RNase Hybridization-Assisted Amplification (RHAM) Technology: A High-Sensitivity, Field-Deployable Alternative to Quantitative Polymerase Chain Reaction for the Rapid Detection of African Swine Fever Virus
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Sample and Sample Preparation
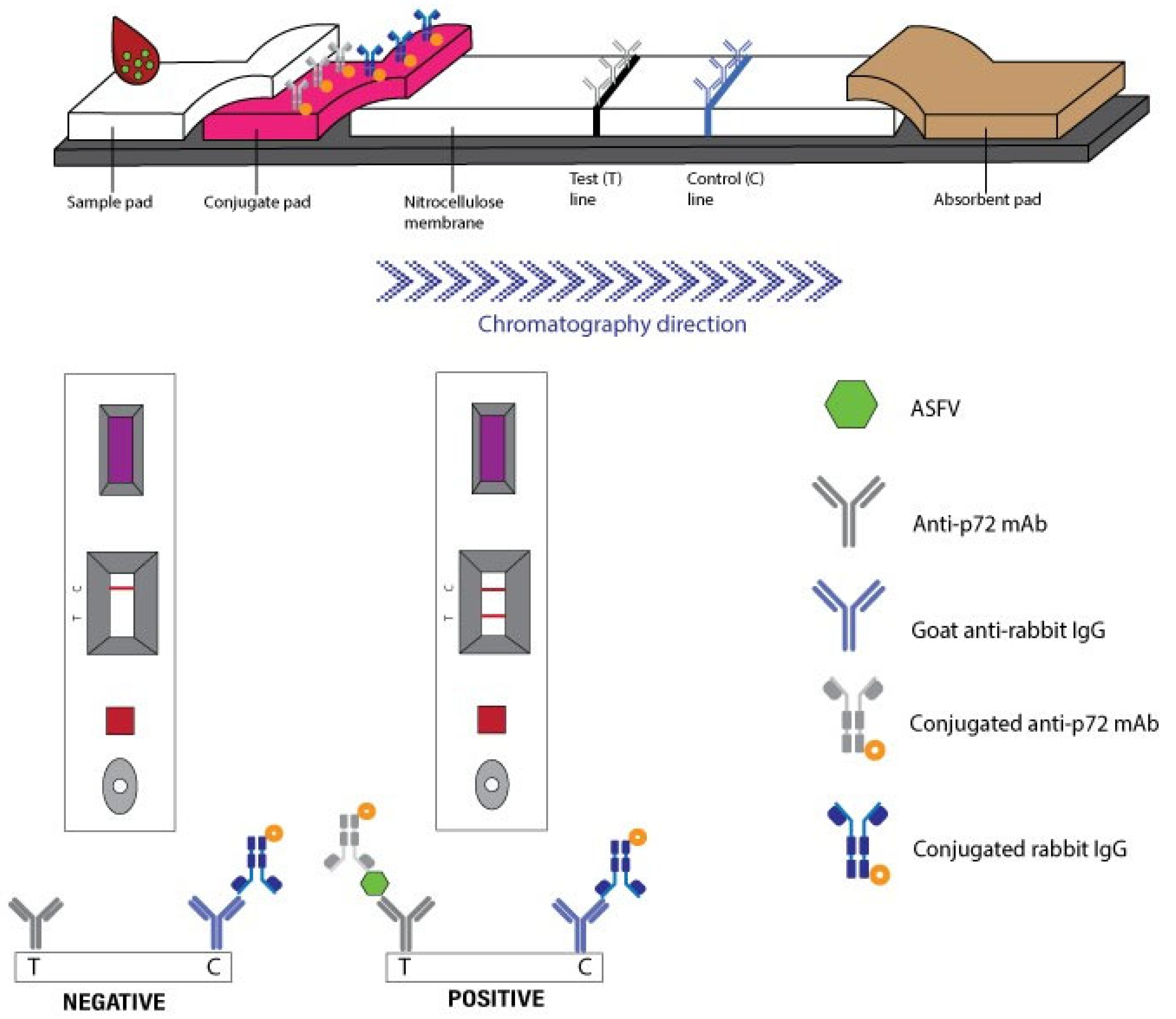
2.3. Immunochromatography Assay
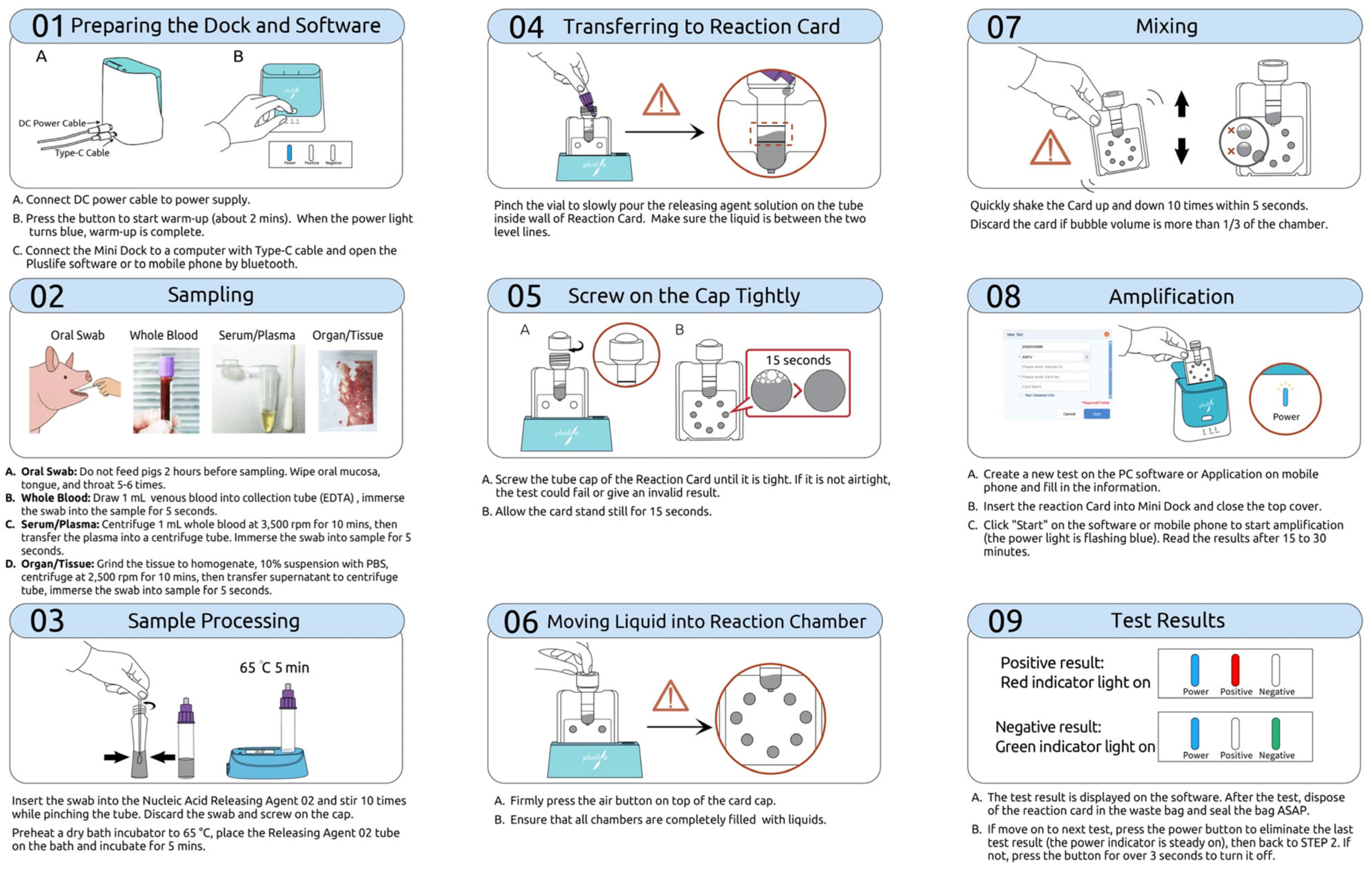
2.4. RNase Hybridization-Assisted Amplification (RHAM) Test
2.5. Quantitative Polymerase Chain Reaction
2.6. Statistical Analysis
3. Results
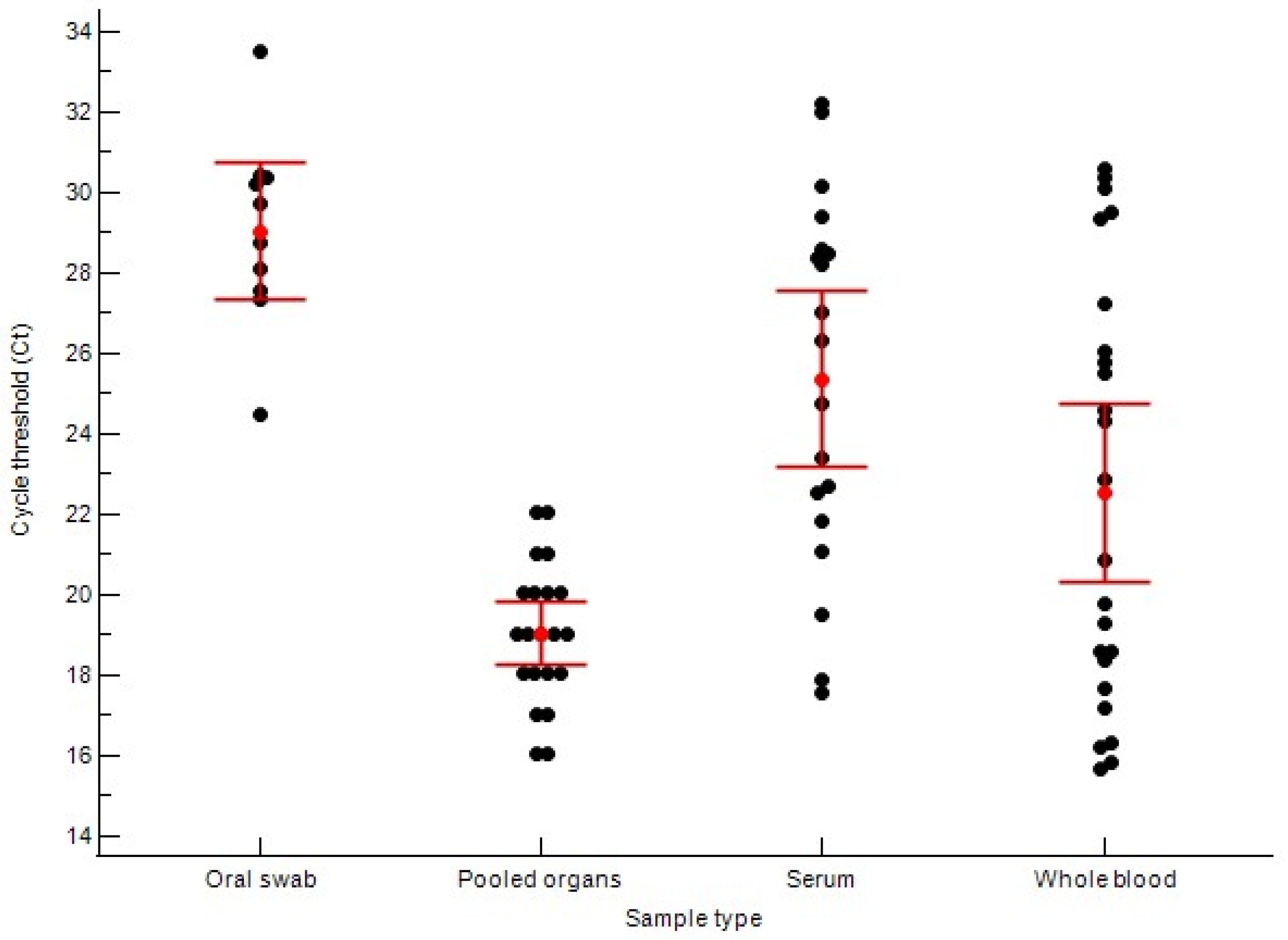
3.1. Distribution of Viral Load Across Sample Types
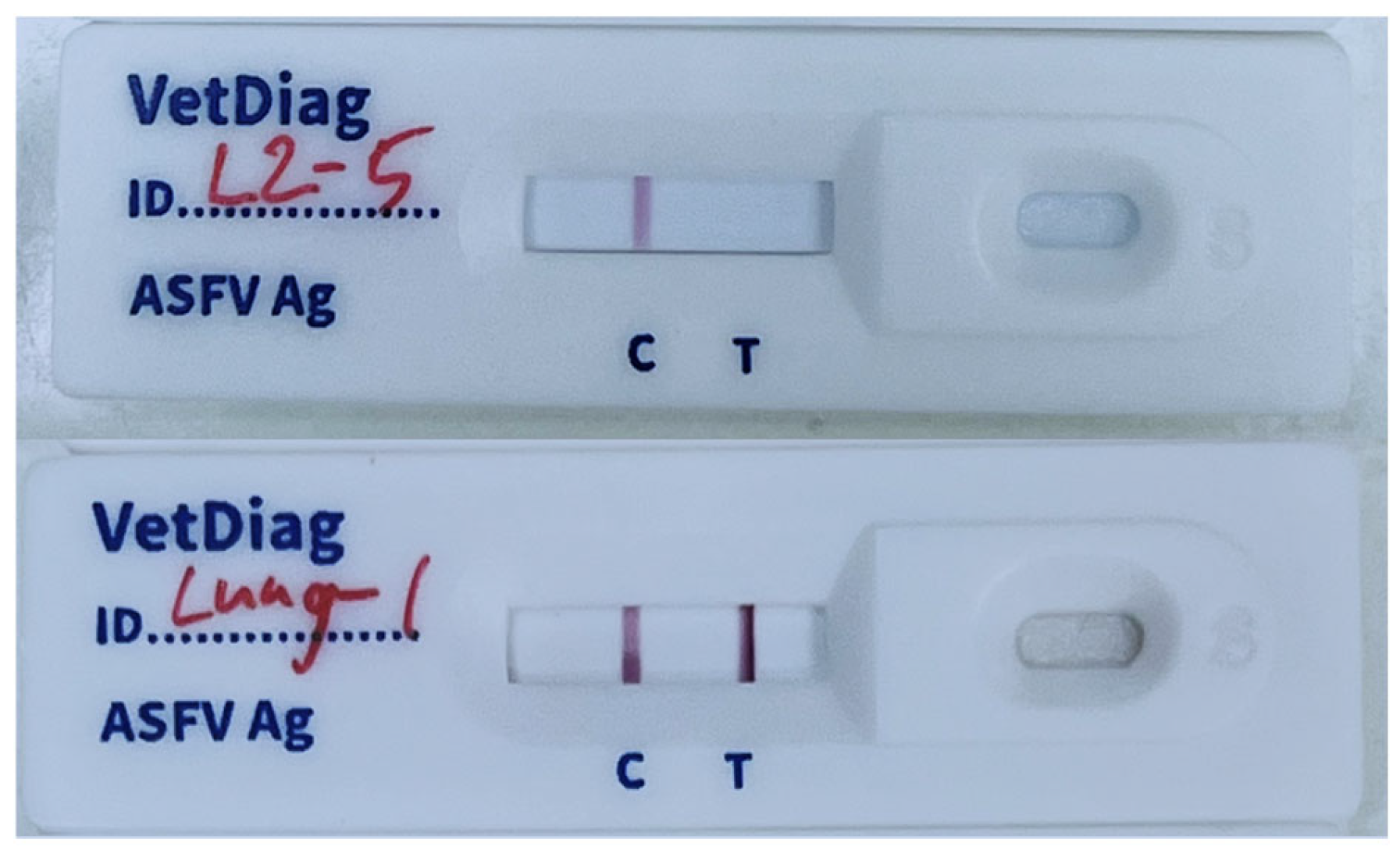
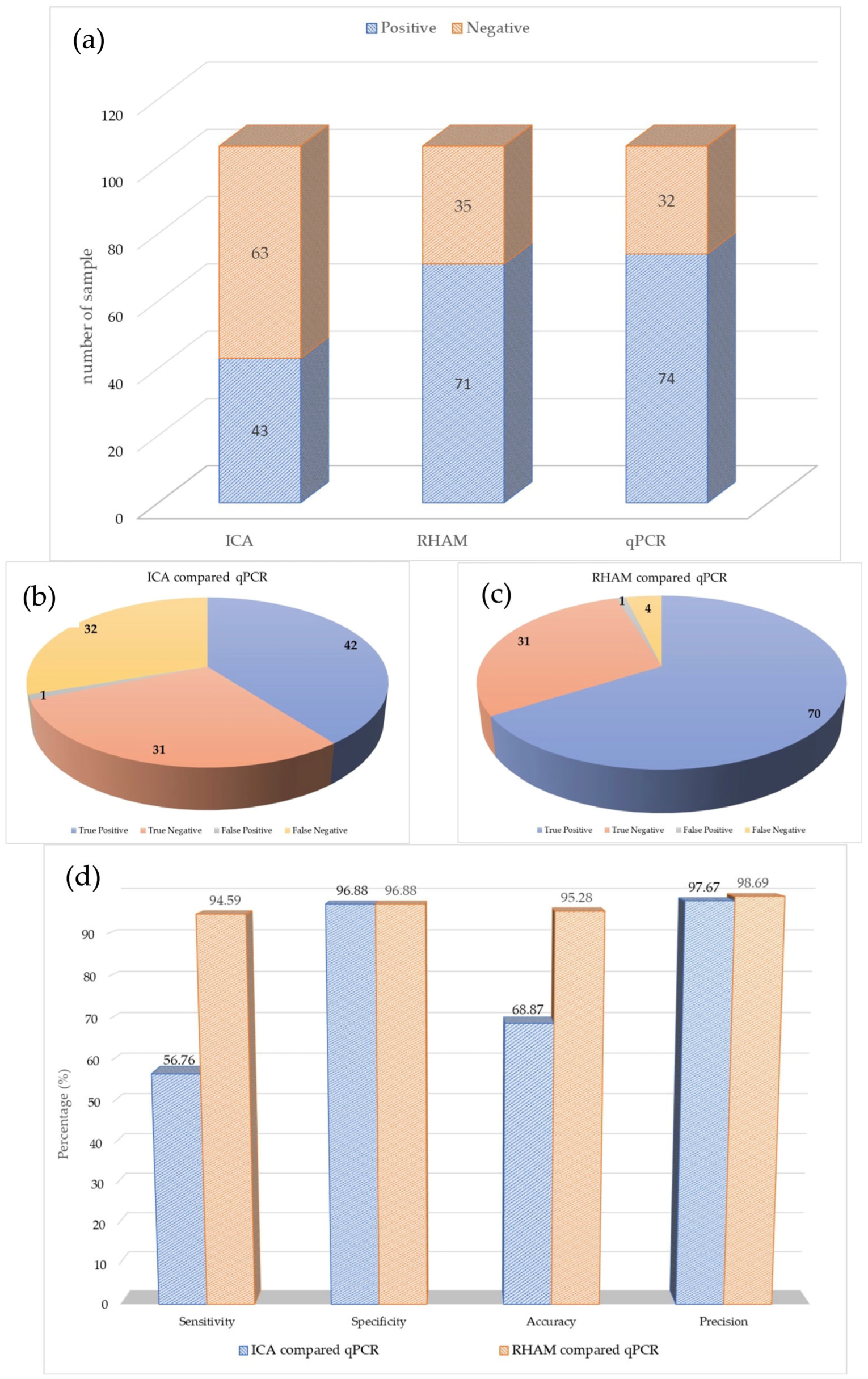
3.2. Performance of the Immunochromatography Assay
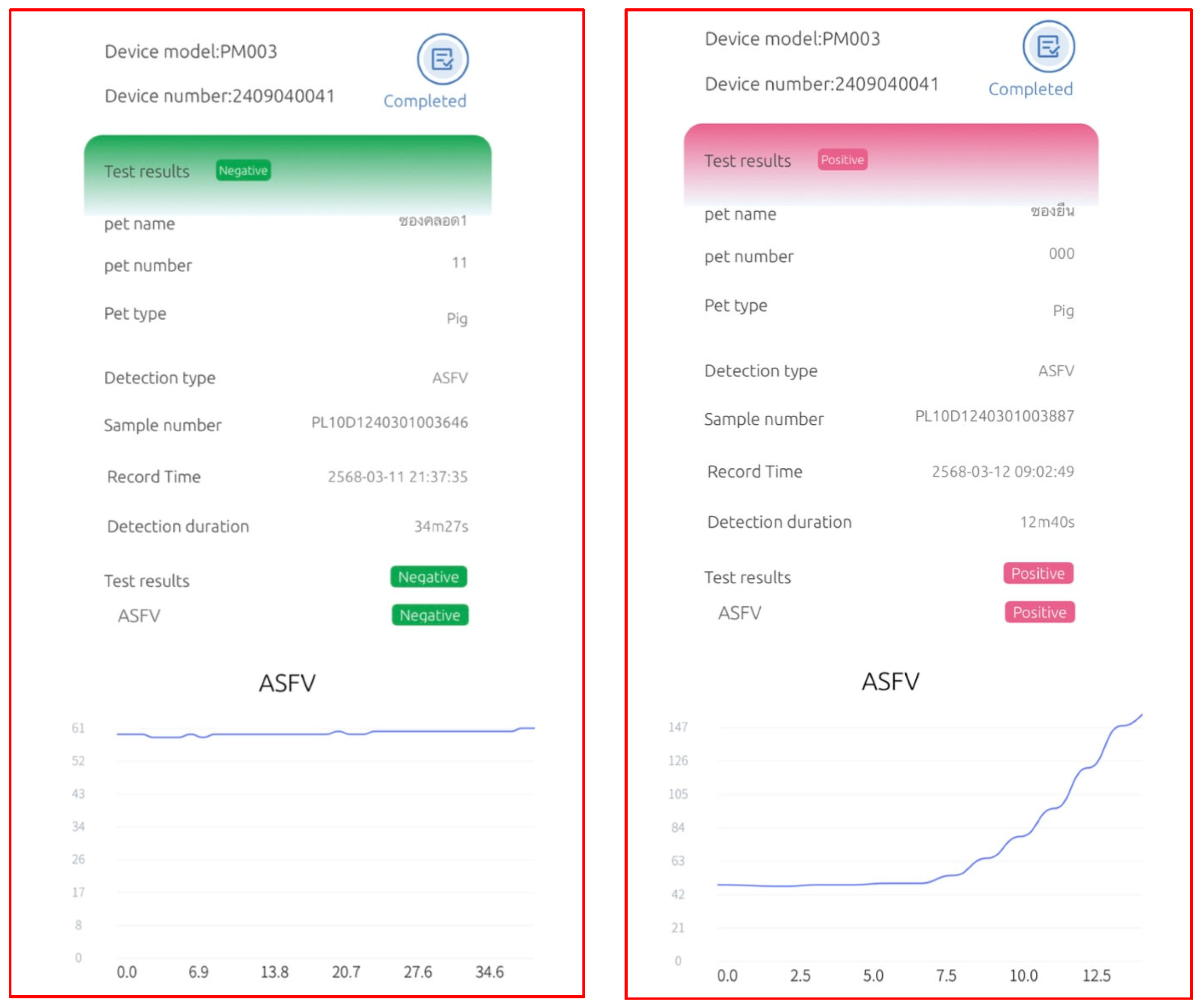
3.3. Performance of the RNase Hybridization-Assisted Amplification Test
