ColorDetect RT-LAMP Assay for the Rapid, Sensitive, and Specific Detection of Porcine Abortion-Associated Pestivirus (PAAPeV)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses Strains and Clinical Samples
2.2. Sequence Alignment and Phylogenetic Analysis
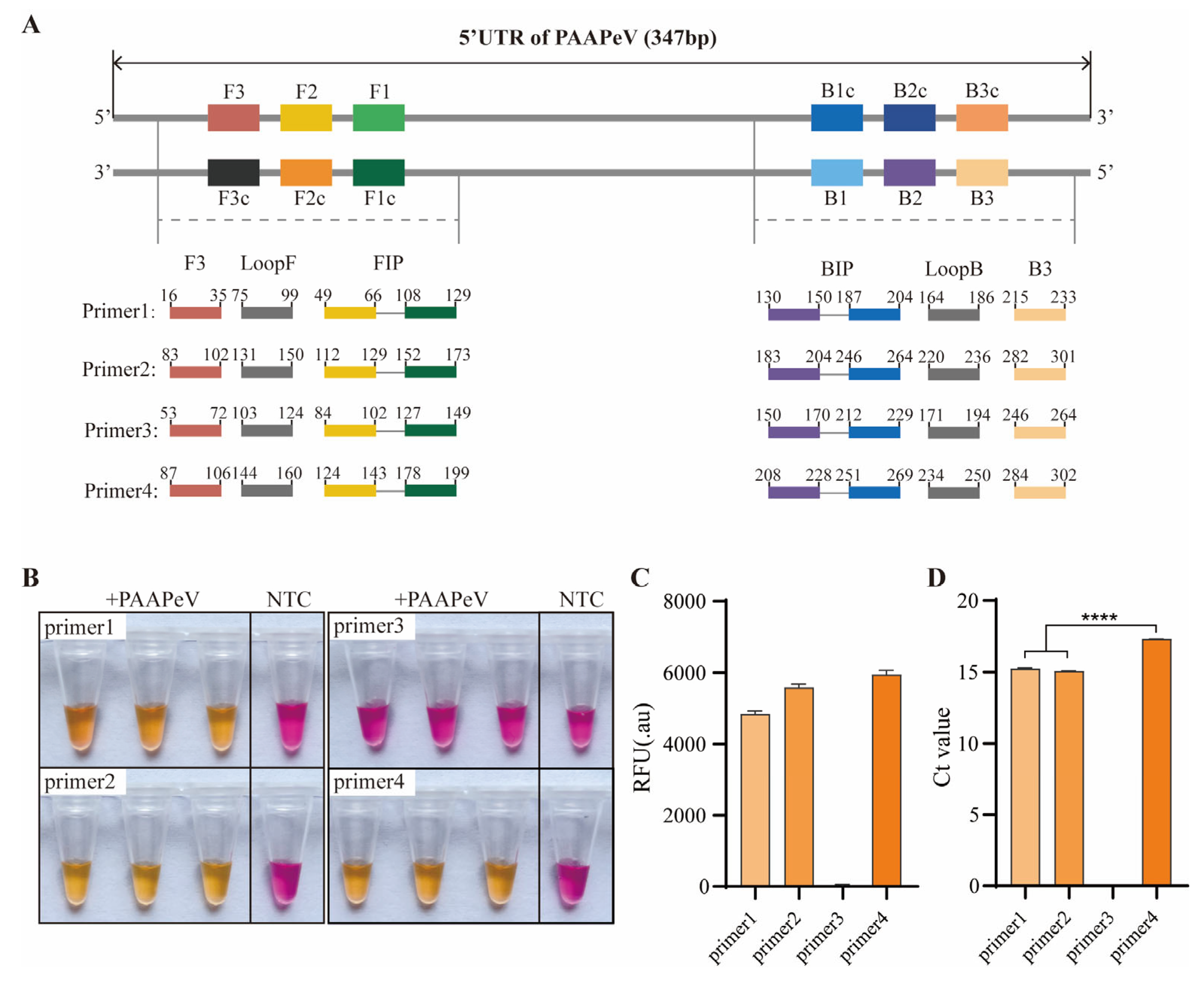
2.3. ColorDetect RT-LAMP Primer Design and Screen
2.4. ColorDetect RT-LAMP Reaction Condition Optimization
2.5. Specificity and Sensitivity Assays
2.6. RT-qPCR
2.7. Clinical Sample Tests
2.8. Statistical Analysis
3. Results
3.1. Phylogenetic and Sequence Identity Analysis of the PAAPeV 5′UTR Region
3.2. Colordetect RT-LAMP Assay Development and Primers Screen
3.3. Optimization of Colordetect RT-LAMP Reaction Conditions
3.4. Specificity of Colordetect RT-LAMP Assay
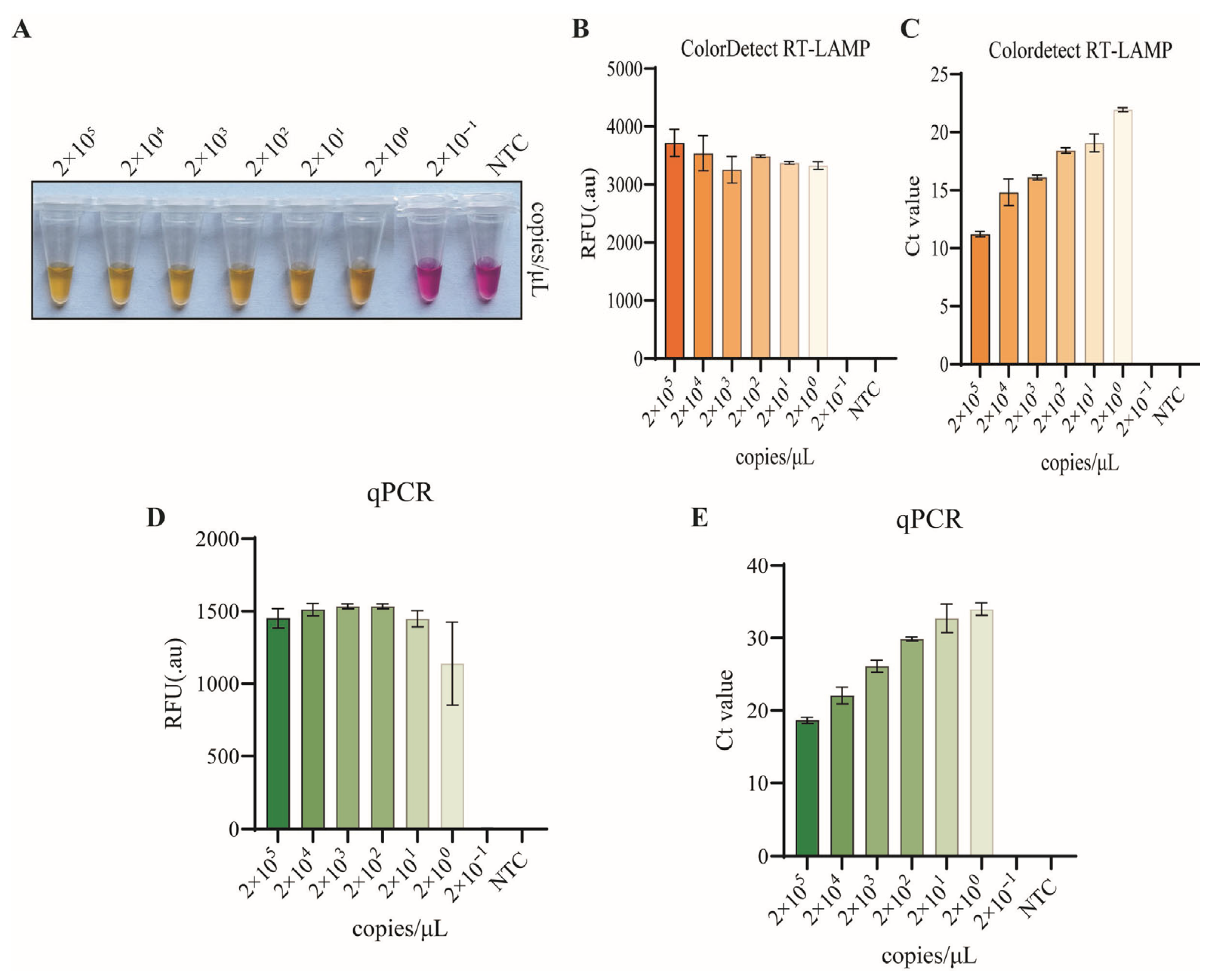
3.5. Sensitivity of Colordetect RT-LAMP Assay
3.6. Clinical Concordance of Colordetect RT-LAMP and RT-qPCR
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Deng, L.S.; Xu, T.; Xu, Z.W.; Zhu, L. Emergence of a novel porcine pestivirus with potential for cross-species transmission in China, 2023. Vet. Res. 2025, 56, 32. [Google Scholar] [CrossRef]
- Meyers, G.; Thiel, H.J. Molecular characterization of pestiviruses. Adv. Virus Res. 1996, 47, 53–118. [Google Scholar]
- Vilcek, S.; Ridpath, J.F.; Van Campen, H.; Cavender, J.L.; Warg, J. Characterization of a novel pestivirus originating from a pronghorn antelope. Virus Res. 2005, 108, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Postel, A.; Smith, D.B.; Becher, P. Proposed Update to the Taxonomy of Pestiviruses: Eight Additional Species within the Genus Pestivirus, Family Flaviviridae. Viruses 2021, 13, 1542. [Google Scholar] [CrossRef]
- Dénes, L.; Biksi, I.; Albert, M.; Szeredi, L.; Knapp, D.G.; Szilasi, A.; Bálint, Á.; Balka, G. Detection and phylogenetic characterization of atypical porcine pestivirus strains in Hungary. Transbound. Emerg. Dis. 2018, 65, 2039–2042. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Xie, Z.; Xie, L.; Liu, J.; Pang, Y.; Deng, X.; Xie, Z.; Peng, Y.; Wang, X. A reverse transcription loop-mediated isothermal amplification method for rapid detection of bovine viral diarrhea virus. J. Virol. Methods 2012, 186, 43–48. [Google Scholar] [CrossRef]
