Evaluation of the Effectiveness of Single-Nucleotide Polymorphisms Versus Microsatellites for Parentage Verification in Horse Breeds
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Genomic DNA Extraction
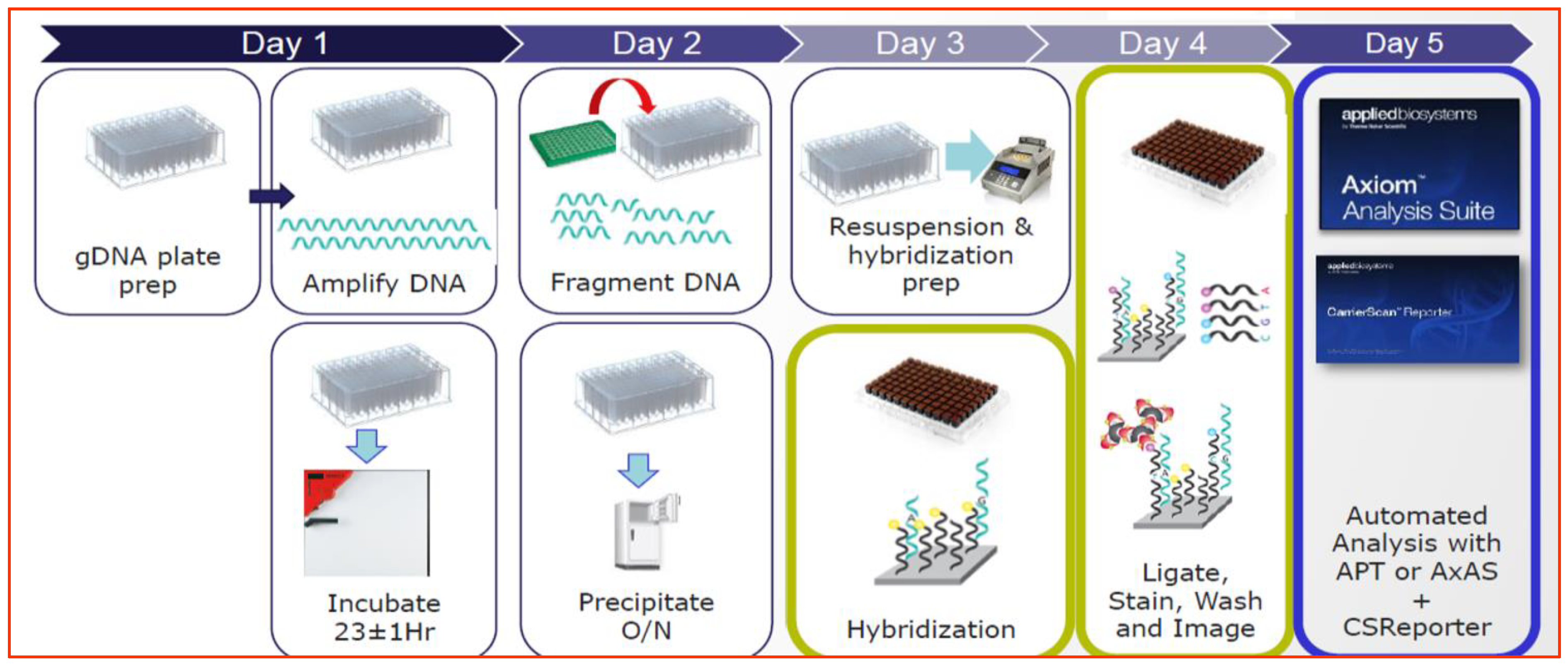
2.2. STR and SNP Analysis
2.3. Genetic Diversity Analysis
3. Results
3.1. Genetic Diversity
3.2. Comparison of Parentage Testing Using STRs and SNPs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tozaki, T.; Kakoi, H.; Mashima, S.; Hirota, K.-I.; Hasegawa, T.; Ishida, N.; Miura, N.; Choi-Miura, N.H.; Tomita, M. Population Study and Validation of Paternity Testing for Thoroughbred Horses by 15 Microsatellite Loci. J. Vet. Med. Sci. 2001, 63, 1191–1197. [Google Scholar] [CrossRef] [PubMed]
- Park, C.S.; Lee, S.Y.; Cho, G.J. Evaluation of recent changes in genetic variability in Thoroughbred horses based on microsatellite markers parentage panel in Korea. Anim. Biosci. 2022, 35, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Al Abri, M.A.; Brooks, S.A.; Al-Saqri, N.; Alkharousi, K.; Johnson, E.H.; Alqaisi, O.; Al-Rawahi, A.; Al Marzooqi, W. Investigating the population structure and genetic diversity of Arabian horses in Oman using SNP markers. Anim. Genet. 2021, 52, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Aminou, O.; Badaoui, B.; Machmoum, M.; Piro, M. Evaluation of the effectiveness of single nucleotide polymorphisms compared to microsatellite markers for parentage verification in Moroccan horses. Anim. Genet. 2024, 55, 404–409. [Google Scholar] [CrossRef]
- Hirota, K.I.; Kakoi, H.; Gawahara, H.; Hasegawa, T.; Tozaki, T. Construction and Validation of Parentage Testing for Thoroughbred Horses by 53 Single Nucleotide Polymorphisms. J. Vet. Med. Sci. 2010, 72, 719–726. [Google Scholar] [CrossRef][Green Version]
- Nolte, W.; Alkhoder, H.; Wobbe, M.; Stock, K.F.; Kalm, E.; Vosgerau, S.; Krattenmacher, N.; Thaller, G.; Tetens, J.; Kühn, C. Replacement of microsatellite markers by imputed medium-density SNP arrays for parentage control in German warmblood horses. J. Appl. Genet. 2022, 63, 783–792. [Google Scholar] [CrossRef]
- Hajihosseinlo, A.; Nejati-Javaremib, A.; Miraei-Ashtian, S.R. Genetic structure analysis in several populations of cattle using SNP genotypes. Anim. Biotechnol. 2023, 34, 288–300. [Google Scholar] [CrossRef]
- Weng, Z.; Yang, Y.; Wang, X.; Wu, L.; Hua, S.; Zhang, H.; Meng, Z. Parentage Analysis in Giant Grouper (Epinephelus lanceolatus) Using Microsatellite and SNP Markers from Genotyping-by-Sequencing Data. Genes 2021, 12, 1042. [Google Scholar] [CrossRef]