3.4. Statistic Comparison of Assays
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Costard, S.; Wieland, B.; De Glanville, W.; Jori, F.; Rowlands, R.; Vosloo, W.; Roger, F.; Pfeiffer, D.U.; Dixon, L.K. African Swine fever: How can global spread be prevented? Philos. Trans. R. Soc. B Biol. Sci. 2009, 364, 2683–2696. [Google Scholar] [CrossRef] [PubMed]
- Sauter-Louis, C.; Conraths, F.J.; Probst, C.; Blohm, U.; Schulz, K.; Sehl, J.; Fischer, M.; Forth, J.H.; Zani, L.; Depner, K.; et al. African Swine fever in wild boar in Europe—A review. Viruses 2021, 13, 1717. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Medina, E.; O’Donnell, V.; Silva, E.; Esoinoza, N.; Velazquez-Salinas, L.; Moran, K.; Daite, D.A.; Barrette, R.; FAburay, B.; Holland, R.; et al. Experimental infection of domestic pigs with an African swine fever virus field strain isolated in 2021 from the Dominican Republic. Viruses 2022, 14, 1090. [Google Scholar] [CrossRef] [PubMed]
- Taesuji, M.; Rattanamas, K.; Kulthonggate, U.; Mamom, T.; Ruenphet, S. Sensitivity and specificity for African horse sickness antibodies detection using monovalent and polyvalent vaccine antigen-based dot blotting. Vet. World 2022, 15, 2760–2763. [Google Scholar] [CrossRef]
- Rattanamas, K.; Taesuji, M.; Kulthonggate, U.; Jantafong, T.; Mamom, T.; Ruenphet, S. Sensitivity of RNA viral nucleic acid-based detection of avian influenza virus, Newcastle disease virus, and African horse sickness virus on flinders technology associates card using conventional reverse-transcription polymerase chain reaction. Vet. World 2022, 15, 2754–2759. [Google Scholar] [CrossRef]
- Nguyen-Thi, T.; Pham-Thi-Ngoc, L.; Nguyen-Ngoc, Q.; Dang-Xuan, S.; Lee, H.S.; Nguyen-Viet, H.; Padungtod, P.; Nguyen-Thu, T.; Nguyen-Thi, T.; Tran-Cong, T.; et al. An assessment of the economic impacts of the 2019 African swine fever outbreaks in Vietnam. Front. Vet. Sci. 2021, 8, 686038. [Google Scholar] [CrossRef]
- Ata, E.B.; Li, Z.J.; Shi, C.W.; Yang, G.L.; Yang, W.T.; Wang, C.F. African Swine Fever Virus: A Raised Global Upsurge and a Continuous Threaten to Pig Husbandry. Microb. Pathog. 2022, 167, 105561. [Google Scholar] [CrossRef]
- World Organisation for Animal Health (WOAH). Chapter 15.1. Infection with African Swine Fever Virus. In Terrestrial Animal Health Code; World Organisation for Animal Health (WOAH): Paris, France, 2016; Available online: https://www.woah.org/fileadmin/Home/eng/Health_standards/tahc/2016/en_chapitre_asf.htm (accessed on 5 November 2025).
