar] [PubMed]
- Poumarat, F.; Le Grand, D.; Solsona, M.; Rosengarten, R.; Citti, C. Vsp antigens and vsp-related DNA sequences in field isolates of Mycoplasma bovis. FEMS Microbiol. Lett. 1999, 173, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Le Grand, D.; Calavas, D.; Brank, M.; Citti, C.; Rosengarten, R.; Bezille, P.; Poumarat, F. Serological prevalence of Mycoplasma bovis infection in suckling beef cattle in France. Vet. Rec. 2002, 150, 268–273. [Google Scholar] [CrossRef]
- Rosengarten, R.; Behrens, A.; Stetefeld, A.; Heller, M.; Ahrens, M.; Sachse, K.; Yogev, D.; Kirchhoff, H. Antigen heterogeneity among isolates of Mycoplasma bovis is generated by high-frequency variation of diverse membrane surface proteins. Infect. Immun. 1994, 62, 5066–5074. [Google Scholar] [PubMed]
- Thomas, C.B.; Jasper, D.E.; Boothby, J.T.; Dellinger, J.D. Detection of bovine serum antibody specific to Mycoplasma bovis and Mycoplasma californicum by enzyme-linked immunosorbent assay (ELISA). Isr. J. Med. Sci. 1987, 23, 723–728. [Google Scholar] [PubMed]
- Nicholas, R.A.; Ayling, R.D.; Stipkovits, L.P. An experimental vaccine for calf pneumonia caused by Mycoplasma bovis: Clinical, cultural, serological and pathological findings. Vaccine 2002, 20, 3569–3575. [Google Scholar] [CrossRef]
- Prysliak, T.; van der Merwe, J.; Lawman, Z.; Wilson, D.; Townsend, H.; van Drunen Littel-van den Hurk, S.; Perez-Casal, J. Respiratory disease caused by Mycoplasma bovis is enhanced by exposure to bovine herpes virus 1 (BHV-1) but not to bovine viral diarrhea virus (BVDV) type 2. Can. Vet. J. 2011, 52, 1195–1202. [Google Scholar] [PubMed]
- Prysliak, T.; van der Merwe, J.; Perez-Casal, J. Vaccination with recombinant Mycoplasma bovis GAPDH results in a strong humoral immune response but does not protect feedlot cattle from an experimental challenge with M. bovis. Microb. Pathog. 2013, 55, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ghadersohi, A.; Fayazi, Z.; Hirst, R.G. Development of a monoclonal blocking ELISA for the detection of antibody to Mycoplasma bovis in dairy cattle and comparison to detection by PCR. Vet. Immunol. Immunopathol. 2005, 104, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Register, K.B.; Sacco, R.E.; Olsen, S.C. Evaluation of enzyme-linked immunosorbent assays for detection of Mycoplasma bovis-specific antibody in bison sera. Clin. Vaccine Immunol. 2013, 20, 1405–1409. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Khan, F.A.; Zhu, X.; Zhang, R.; Mustafa, R.; Hu, C.; Chen, Y.; Chen, H.; Guo, A. Establishment of an antibody avidity test to differentiate vaccinated cattle from those naturally infected with Mycoplasma bovis. Vet. J. 2015, 203, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Robino, P.; Alberti, A.; Pittau, M.; Chessa, B.; Miciletta, M.; Nebbia, P.; Le Grand, D.; Rosati, S. Genetic and antigenic characterization of the surface lipoprotein p48 of Mycoplasma bovis. Vet. Microbiol. 2005, 109, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Wawegama, N.K.; Browning, G.F.; Kanci, A.; Marenda, M.S.; Markham, P.F. Development of a recombinant protein-based enzyme-linked immunosorbent assay for diagnosis of Mycoplasma bovis infection in cattle. Clin. Vaccine Immunol. 