- Pakstisa, J.; Speedw, C.; Kiddj, R.; Kiddk, K. Candidate SNPs for a universal individual identification panel. Hum. Genet. 2007, 121, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Thermo Fisher Scientific. KingFisher Presto User Manual; Thermo Fisher Scientific: Waltham, MA, USA, 2016. [Google Scholar]
- Dimsoski, P. Development of a 17plex microsatellite polymerase chain reaction kit for genotyping horses. Croat. Med. J. 2003, 44, 332–335. [Google Scholar]
- Kakoi, H.; Nagata, S.; Kurosawa, M. DNA Typing with 17 microsatellites for parentage verification of racehorses in Japan. Nihon Chikusan Gakkaiho 2001, 72, 453–460. [Google Scholar] [CrossRef]
- Bellone, R.R.; Mansour, T.A.; Esdaile, E.; Wallner, B.; Raudsepp, T.; Till, B.; Kallenberg, A.; Hughes, S.; Chadaram, S.; Shrestha, S.; et al. Development of a Robust Across Breed Equine Parentage SNP Panel for ISAG Approval. In Proceedings of the 39th ISAG Conference, Cape Town, South Africa, 2–7 July 2023. [Google Scholar]
- Santos, W.B.; Schettini, G.P.; Maiorano, A.M.; Bussiman, F.O.; Balieiro, J.C.C.; Ferraz, G.C.; Pereira, G.I.; Baldassini, W.A.; Neto, O.R.M.; Oliveira, H.N.; et al. Genome-wide scans for signatures of selection in Mangalarga Marchador horses using high-throughput SNP genotyping. BMC Genom. 2021, 22, 737. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Kim, S.M.; Oyungerel, B.; Cho, G.J. Single nucleotide polymorphisms for parentage testing of horse breeds in Korea. Anim. Biosci. 2024, 37, 600–608. [Google Scholar] [CrossRef] [PubMed]
- Thermo Fisher Scientific. Axiom™ Genotyping Solution Data Analysis USER GUIDE; Thermo Fisher Scientific: Waltham, MA, USA, 2022. [Google Scholar]
- Sekino, M.; Kakehi, S. PARFEX v1.0: An EXCEL™-based software package for parentage allocation. Cons. Genet. Res. 2012, 4, 275–278. [Google Scholar] [CrossRef]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Raymond, M.; Rousset, F. GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism. J. Hered. 1995, 86, 248–249. [Google Scholar] [CrossRef]
- Binns, M.M.; Holmes, N.G.; Rolliman, A.; Scott, A.M. The identification of polymorphic microsatellite loci in the horse and their use in thoroughbred parentage testing. Br. Vet. J. 1995, 151, 9–15. [Google Scholar] [CrossRef]
- de Groot, M.; van Haeringen, W.A. An evaluation of the International Society for Animal Genetics recommended parentage and identification panel for the domestic pigeon (Columba livia domestica). Anim. Genet. 2017, 48, 431–435. [Google Scholar] [CrossRef]