- Beltrán-Alcrudo, D.; Arias, M.; Gallardo, C.; Kramer, S.; Penrith, M.L. African Swine Fever: Detection and Diagnosis—A Manual for Veterinarians. In FAO Animal Production and Health Manual No. 19; Food and Agriculture Organization of the United Nations (FAO): Rome, Italy, 2017; 88p. [Google Scholar]
- Dixon, L.K.; Stahl, K.; Jori, F.; Vial, L.; Pfeiffer, D.U. African Swine Fever Epidemiology and Control. Annu. Rev. Anim. Biosci. 2020, 8, 221–246. [Google Scholar] [CrossRef]
- Fernández-Pinero, J.; Gallardo, C.; Elizalde, M.; Robles, A.; Gomez, C.; Bishop, R.; Heath, L.; Couacy-Hymann, E.; Fasina, F.O.; Pelayo, V.; et al. Molecular diagnosis of African swine fever by a new real-time PCR using universal probe library. Transbound. Emerg. Dis. 2013, 60, 48–58. [Google Scholar] [CrossRef]
- Gallardo, C.; Soler, A.; Nurmoja, I.; Cano-Gomez, C.; Cvetkova, S.; Frant, M.; Wozniakowski, G.; Simon, A.; Perez, C.; Nieto, R.; et al. Dynamics of African swine fever virus (ASFV) infection in domestic pigs infected with virulent, moderate virulent and attenuated genotype II ASFV European isolates. Transbound. Emerg. Dis. 2021, 68, 2826–2841. [Google Scholar] [CrossRef]
- Wang, S.; Shen, H.; Lin, Q.; Huang, J.; Zhang, C.; Liu, Z.; Sun, M.; Zhang, J.; Liao, M.; Li, Y.; et al. Development of a. cleaved probe-based loop-mediated isothermal amplification assay for rapid detection of African swine fever virus. Front. Cell. Infect. Microbiol. 2022, 12, 884430. [Google Scholar] [CrossRef]
- Prasitsuwan, W.; Suwannachote, T.; Sumalai, T.; Lertpatarakomol, R.; Trairatapiwan, T.; Ruenphart, S. A comparison of diagnostic methods for canine Ehrlichiosis: Microscopy and RNases hybridization-assisted amplification technology compared with the quantitative polymerase chain reaction. Vet. World 2025, 18, 1214–1223. [Google Scholar] [CrossRef] [PubMed]
- Punyadarsaniya, D.; Taesuji, M.; Rattanamas, K.; Ruenphet, S. Establishment of an In-House Indirect Enzyme-Linked Immunosorbent Assay to Detect Antibodies Against African Horse Sickness Based on Monovalent and Polyvalent Live Attenuated Vaccines During the First Outbreak in Thailand. Animals 2025, 15, 1433. [Google Scholar] [CrossRef] [PubMed]
- Suwannachote, T.; Prasitsuwan, W.; Sumalai, T.; Ruenphet, S. A Comparison of Diagnostic Methods for Feline Leukemia Virus and Feline Immunodeficiency Virus: Immunochromatographic Assay and RNases Hybridization-Assisted Amplification Test Kit Compared to Reverse Transcription Quantitative Polymerase Chain Reaction. Animals 2025, 15, 1484. [Google Scholar] [CrossRef] [PubMed]
- Tran, X.H.; Phuong, L.T.; Huy, N.Q.; Thuy, D.T.; Nguyen, V.D.; Quang, P.H.; Ngon, Q.V.; Rai, A.; Gay, C.G.; Gladue, D.P.; et al. Evaluation of the Safety Profile of the ASFV Vaccine Candidate ASFV-G-∆I177L. Viruses 2022, 14, 896. [Google Scholar] [CrossRef]
- Gallardo, C.; Soler, A.; Nieto, R.; Cano, C.; Pelayo, V.; Sanchez, M.A.; Pridotkas, G.; Fernandez-Pinero, J.; Briones, V.; Arias, M. Experimental infection of domestic pigs with African swine fever virus Lithuania 2014 genotype II field isolate. Transbound. Emerg. Dis. 2017, 64, 300–304. [Google Scholar] [CrossRef]
- Salguero, F.J.; Frossard, J.P.; Rebel, J.M.; Stadejek, T.; Morgan, S.B.; Graham, S.P.; Steinbach, F. Host-pathogen interactions during porcine reproductive and respiratory syndrome virus 1 infection of piglets. Virus Res. 2015, 202, 135–143. [Google Scholar] [CrossRef]
- Morgan, S.B.; Frossard, J.P.; Pallares, F.J.; Gough, J.; Stadejek, T.; Graham, S.P.; Steinbach, F.; Drew, T.W.; Salguero, F.J. Pathology and virus distribution in the lung and lymphoid tissues of pigs experimentally inoculated with three distinct type 1 PRRS virus isolates of varying pathogenicity. Transbound. Emerg. Dis. 2016, 63, 285–295. [Google Scholar] [CrossRef]
- Huong Giang, N.T.; Lan, N.T.; Nam, N.H.; Hirai, T.; Yamaguchi, R. Pathological characterization of an outbreak of porcine reproductive and respiratory syndrome in Northern Vietnam. J. Comp. Pathol. 2016, 154, 135–149. [Google Scholar] [CrossRef]
- Choe, S.; Le, V.P.; Shin, J.; Kim, J.H.; Kim, K.S.; Song, S.; Cha, R.M.; Park, G.N.; Nguyen, T.L.; Hyun, B.H.; et al. Pathogenicity and genetic characterization of Vietnamese classical swine fever virus: 2014–2018. Pathogens 2020, 9, 169. [Google Scholar] [CrossRef]
- King, D.P.; Reid, S.M.; Hutchings, G.H.; Grierson, S.S.; Wilkinson, P.J.; Dixon, L.K.; Bastos, A.D.S.; Drew, T.W. Development of a TaqMan® PCR assay with internal amplification control for the detection of African swine fever virus. J. Virol. Methods 2003, 107, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [PubMed]