- Chen, H.T.; Zhang, J.; Ma, L.N.; Ma, Y.P.; Ding, Y.Z.; Liu, X.T.; Chen, L.; Ma, L.Q.; Zhang, Y.G.; Liu, Y.S. Rapid pre-clinical detection of classical swine fever by reverse transcription loop-mediated isothermal amplification. Mol. Cell. Probes 2009, 23, 71–74. [Google Scholar] [CrossRef]
- Wang, L.; Madera, R.; Li, Y.; McVey, D.S.; Drolet, B.S.; Shi, J. Recent Advances in the Diagnosis of Classical Swine Fever and Future Perspectives. Pathogens 2020, 9, 658. [Google Scholar] [CrossRef] [PubMed]
- Deregt, D.; Carman, P.S.; Clark, R.M.; Burton, K.M.; Olson, W.O.; Gilbert, S.A. A comparison of polymerase chain reaction with and without RNA extraction and virus isolation for detection of bovine viral diarrhea virus in young calves. J. Vet. Diagn. Investig. 2002, 14, 433–437. [Google Scholar] [CrossRef]
- Ridpath, J.F.; Hietala, S.K.; Sorden, S.; Neill, J.D. Evaluation of the reverse transcription-polymerase chain reaction/probe test of serum samples and immunohistochemistry of skin sections for detection of acute bovine viral diarrhea infections. J. Vet. Diagn. Investig. 2002, 14, 303–307. [Google Scholar] [CrossRef]
- Fulton, R.W.; Hessman, B.; Johnson, B.J.; Ridpath, J.F.; Saliki, J.T.; Burge, L.J.; Sjeklocha, D.; Confer, A.W.; Funk, R.A.; Payton, M.E. Evaluation of diagnostic tests used for detection of bovine viral diarrhea virus and prevalence of subtypes 1a, 1b, and 2a in persistently infected cattle entering a feedlot. J. Am. Vet. Med. Assoc. 2006, 228, 578–584. [Google Scholar] [CrossRef] [PubMed]
- Youssef, B.Z. Comparative study between ELISA, immuno-diffusion and cell bound immuno assay techniques for detection of anti-bovine viral diarrhea antibodies in calves of some farms in alexandria and behira governorates. J. Egypt. Public Health Assoc. 2006, 81, 29–41. [Google Scholar]
- Wang, X.; Zhang, X.; Sun, Y.; Li, N.; He, F.; Chang, T.; Li, H.; Yang, Z.; Qiu, H. Comparison of six detection methods for classical swine fever virus. Wei Sheng Wu Xue Bao 2010, 50, 1087–1093. [Google Scholar]
- Parida, M.; Posadas, G.; Inoue, S.; Hasebe, F.; Morita, K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J. Clin. Microbiol. 2004, 42, 257–263. [Google Scholar] [CrossRef]
- Enomoto, Y.; Yoshikawa, T.; Ihira, M.; Akimoto, S.; Miyake, F.; Usui, C.; Suga, S.; Suzuki, K.; Kawana, T.; Nishiyama, Y.; et al. Rapid diagnosis of herpes simplex virus infection by a loop-mediated isothermal amplification method. J. Clin. Microbiol. 2005, 43, 951–955. [Google Scholar] [CrossRef]