- Heaton, M.P.; Harhay, G.P.; Bennett, G.L.; Stone, R.T.; Grosse, W.M.; Casas, E.; Keele, J.W.; Smith, T.P.; Chitko-McKown, C.G.; Laegreid, W.W. Selection and use of SNP markers for animal identification and paternity analysis in US beef cattle. Mamm. Genome 2002, 13, 272–281. [Google Scholar] [CrossRef]
- Jones, A.G.; Small, C.M.; Paczolt, K.A.; Ratterman, N.L. A practical guide to methods of parentage analysis. Mol. Ecol. Resour. 2010, 10, 6–30. [Google Scholar] [CrossRef]
- Perlin, M.W.; Lancia, G.; Ng, S.K. Toward fully automated genotyping: Genotyping microsatellite markers by deconvolution. Am. J. Hum. Genet. 1995, 57, 1199–1210. [Google Scholar]
- Holl, H.M.; Vanhnasy, J.; Everts, R.E.; Hoefs-Martin, K.; Cook, D.; Brooks, S.A.; Carpenter, M.L.; Bustamante, C.D.; Lafayette, C. Single nucleotide polymorphisms for DNA typing in the domestic horse. Anim. Genet. 2017, 48, 669–676. [Google Scholar] [CrossRef]
- Wu, X.L.; Xu, J.; Li, H.; Ferretti, R.; He, J.; Qiu, J.; Xiao, Q.; Simpson, B.; Michell, T.; Kachman, S.D.; et al. Evaluation of genotyping concordance for commercial bovine SNP arrays using quality-assurance samples. Anim. Genet. 2019, 50, 367–371. [Google Scholar] [CrossRef]
- Bertolini, F.; Galimberti, G.; Calò, D.G.; Schiavo, G.; Matassino, D.; Fontanesi, L. Combined use principal component analysis and random forests identify population-informative single nucleotide polymorphisms: Application in cattle breeds. J. Anim. Breed. Genet. 2015, 132, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, F.; Galimberti, G.; Schiavo, G.; Mastrangelo, S.; Di Gerlando, R.; Strillacci, M.G.; Bagnato, A.; Portolano, B.; Fontanesi, L. Preselection statistics and Random Forest classification identify population informative single nucleotide polymorphisms in cosmopolitan and autochthonous cattle breeds. Animal 2018, 12, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Väli, U.; Einarsson, A.; Waits, L.; Ellegren, H. To what extent do microsatellite markers reflect genome-wide genetic diversity in natural populations? Mol. Ecol. 2008, 17, 3808–3817. [Google Scholar] [CrossRef] [PubMed]
- Tokarska, M.; Marshall, T.; Kowalczyk, R.; Wojcik, J.M.; Pertoldi, C.; Kristensen, T.N.; Loeschcke, V.; Gregersen, V.R.; Bendixen, C. Effectiveness of microsatellite and SNP markers for parent age and identity analysis in species with low genetic diversity: The case of European bison. Heredity 2009, 103, 326–332. [Google Scholar] [CrossRef]
- de Groot, M.; Anderson, H.; Bauer, H.; Bauguil, C.; Bellone, R.R.; Brugidou, R.; Buckley, R.M.; Dovč, P.; Forman, O.; Grahn, R.A.; et al. Standardization of a SNP panel for parentage verification and identification in the domestic cat (Felis silvestris catus). Anim. Genet. 2021, 52, 675–682. [Google Scholar] [CrossRef]