- Rowlands, R.J.; Duarte, M.M.; Boinas, F.; Hutchings, G.; Dixon, L.K. The CD2v protein enhances African swine fever virus replication in the tick vector, Ornithodoros erraticus. Virology 2009, 393, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Taesuji, M.; Rattanamas, K.; Punyadarsaniya, D.; Mamom, T.; Nguyen, H.T.; Ruenphet, S. In vitro primary porcine alveolar macrophage cell toxicity and African swine fever virus inactivation using five commercially supply compound disinfectants under various condition. J. Vet. Med. Sci. 2021, 83, 1800–1804. [Google Scholar] [CrossRef]
- Sovijit, W.; Taesuji, W.; Rattanamas, K.; Punyadarsaniya, D.; Mamom, T.; Nguyen, H.T.; Ruenphet, S. In vitro cytotoxicity and virucidal efficacy against African swine fever using two potassium hydrogen peroxymonosulfate compared to a quaternary ammonium compound under various concentrations, exposure times and temperatures. Vet. World 2021, 14, 2936–2940. [Google Scholar] [CrossRef]
- Petrovan, V.; Buburuzan, L.; Zaulet, M. False positive results using PCR detection method for African swine fever virus in wild boars from northern Romanian hunting zones. Turk. J. Vet. Anim. Sci. 2015, 39, 3. [Google Scholar] [CrossRef]
- Zhu, D.; Huang, J.; Hu, B.; Cao, D.; Chen, D.; Song, X.; Chen, J.; Zhou, H.; Cen, A.; Hou, T. Trial of the Pluslife SARS-CoV-2 Nucleic Acid Rapid Test Kit: Prospective Cohort Study. JMIR Public Health Surveill. 2023, 9, e48107. [Google Scholar] [CrossRef]
- Xiao, Z.; Liu, X.; Kang, X.; Feng, Y.; Zheng, L.; Chen, C. Rapid and accurate detection of SARS-CoV-2 using the RHAM technology. Sci. Rep. 2023, 13, 22798. [Google Scholar] [CrossRef]
- Herrmann, L.; Breuer, J.; Duc, T.N.; Thomé, N.; Ghazaani, F.; Kamhieh-Milz, S.; Pfützner, A. Comparison of the diagnostic accuracy of the Pluslife Mini Dock RHAM technology with Abbott ID Now and Cepheid GenXpert: A retrospective evaluation study. Sci. Rep. 2024, 14, 13978. [Google Scholar] [CrossRef]
- Zhong, G.; Zhao, Z.; He, Y.; Yu, X.; Yang, T.; Liu, H.; Huang, B. Evaluation of the RHAM-based Pluslife Rapid SARS-CoV-2 assay and comparison with EasyNAT Rapid SARS-CoV-2 assay and Wondfo 2019-nCoV antigen assay. Microbiol. Spectr. 2025, e03418-24. [Google Scholar] [CrossRef]
- Zsak, L.; Borca, M.V.; Risatti, G.R.; Zsak, A.; French, R.A.; Kutish, G.F.; Neilan, J.G.; Callahan, J.D.; Nelson, W.M.; Rock, D.L. Preclinical diagnosis of African swine fever in contact-exposed swine by a real-time PCR assay. J. Clin. Microbiol. 2005, 43, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Bohorquez, J.A.; Lanka, S.; Rosell, R.; Perez-Simo, M.; Alberch, M.; Rodriguez, F.; Ganges, L.; Maddox, C.W. Efficient detection of African Swine Fever Virus using minimal equipment through a LAMP PCR method. Front. Cell. Infect. Microbiol. 2023, 13, 1114772. [Google Scholar] [CrossRef] [PubMed]
- Mee, P.T.; Wong, S.; O’Riley, K.J.; da Conceicao, F.; da Costa Jong, J.B.; Phillips, D.E.; Rodoni, B.C.; Rawlin, G.T.; Lynch, S.E. Field Verification of an African Swine Fever Virus Loop-Mediated Isothermal Amplification (LAMP) Assay during an Outbreak in Timor-Leste. Viruses 2020, 12, 1444. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Dai, J.F.; Liu, Y.S.; Yang, J.; Hou, Q.; Ou, Y.; Ding, Y.; Ma, B.; Chen, H.; Li, M.M.; et al. Development of a Potential Penside Colorimetric LAMP Assay Using Neutral Red for Detection of African Swine Fever Virus. Front. Microbiol. 2021, 12, 609821. [Google Scholar] [CrossRef]
- Zuo, L.; Song, Z.; Zhang, Y.; Zhai, X.; Zhai, Y.; Mei, X.; Yang, X.; Wang, H. Loop-Mediated Isothemal Amplification Combined with Lateral Flow Dipstrick for On-Site Diagnosis of African Swine Fever Virus. Virol. Sin. 2020, 36, 325–328. [Google Scholar] [CrossRef]
- Ji, C.; Zhou, L.; Chen, Y.; Fang, X.; Liu, Y.; Du, M.; Lu, X.; Li, Q.; Wang, H.; Sun, Y.; et al. Microfluidic-LAMP chip for the point-of-care detection of gene-deleted and wild-type African swine fever viruses and other four swine pathogens. Front. Vet. Sci. 2023, 10, 1116352. [Google Scholar] [CrossRef]
- Gallardo, C.; Nieto, R.; Soler, A.; Pelayo, V.; Fernández-Pinero, J.; Markowska-Daniel, I.; Arias, M. Assessment of African swine fever diagnostic techniques as a response to the epidemic outbreaks in eastern european union countries: How to improve surveillance and control programs. J. Clin. Microbiol. 2015, 53, 2555–2565. [Google Scholar] [CrossRef]
- Oura, C.A.L.; Edwards, L.; Batten, C.A. Virological diagnosis of African swine fever—Comparative study of available tests. Virus Res. 2013, 173, 150–158. [Google Scholar] [CrossRef]
| No. | ICA | qPCR | No. | ICA | qPCR | No. | ICA | qPCR | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Result | Result | Ct | Result | Result | Ct | Result | Result | Ct | |||
| 1 | positive | positive | 24.51 | 37 | positive | positive | 23.59 | 73 | negative | negative | - |
| 2 | negative | positive | 30.21 | 38 | positive | positive | 24.25 | 74 | negative | negative | - |