- Cho, H.S.; Kang, J.I.; Park, N.Y. Detection of canine parvovirus in fecal samples using loop-mediated isothermal amplification. J. Vet. Diagn. Investig. 2006, 18, 81–84. [Google Scholar]
- Chen, H.T.; Zhang, J.; Sun, D.H.; Ma, L.N.; Liu, X.T.; Cai, X.P.; Liu, Y.S. Development of reverse transcription loop-mediated isothermal amplification for rapid detection of H9 avian influenza virus. J. Virol. Methods 2008, 151, 200–203. [Google Scholar] [CrossRef]
- Komiyama, C.; Suzuki, K.; Miura, Y.; Sentsui, H. Development of loop-mediated isothermal amplification method for diagnosis of bovine leukemia virus infection. J. Virol. Methods 2009, 157, 175–179. [Google Scholar] [CrossRef]
- Rovira, A.; Abrahante, J.; Murtaugh, M.; Muñoz-Zanzi, C. Reverse transcription loop-mediated isothermal amplification for the detection of Porcine reproductive and respiratory syndrome virus. J. Vet. Diagn. Investig. 2009, 21, 350–354. [Google Scholar] [CrossRef]
- Yin, S.; Shang, Y.; Zhou, G.; Tian, H.; Liu, Y.; Cai, X.; Liu, X. Development and evaluation of rapid detection of classical swine fever virus by reverse transcription loop-mediated isothermal amplification (RT-LAMP). J. Biotechnol. 2010, 146, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Mansour, S.M.; Ali, H.; Chase, C.C.; Cepica, A. Loop-mediated isothermal amplification for diagnosis of 18 World Organization for Animal Health (OIE) notifiable viral diseases of ruminants, swine and poultry. Anim. Health Res. Rev. 2015, 16, 89–106. [Google Scholar] [CrossRef]
- Pan, S.; Mou, C.; Chen, Z. An emerging novel virus: Atypical porcine pestivirus (APPV). Rev. Med. Virol. 2019, 29, e2018. [Google Scholar] [CrossRef]
- Blome, S.; Beer, M.; Wernike, K. New Leaves in the Growing Tree of Pestiviruses. Adv. Virus Res. 2017, 99, 139–160. [Google Scholar]
- Chen, L.; Fan, X.Z.; Wang, Q.; Xu, L.; Zhao, Q.Z.; Zhou, Y.C.; Liu, J.; Tang, B.; Zou, X.Q. A novel RT-LAMP assay for rapid and simple detection of classical swine fever virus. Virol. Sin. 2010, 25, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [PubMed]