2014, 21, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Fu, P.; Sun, Z.; Zhang, Y.; Yu, Z.; Zhang, H.; Su, D.; Jiang, F.; Wu, W. Development of a direct competitive elisa for the detection of Mycoplasma bovis infection based on a monoclonal antibody of p48 protein. BMC Vet. Res. 2014, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Fu, P.; Wei, K.; Zhang, H.; Zhang, Y.; Xu, J.; Jiang, F.; Liu, X.; Xu, W.; Wu, W. Identification of novel immunogenic proteins from Mycoplasma bovis and establishment of an indirect ELISA based on recombinant E1 beta subunit of the pyruvate dehydrogenase complex. PLoS ONE 2014, 9, e88328. [Google Scholar] [CrossRef] [PubMed]
- Wawegama, N.K.; Markham, P.F.; Kanci, A.; Schibrowski, M.; Oswin, S.; Barnes, T.S.; Firestone, S.M.; Mahony, T.J.; Browning, G.F. Evaluation of an IgG enzyme-linked immunosorbent assay as a serological assay for detection of Mycoplasma bovis infection in feedlot cattle. J. Clin. Microbiol. 2016, 54, 1269–1275. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.K.; Petersen, M.B.; Nielsen, L.R.; Halasa, T.; Toft, N. Latent class analysis of bulk tank milk PCR and ELISA testing for herd level diagnosis of Mycoplasma bovis. Prev. Vet. Med. 2015, 121, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Byrne, W.J.; Ball, H.J.; Brice, N.; McCormack, R.; Baker, S.E.; Ayling, R.D.; Nicholas, R.A. Application of an indirect ELISA to milk samples to identify cows with Mycoplasma bovis mastitis. Vet. Rec. 2000, 146, 368–369. [Google Scholar] [CrossRef] [PubMed]
- Wawegama, N.K.; Kanci, A.; Marenda, M.S.; Mansell, P.D.; Browning, G.F.; Markham, P.F. Histochemical and morphometric characterization of broncho-pneumonia in calves caused by infection with Mycoplasma bovis. Vet. Microbiol. 2012, 158, 220–224. [Google Scholar] [CrossRef] [PubMed]
- Lysnyansky, I.; Rosengarten, R.; Yogev, D. Phenotypic switching of variable surface lipoproteins in Mycoplasma bovis involves high-frequency chromosomal rearrangements. J. Bacteriol. 1996, 178, 5395–5401. [Google Scholar] [CrossRef] [PubMed]
- Lysnyansky, I.; Ron, Y.; Yogev, D. Juxtaposition of an active promoter to vsp genes via site-specific DNA inversions generates antigenic variation in Mycoplasma bovis. J. Bacteriol. 2001, 183, 5698–5708. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lin, J.S.; Albrecht, J.C.; Meagher, R.J.; Wang, X.; Barron, A.E. Completely monodisperse, highly repetitive proteins for bioconjugate capillary electrophoresis: Development and characterization. Biomacromolecules 2011, 12, 2275–2284. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Parker, A.M.; Shukla, A.; House, J.K.; Hazelton, M.S.; Bosward, K.L.; Kokotovic, B.; Sheehy, P.A. Genetic characterization of Australian Mycoplasma bovis isolates through whole genome sequencing analysis. Vet. Microbiol. 2016, 196, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Caswell, J.L.; Bateman, K.G.; Cai, H.Y.; Castillo-Alcala, F. Mycoplasma bovis in respiratory disease of feedlot cattle. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Nicholas, R.A.; Ayling, R.D. Mycoplasma bovis: Disease, diagnosis, and control. Res. Vet. Sci. 2003, 74, 105–112. [Google Scholar] [CrossRef]
- Jacobson, R.H. Validation of serological assays for diagnosis of infectious diseases. Rev. Sci. Tech. 1998, 17, 469–526. [Google Scholar] [CrossRef] [PubMed]
| Serological Assay | ||||||||
|---|---|---|---|---|---|---|---|---|
| B302 | B260 1 | WB | wcE 2 | |||||