| 3 | negative | positive | 28.77 | 39 | negative | positive | 28.13 | 75 | negative | negative | - |
| 4 | negative | positive | 27.55 | 40 | negative | positive | 31.43 | 76 | negative | negative | - |
| 5 | negative | positive | 27.33 | 41 | negative | positive | 35.01 | 77 | negative | negative | - |
| 6 | negative | positive | 29.75 | 42 | negative | negative | - | 78 | negative | negative | - |
| 7 | negative | positive | 28.11 | 43 | negative | negative | - | 79 | negative | negative | - |
| 8 | negative | positive | 30.37 | 44 | negative | negative | - | 80 | negative | negative | - |
| 9 | negative | positive | 33.51 | 45 | positive | positive | 19.81 | 81 | negative | negative | - |
| 10 | negative | positive | 30.41 | 46 | positive | positive | 18.61 | 82 | negative | negative | - |
| 11 | negative | negative | - | 47 | positive | positive | 24.32 | 83 | positive | positive | 24.74 |
| 12 | negative | negative | - | 48 | positive | positive | 25.53 | 84 | positive | positive | 23.42 |
| 13 | positive | positive | 17.26 | 49 | positive | positive | 17.18 | 85 | negative | positive | 28.51 |
| 14 | positive | positive | 20.52 | 50 | negative | negative | - | 86 | positive | positive | 22.54 |
| 15 | positive | positive | 23.98 | 51 | positive | positive | 16.24 | 87 | negative | positive | 29.39 |
| 16 | negative | positive | 28.02 | 52 | positive | positive | 19.3 | 88 | negative | positive | 28.37 |
| 17 | positive | negative | - | 53 | positive | positive | 18.59 | 89 | positive | positive | 26.31 |
| 18 | negative | negative | - | 54 | positive | positive | 16.35 | 90 | negative | positive | 30.17 |
| 19 | negative | negative | - | 55 | positive | positive | 18.37 | 91 | negative | positive | 28.58 |
| 20 | negative | negative | - | 56 | positive | positive | 15.85 | 92 | negative | positive | 28.19 |
| 21 | positive | positive | 18.03 | 57 | positive | positive | 15.68 | 93 | negative | positive | 32.21 |
| 22 | positive | positive | 19.06 | 58 | positive | positive | 17.66 | 94 | negative | negative | - |
| 23 | positive | positive | 22.40 | 59 | positive | positive | 26.08 | 95 | negative | negative | - |
| 24 | positive | positive | 26.51 | 60 | positive | positive | 20.85 | 96 | negative | negative | - |
| 25 | negative | positive | 30.14 | 61 | negative | negative | - | 97 | negative | negative | - |
| 26 | negative | positive | 33.77 | 62 | negative | negative | - | 98 | negative | negative | -s |
| 27 | negative | negative | - | 63 | positive | positive | 25.79 | 99 | positive | positive | 21.08 |
| 28 | negative | negative | - | 64 | positive | positive | 24.61 | 100 | positive | positive | 19.49 |
| 29 | positive | positive | 17.29 | 65 | negative | positive | 30.38 | 101 | positive | positive | 17.58 |
| 30 | positive | positive | 18.34 | 66 | positive | positive | 22.85 | 102 | positive | positive | 31.98 |
| 31 | positive | positive | 21.04 | 67 | negative | positive | 30.59 | 103 | positive | positive | 17.91 |
| 32 | positive | positive | 24.28 | 68 | negative | positive | 29.35 | 104 | positive | positive | 22.68 |
| 33 | negative | positive | 28.27 | 69 | negative | positive | 27.22 | 105 | negative | positive | 27.02 |
| 34 | negative | positive | 31.86 | 70 | negative | positive | 30.09 | 106 | positive | positive | 21.83 |
| 35 | negative | negative | - | 71 | negative | positive | 29.50 | ||||
| 36 | negative | negative | - | 72 | negative | negative | - | ||||
| No. | RHAM | qPCR | No. | RHAM | qPCR | No. | RHAM | qPCR | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Result | Min | Result | Ct | Result | Min | Result | Ct | Result | Min | Result | Ct | |||
| 1 | positive | 13.37 | positive | 24.51 | 37 | positive | 13.34 | positive | 23.59 | 73 | negative | 35.00 | negative | - |
| 2 | positive | 13.35 | positive | 30.21 | 38 | positive | 13.34 | positive | 24.25 | 74 | negative | 35.00 | negative | - |
| 3 | positive | 13.42 | positive | 28.77 | 39 | positive | 14.34 | positive | 28.13 | 75 | negative | 35.00 | negative | - |
| 4 | positive | 13.35 | positive | 27.55 | 40 | positive | 16.33 | positive | 31.43 | 76 | negative | 35.00 | negative | - |
| 5 | positive | 13.37 | positive | 27.33 | 41 | positive | 22.29 | positive | 35.01 | 77 | negative | 35.00 | negative | - |
| 6 | positive | 14.34 | positive | 29.75 | 42 | negative | 35.00 | negative | - | 78 | negative | 35.00 | negative | - |
| 7 | positive | 13.35 | positive | 28.11 | 43 | negative | 35.00 | negative | - | 79 | negative | 35.00 | negative | - |
| 8 | positive | 20.40 | positive | 30.37 | 44 | negative | 35.00 | negative | - | 80 | negative | 35.00 | negative | - |
| 9 | negative | 35.00 | positive | 33.51 | 45 | positive | 10.37 | positive | 19.81 | 81 | negative | 35.00 | negative | - |
| 10 | negative | 35.00 | positive | 30.41 | 46 | positive | 11.36 | positive | 18.61 | 82 | negative | 35.00 | negative | - |