- Coronado, L.; Muñoz-Aguilera, A.; Wang, M.; Muñoz, I.; Riquelme, C.; Heredia, S.; Stępniewska, K.; Gallardo, C.; Ganges, L. Simultaneous Detection of Classical and African Swine Fever Viruses by Duplex Taqman Real-Time PCR Assay in Pigs Infected with Both Diseases. Pathogens 2025, 14, 473. [Google Scholar] [CrossRef]
- Bohórquez, J.A.; Muñoz-Aguilera, A.; Lanka, S.; Coronado, L.; Rosell, R.; Alberch, M.; Maddox, C.W.; Ganges, L. Development of a new loop-mediated isothermal amplification test for the sensitive, rapid, and economic detection of different genotypes of Classical swine fever virus. Front. Cell. Infect. Microbiol. 2024, 14, 1372166. [Google Scholar] [CrossRef]
- Ridpath, J.F.; Bolin, S.R. Comparison of the complete genomic sequence of the border disease virus, BD31, to other pestiviruses. Virus Res. 1997, 50, 237–243. [Google Scholar] [CrossRef]
- Park, J.Y.; Park, S.; Park, Y.R.; Kang, D.Y.; Kim, E.M.; Jeon, H.S.; Kim, J.J.; Kim, W.I.; Lee, K.T.; Kim, S.H.; et al. Reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the visual detection of European and North American porcine reproductive and respiratory syndrome viruses. J. Virol. Methods 2016, 237, 10–13. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Cui, J.; Ren, Y.; Suo, S.; Li, G.; Sun, X.; Su, D.; Opriessnig, T.; Ren, X. Development and evaluation of a novel reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay for detection of type II porcine reproductive and respiratory syndrome virus. J. Virol. Methods 2012, 185, 18–23. [Google Scholar] [CrossRef]
- Tran, D.H.; Ngoc, N.A.; Tran, H.T.; Minh, T.N.; Ngoc, T.B.; Nguyen, V.T.; Le, V.P.; Thu, H.T. Instrument-free, visual and direct detection of porcine reproductive and respiratory syndrome viruses in resource-limited settings. Acta Virol. 2023, 67, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Shao, Y.; Zhang, Y.; Cheng, F.; Fang, P.; Tu, J.; Song, X.; Qi, K.; Wang, Z. LAMP Assay Coupled with a Pyrococcus furiosus Argonaute System for the Rapid Detection of Porcine Epidemic Diarrhea Virus. ACS Synth. Biol. 2025, 14, 689–698. [Google Scholar] [CrossRef] [PubMed]



| Primers | Sequences (5′-3′) |
|---|---|
| 1-F3 | TGGGGATGCCAAAAACTGAA |
| 1-B3 | CGTCCCTAACACTGTGTCG |
| 1-FIP | CAATGCAGCCTGCTCACCTACTAGGCCGAGGAAAGGTAGC |
| 1-BIP | TGTAAGCGGTGAGTACACCGCCGTCCACGAGGCATAGCT |
| 1-LoopF | CCACCTATCGAGTTCGTCCTACTAA |
| 1-LoopB | GCTACTGGTAAGGATCACCCACT |
| 2-F3 | ACGAACTCGATAGGTGGACT |
| 2-B3 | CCGGCATCCTATCAGACTGT |