| Animal | Day 0 | Day 68 | Day 0 | Day 68 | Day 0 | Day 68 | Day 0 | Day 68 |
| VIDO18 | Pos | Pos (5) | Pos | Pos (H) | ||||
| VIDO19 | Neg | Neg | Neg (0) | Neg (0) | Neg | Pos | Neg | Pos (L) |
| VIDO20 | Neg | Pos | Neg (0) | Pos (2) | Pos | Pos | Neg | Pos (H) |
| VIDO21 | Neg | Neg | Neg (0) | Neg (0) | Neg | Pos | Neg | Pos (L) |
| VIDO25 | Pos | Pos (5) | Pos | Pos (H) | ||||
| VIDO26 | Pos | Neg (0) | Pos | Pos (L) | ||||
| VIDO29 | Pos | Pos (4) | Pos | Pos (H) | ||||
| VIDO30 | Neg | Pos | Neg (0) | Pos (3) | Pos | Pos | Neg | Pos (H) |
| VIDO34 | Pos | Pos (3) | Pos | Pos (H) | ||||
| VIDO38 | Neg | Neg (0) | Neg | Neg | ||||
| Serological Assay | ||||||||
|---|---|---|---|---|---|---|---|---|
| B302 | B260 1 | WB | wcE 2 | |||||
| Animal | Day 0 | Day 28 | Day 0 | Day 28 | Day 0 | Day 28 | Day 0 | Day 28 |
| APHA370 3 | Neg | Neg | Neg (0) | Neg (0) | Neg | Neg | Neg | Neg |
| APHA304 4 | Neg | Neg | Neg (0) | Neg (0) | Neg | Pos | Neg | Pos |
| APHA339 4 | Neg | Pos | Neg (0) | Pos (1) | Neg | Pos | Neg | Pos |
| C_Cont 2 | Neg | Neg (0) | Pos | Pos | ||||
| P_Cont 2 | Pos | Pos (4) | Pos | Pos | ||||
| Serological Assay | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| B302 | B260 1 | WB1 2 | WB2 | MilA 3 | ||||||
| Animal | D0 | D24 | D0 | D24 | D0 | D24 | D0 | D24 | D0 | D24 |
| Unexposed | ||||||||||
| APCAH13 | N | N | N (0) | N (0) | N | N | P | P | N | N |
| APCAH24 | N | N | N (0) | N (0) | N | N | P | N | N | N |
| APCAH30 | N | N | N (0) | N (0) | N | N | N | N | N | N |
| APCAH43 | N | N | N (0) | N (0) | N | N | N | N | ||
| APCAH71 | N | N | N (0) | N (0) | N | N | N | N | ||
| Exposed | ||||||||||
| APCAH3 | N | N | N (0) | N (0) | N | N | N | N | N | P |
| APCAH5 | N | P | N (0) | N (0) | N | P | N | P | N | P |
| APCAH7 | N | N | N (0) | N (0) | N | P | N | N | N | N |
| APCAH8 | N | P | N (0) | N (0) | N | P | N | N | N | N |
| APCAH9 | N | P | N (0) | P (1) | N | P | P | P | N | P |
| APCAH10 | N | N | N (0) | N (0) | N | P | P | P | N | P |
| APCAH18 | N | N | N (0) | N (0) | N | P | P | P | N | N |
| APCAH20 | N | P | N (0) | N (0) | N | P | N | P | N | P |
| APCAH21 | N | N | N (0) | N (0) | N | P | N | P | N | P |
| APCAH22 | N | P | N (0) | P (2) | N | P | N | N | N | P |
| APCAH23 | N | N | N (0) | N (0) | P | P | P | P | N | P |
| APCAH26 | N | P | N (0) | N (0) | N | P | P | P | N | P |
| APCAH31 | N | N | N (0) | N (0) | N | N | N | P | N | P |
| APCAH32 | P | P | N (0) | P (1) | P | P | N | P | ||
| APCAH34 | N | N | N (0) | N (0) | N | N | N | P | ||
| APCAH35 | N | P | N (0) | P (1) | N | P | N | P | ||
| APCAH36 | N | P | N (0) | N (0) | N | N | N | P | ||
| APCAH45 | N | P | N (0) | N (0) | N | N | N | P | ||
| APCAH46 | N | N | N (0) | N (0) | N | N | N | P | ||
| APCAH48 | P | P | N (0) | N (0) | P | P | N | P | ||
| APCAH49 | N | N | N (0) | N (0) | N | N | N | P | ||
| APCAH51 | N | N | N (0) | N (0) | N | N | N | P | ||
| APCAH52 | N | N | N (0) | N (0) | N | N | N | P | ||
| APCAH53 | N | N | N (0) | N (0) | N | N | N | P | ||
| APCAH54 | N | N | N (0) | N (0) | N | P | N | P | ||
| APCAH57 | N | N | N (0) | N (0) | N | P | N | P | ||
| APCAH60 | N | N | N (0) | N (0) | N | N | N | P | ||
| APCAH62 | N | N | N (0) | N (0) | N | P | N | P | ||
| APCAH63 | N | N | N (0) | N (0) | N | N | N | P | ||
| APCAH64 | N | N | N (0) | N (0) | N | P | N | P | ||
