Detection of Leptospiral DNA in the Urine of Donkeys on the Caribbean Island of Saint Kitts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Samples
2.2. DNA Extraction
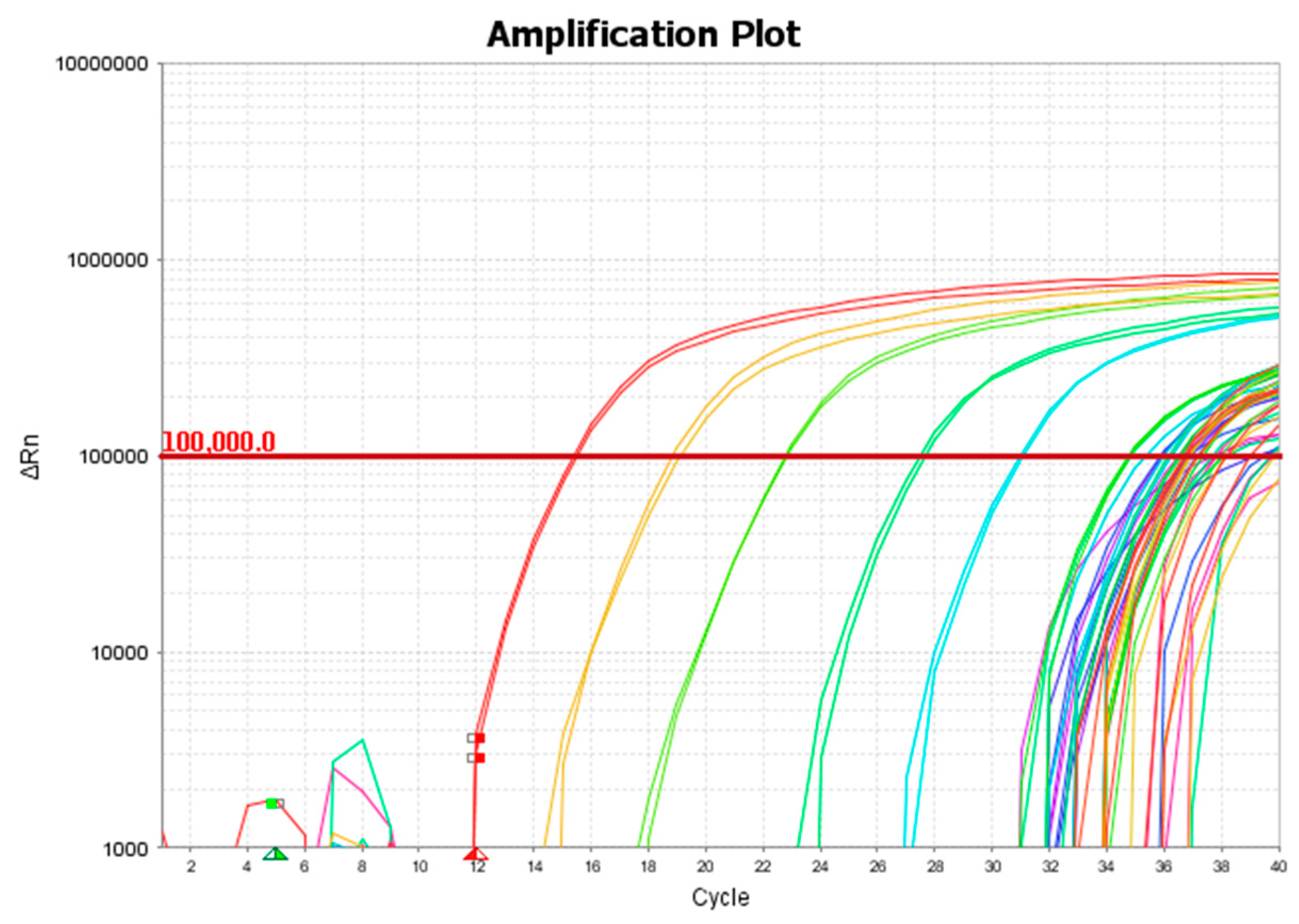
2.3. Quantitative Polymerase Chain Reaction (qPCR)
3. Results and Discussion
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Adler, B.; de la Pena Moctezuma, A. Leptospira and leptospirosis. Vet. Microbiol. 2010, 140, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Levett, P.N. Leptospirosis. Clin. Microbiol. Rev. 2001, 14, 296–326. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Stevenson, B.; Adler, B. Leptospirosis in horses. Vet. Microbiol. 2013, 167, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Ko, A.I.; Goarant, C.; Picardeau, M. Leptospira: The dawn of the molecular genetics era for an emerging zoonotic pathogen. Nat. Rev. Microbiol. 2009, 7, 736–747. [Google Scholar] [CrossRef] [PubMed]
- Pappas, G.; Papdimitriou, P.; Siozopoulou, V.; Christou, L.; Akritidis, N. The globalization of leptospirosis: Worldwide incidence trends. Int. J. Infect. Dis. 2008, 12, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Rawlins, J.; Portanova, A.; Zuckerman, I.; Loftis, A.; Ceccato, P.; Willingham, A.L.; Verma, A. Molecular detection of leptospiral DNA in environmental water on St. Kitts. Int. J. Environ. Res. Public Health 2014, 11, 7953–7960. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Soto, E.; Illanes, O.; Ghosh, S.; Fuentealba, C. Detection and genotyping of Leptospira spp. from the kidneys of a seemingly healthy pig slaughtered for human consumption. J. Infect. Dev. Ctries. 2015, 9, 530–532. [Google Scholar] [CrossRef] [PubMed]
- Vein, J.; Perrin, A.; Berny, P.J.; Benoit, E.; Leblond, A.; Kodjo, A. Adaptation of a real-time PCR method for the detection and quantification of pathogenic leptospires in environmental water. Can. J. Microbiol. 2012, 58, 828–835. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.J.; McWhorter, A.C.; Knutson, J.K.; Steigerwalt, A.G. Escherichia vulneris: A new species of Enterobacteriaceae associated with human wounds. J. Clin. Microbiol. 1982, 15, 1133–1140. [Google Scholar] [PubMed]