| 2-FIP | TACCAGTAGCACCTCTGACGGCGGTGAGCAGGCTGCATTG |
| 2-BIP | CACTAGCTATGCCTCGTGGACGTCACCCCCACTTTCAGGAC |
| 2-LoopF | GCGGTGTACTCACCGCTTAC |
| 2-LoopB | CAGTGTTAGGGACGGCG |
| 3-F3 | CGAGGAAAGGTAGCCAATCC |
| 3-B3 | TCACCCCCACTTTCAGGAC |
| 3-FIP | CGGTGTACTCACCGCTTACACAACGAACTCGATAGGTGGACT |
| 3-BIP | CAGCCGTCAGAGGTGCTACTGCCTAACACTGTGTCGGGC |
| 3-LoopF | CAGCCTGCTCACCTACTAAACC |
| 3-LoopB | GTAAGGATCACCCACTAGCTATGC |
| 4-F3 | ACTCGATAGGTGGACTGGTT |
| 4-B3 | GCCGGCATCCTATCAGACT |
| 4-FIP | ACGAGGCATAGCTAGTGGGTGAGCATTGTGTAAGCGGTGAGT |
| 4-BIP | GCGTGCCCGACACAGTGTTAGGGTATTCACCCCCACTTTC |
| 4-LoopF | TCTGACGGCTGCGGTGT |
| 4-LoopB | GCGGGGGCTGTCGTCCT |
| Sample Source | ColorDetect RT-LAMP (Positive/Total) | RT-qPCR (Positive/Total) |
|---|---|---|
| Clinical PAAPeV-positive samples | 24/24 | 24/24 |
| Clinical unknown samples | 0/30 | 0/30 |
| Positive control | 3/3 | 3/3 |
| Negative control | 0/3 | 0/3 |
| Total | 60 | 60 |
| Diagnostic accuracy | 100% | 100% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2026 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Yang, X.; Li, Y.; Yin, W.; Tang, W.; Diao, H.; Zhou, M.; Yang, H.; Fu, W.; Yong, L.; Luo, X.; et al. ColorDetect RT-LAMP Assay for the Rapid, Sensitive, and Specific Detection of Porcine Abortion-Associated Pestivirus (PAAPeV). Vet. Sci. 2026, 13, 74. https://doi.org/10.3390/vetsci13010074
Yang X, Li Y, Yin W, Tang W, Diao H, Zhou M, Yang H, Fu W, Yong L, Luo X, et al. ColorDetect RT-LAMP Assay for the Rapid, Sensitive, and Specific Detection of Porcine Abortion-Associated Pestivirus (PAAPeV). Veterinary Sciences. 2026; 13(1):74. https://doi.org/10.3390/vetsci13010074
Chicago/Turabian StyleYang, Xu, Ying Li, Wenqi Yin, Wenjie Tang, Hui Diao, Mengjia Zhou, Hao Yang, Wenyi Fu, Lu Yong, Xu Luo, and et al. 2026. "ColorDetect RT-LAMP Assay for the Rapid, Sensitive, and Specific Detection of Porcine Abortion-Associated Pestivirus (PAAPeV)" Veterinary Sciences 13, no. 1: 74. https://doi.org/10.3390/vetsci13010074
APA StyleYang, X., Li, Y., Yin, W., Tang, W., Diao, H., Zhou, M., Yang, H., Fu, W., Yong, L., Luo, X., Liao, G., & Zhou, Y. (2026). ColorDetect RT-LAMP Assay for the Rapid, Sensitive, and Specific Detection of Porcine Abortion-Associated Pestivirus (PAAPeV). Veterinary Sciences, 13(1), 74. https://doi.org/10.3390/vetsci13010074