| 11 | negative | 35.00 | negative | - | 47 | positive | 13.35 | positive | 24.32 | 83 | positive | 26.37 | positive | 24.74 |
| 12 | negative | 35.00 | negative | - | 48 | positive | 12.36 | positive | 25.53 | 84 | positive | 17.31 | positive | 23.42 |
| 13 | positive | 11.45 | positive | 17.26 | 49 | positive | 16.31 | positive | 17.18 | 85 | positive | 15.33 | positive | 28.51 |
| 14 | positive | 19.30 | positive | 20.52 | 50 | negative | 35.00 | negative | - | 86 | positive | 22.32 | positive | 22.54 |
| 15 | positive | 15.33 | positive | 23.98 | 51 | positive | 12.35 | positive | 16.24 | 87 | positive | 17.32 | positive | 29.39 |
| 16 | positive | 15.33 | positive | 28.02 | 52 | positive | 20.33 | positive | 19.3 | 88 | positive | 18.31 | positive | 28.37 |
| 17 | positive | 18.33 | negative | - | 53 | positive | 14.35 | positive | 18.59 | 89 | positive | 15.33 | positive | 26.31 |
| 18 | negative | 35.00 | negative | - | 54 | positive | 11.42 | positive | 16.35 | 90 | positive | 20.28 | positive | 30.17 |
| 19 | negative | 35.00 | negative | - | 55 | positive | 15.33 | positive | 18.37 | 91 | positive | 15.33 | positive | 28.58 |
| 20 | negative | 35.00 | negative | - | 56 | positive | 12.38 | positive | 15.85 | 92 | positive | 15.33 | positive | 28.19 |
| 21 | positive | 15.14 | positive | 18.03 | 57 | positive | 12.36 | positive | 15.68 | 93 | positive | 18.30 | positive | 32.21 |
| 22 | positive | 11.37 | positive | 19.06 | 58 | positive | 16.41 | positive | 17.66 | 94 | negative | 35.00 | negative | - |
| 23 | positive | 11.39 | positive | 22.4 | 59 | positive | 24.25 | positive | 26.08 | 95 | negative | 35.00 | negative | - |
| 24 | positive | 12.36 | positive | 26.51 | 60 | positive | 20.33 | positive | 20.85 | 96 | negative | 35.00 | negative | - |
| 25 | positive | 12.36 | positive | 30.14 | 61 | negative | 35.00 | negative | - | 97 | negative | 35.00 | negative | - |
| 26 | negative | 35.00 | positive | 33.77 | 62 | negative | 35.00 | negative | - | 98 | negative | 35.00 | negative | - |
| 27 | negative | 35.00 | negative | - | 63 | positive | 16.32 | positive | 25.79 | 99 | positive | 14.34 | positive | 21.08 |
| 28 | negative | 35.00 | negative | - | 64 | positive | 19.34 | positive | 24.61 | 100 | positive | 12.36 | positive | 19.49 |
| 29 | positive | 14.35 | positive | 17.29 | 65 | negative | 35.00 | positive | 30.38 | 101 | positive | 11.37 | positive | 17.58 |
| 30 | positive | 18.31 | positive | 18.34 | 66 | positive | 17.36 | positive | 22.85 | 102 | positive | 15.33 | positive | 31.98 |
| 31 | positive | 13.36 | positive | 21.04 | 67 | positive | 20.29 | positive | 30.59 | 103 | positive | 13.35 | positive | 17.91 |
| 32 | positive | 13.36 | positive | 24.28 | 68 | positive | 30.21 | positive | 29.35 | 104 | positive | 13.35 | positive | 22.68 |
| 33 | positive | 15.34 | positive | 28.27 | 69 | positive | 17.40 | positive | 27.22 | 105 | positive | 16.33 | positive | 27.02 |
| 34 | positive | 21.29 | positive | 31.86 | 70 | positive | 20.32 | positive | 30.09 | 106 | positive | 13.35 | positive | 21.83 |
| 35 | negative | 35.00 | negative | - | 71 | positive | 19.30 | positive | 29.50 | |||||
| 36 | negative | 35.00 | negative | - | 72 | negative | 35.00 | negative | - | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruenphet, S.; Srionrod, N.; Nipakornpun, T.; Wutthiwitthayaphong, S. RNase Hybridization-Assisted Amplification (RHAM) Technology: A High-Sensitivity, Field-Deployable Alternative to Quantitative Polymerase Chain Reaction for the Rapid Detection of African Swine Fever Virus. Vet. Sci. 2025, 12, 1068. https://doi.org/10.3390/vetsci12111068
Ruenphet S, Srionrod N, Nipakornpun T, Wutthiwitthayaphong S. RNase Hybridization-Assisted Amplification (RHAM) Technology: A High-Sensitivity, Field-Deployable Alternative to Quantitative Polymerase Chain Reaction for the Rapid Detection of African Swine Fever Virus. Veterinary Sciences. 2025; 12(11):1068. https://doi.org/10.3390/vetsci12111068
Chicago/Turabian StyleRuenphet, Sakchai, Nitipon Srionrod, Teera Nipakornpun, and Supphathat Wutthiwitthayaphong. 2025. "RNase Hybridization-Assisted Amplification (RHAM) Technology: A High-Sensitivity, Field-Deployable Alternative to Quantitative Polymerase Chain Reaction for the Rapid Detection of African Swine Fever Virus" Veterinary Sciences 12, no. 11: 1068. https://doi.org/10.3390/vetsci12111068
APA StyleRuenphet, S., Srionrod, N., Nipakornpun, T., & Wutthiwitthayaphong, S. (2025). RNase Hybridization-Assisted Amplification (RHAM) Technology: A High-Sensitivity, Field-Deployable Alternative to Quantitative Polymerase Chain Reaction for the Rapid Detection of African Swine Fever Virus. Veterinary Sciences, 12(11), 1068. https://doi.org/10.3390/vetsci12111068