- Levett, P.N.; Morey, R.E.; Galloway, R.L.; Turner, D.E.; Steigerwalt, A.G.; Mayer, L.W. Detection of pathogenic leptospires by real-time quantitative PCR. J. Med. Microbiol. 2005, 54, 45–49. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, R.A.; Gee, J.E.; Wilkins, P.P.; McCaustland, K.; Hoffmaster, A.R. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn. Microbiol. Infect. Dis. 2009, 64, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Benkirane, A.; Noury, S.; Hartskeerl, R.A.; Goris, M.G.; Ahmed, A.; Nally, J.E. Preliminary investigations on the distribution of leptospira serovars in domestic animals in north-west Morocco. Transbound. Emerg. Dis. 2016, 63, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Damude, D.F.; Jones, C.J.; Myers, D.M. A study of leptospirosis among animals in Barbados W.I. Trans. R. Soc. Trop. Med. Hyg. 1979, 73, 161–168. [Google Scholar] [CrossRef]
- Everard, C.O.; Fraser-Chanpong, G.M.; James, A.C.; Butcher, L.V. Serological studies on leptospirosis in livestock and chickens from Grenada and Trinidad. Trans. R. Soc. Trop. Med. Hyg. 1985, 79, 859–864. [Google Scholar] [CrossRef]

© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grevemeyer, B.; Vandenplas, M.; Beigel, B.; Cho, E.; Willingham, A.L.; Verma, A. Detection of Leptospiral DNA in the Urine of Donkeys on the Caribbean Island of Saint Kitts. Vet. Sci. 2017, 4, 2. https://doi.org/10.3390/vetsci4010002
Grevemeyer B, Vandenplas M, Beigel B, Cho E, Willingham AL, Verma A. Detection of Leptospiral DNA in the Urine of Donkeys on the Caribbean Island of Saint Kitts. Veterinary Sciences. 2017; 4(1):2. https://doi.org/10.3390/vetsci4010002
Chicago/Turabian StyleGrevemeyer, Bernard, Michel Vandenplas, Brittney Beigel, Ellen Cho, Arve Lee Willingham, and Ashutosh Verma. 2017. "Detection of Leptospiral DNA in the Urine of Donkeys on the Caribbean Island of Saint Kitts" Veterinary Sciences 4, no. 1: 2. https://doi.org/10.3390/vetsci4010002
APA StyleGrevemeyer, B., Vandenplas, M., Beigel, B., Cho, E., Willingham, A. L., & Verma, A. (2017). Detection of Leptospiral DNA in the Urine of Donkeys on the Caribbean Island of Saint Kitts. Veterinary Sciences, 4(1), 2. https://doi.org/10.3390/vetsci4010002