| Serological Assay | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| B302 | B260 1 | WB1 2 | WB2 2 | ||||||
| Assay | Batch | NegC | PosC | NegC | PosC | NegC | PosC | NegC | PosC |
| B302 | MYC14C04 | N | P | P (3) | N | P | N | N | |
| MYC13K26 | N | P | N (0) | P (3) | N | P | N | N | |
| MYC13J16 | N | P | N (0) | P (3) | N | P | N | N | |
| B260 | SMYC14C04 | N | P | N (0) | P (3) | N | P | N | N |
| SMYC13K26 | N | P | N (0) | P (4) | N | P | N | N | |
| SMYC13J16 | N | P | N (0) | P (3) | N | P | N | N | |
| SMYC09I28 | N | P | N (0) | P (3) | N | P | N | N | |
| B284 | IBRPM12F04 | N | N (0) | P | P | ||||
| IBRPM12K08 | N | N (0) | P | P | |||||
| IBRPM12F28 | N | N (0) | P | P | |||||
| IBRPM09L01 | N | N (0) | P | P | |||||
| BIO K302 | BIO K260 | Western Blot | ||
|---|---|---|---|---|
| Source | n | No. Positive (%) | No. Positive (%) | No. Positive (%) |
| Sensitivity | ||||
| Canada | 9 | 7 (78) | 6 (67) | 9 (100) |
| England | 4 | 2 (50) | 2 (50) | 4 (100) |
| Australia | 30 | 11 (37) | 4 (13) | 18 (60) |
| Total | 43 | 20 (47%; 10–87%) | 12 (28%; 1–92%) | 31 (72%; 16–98%) |
| Specificity | N Negative (%) | N Negative (%) | N Negative (%) | |
| Canada | 5 | 5 (100) | 5 (100) | 3 (60.0) |
| England | 4 | 4 (100) | 4 (100) | 4 (100) |
| Australia | 40 | 38 (95.0) | 40 (100) | 37 (92.5) |
| Total | 49 | 47 (96%; 87–99%) | 49 (100%; 93–100%) | 44 (90%; 56–98%) |
| BIO K302 | BIO K260 | Western Blot | ||||
|---|---|---|---|---|---|---|
| Source | SNeg | SConv | SNeg | SConv | SNeg | SConv |
| Canada | 4 | 2 (50) | 4 | 2 (50) | 2 | 2 (100) |
| England | 2 | 1 (50) | 2 | 0 (0) | 2 | 2 (100) |
| Australia | 28 | 9 (32) | 30 | 1 (13) | 27 | 15 (56) |
| Total | 34 | 12 | 36 | 3 | 31 | 19 |
| 35% (CI: 21–54%) | 8% (CI: 0–87%) | 61% (CI: 29–86%) | ||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schibrowski, M.L.; Barnes, T.S.; Wawegama, N.K.; Vance, M.E.; Markham, P.F.; Mansell, P.D.; Marenda, M.S.; Kanci, A.; Perez-Casal, J.; Browning, G.F.; et al. The Performance of Three Immune Assays to Assess the Serological Status of Cattle Experimentally Exposed to Mycoplasma bovis. Vet. Sci. 2018, 5, 27. https://doi.org/10.3390/vetsci5010027
Schibrowski ML, Barnes TS, Wawegama NK, Vance ME, Markham PF, Mansell PD, Marenda MS, Kanci A, Perez-Casal J, Browning GF, et al. The Performance of Three Immune Assays to Assess the Serological Status of Cattle Experimentally Exposed to Mycoplasma bovis. Veterinary Sciences. 2018; 5(1):27. https://doi.org/10.3390/vetsci5010027
Chicago/Turabian StyleSchibrowski, Meghan L., Tamsin S. Barnes, Nadeeka K. Wawegama, Megan E. Vance, Philip F. Markham, Peter D. Mansell, Marc S. Marenda, Anna Kanci, José Perez-Casal, Glenn F. Browning, and et al. 2018. "The Performance of Three Immune Assays to Assess the Serological Status of Cattle Experimentally Exposed to Mycoplasma bovis" Veterinary Sciences 5, no. 1: 27. https://doi.org/10.3390/vetsci5010027
APA StyleSchibrowski, M. L., Barnes, T. S., Wawegama, N. K., Vance, M. E., Markham, P. F., Mansell, P. D., Marenda, M. S., Kanci, A., Perez-Casal, J., Browning, G. F., Gibson, J. S., & Mahony, T. J. (2018). The Performance of Three Immune Assays to Assess the Serological Status of Cattle Experimentally Exposed to Mycoplasma bovis. Veterinary Sciences, 5(1), 27. https://doi.org/10.3390/vetsci5010027