- Tozaki, T.; Ohnuma, A.; Kikuchi, M.; Ishige, T.; Kakoi, H.; Hirota, K.; Kusano, K.; Nagata, S. Rare and common variant discovery by whole-genome sequencing of 101 thoroughbred racehorses. Sci. Rep. 2021, 11, 16057. [Google Scholar] [CrossRef]
- Wade, C.M.; Giulotto, E.; Sigurdsson, S.; Zoli, M.; Gnerre, S.; Imsland, F.; Lear, T.L.; Adelson, D.L.; Bailey, E.; Bellone, R.R.; et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 2009, 326, 865–867. [Google Scholar] [CrossRef]
- Flynn, P.; Morrin-O’Donnell, R.; Weld, R.; Gargan, L.M.; Carlsson, J.; Daly, S.; Suren, H.; Siddavatam, P.; Gujjula, K.R. Comparative analysis of single nucleotide polymorphisms and microsatellite markers for parentage verification and discovery within the equine Thoroughbred breed. bioRxiv 2021. [Google Scholar] [CrossRef]
- Bayarsaikhan, M.; Purevdorj, N.O.; Kim, B.H.; Jung, J.H.; Cho, G.J. Evaluation of the Microbiological Status of Cattle Carcasses in Mongolia: Considering the Hygienic Practices of Slaughter Establishments. Vet. Sci. 2023, 10, 563. [Google Scholar] [CrossRef]
- Al Abri, M.A.; von Borstel, U.K.; Strecker, V.; Brooks, S.A. Application of Genomic Estimation Methods of Inbreeding and Population Structure in an Arabian Horse Herd. J. Hered. 2017, 108, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.L.; Mickelson, J.R.; Cothran, E.G.; Andersson, L.S.; Axelsson, J.; Bailey, E.; Bannasch, D.; Binns, M.M.; Borges, A.S.; Brama, P.; et al. Genetic diversity in the modern horse illustrated from genome-wide SNP data. PLoS ONE 2013, 8, e54997. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Ermini, L.; Der Sarkissian, C.; Jónsson, H.; Ginolhac, A.; Schaefer, R.; Martin, M.D.; Fernández, R.; Kircher, M.; McCue, M.E.; et al. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat. Protoc. 2014, 9, 1056–1082. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, W.; Wang, W.L.; Yin, T.T.; Dong, W.Q.; Xu, C.J. A high-throughput SNP discovery strategy for RNA-seq data. BMC Genom. 2019, 20, 1471–2164. [Google Scholar] [CrossRef]
| Marker | Ho | He | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TB * | JH | QH | AM | MH | TB | JH | QH | AM | MH | |
| AHT4 | 0.737 | 0.824 | 0.500 | 0.762 | 0.850 | 0.744 | 0.849 | 0.659 | 0.764 | 0.845 |
| AHT5 | 0.868 | 0.824 | 0.700 | 0.714 | 0.817 | 0.690 | 0.754 | 0.775 | 0.764 | 0.788 |
| ASB2 | 0.816 | 0.529 | 0.900 | 0.667 | 0.850 | 0.790 | 0.620 | 0.850 | 0.811 | 0.836 |
| ASB17 | 0.816 | 0.882 | 0.700 | 0.810 | 0.893 | 0.761 | 0.803 | 0.829 | 0.832 | 0.889 |
| ASB23 | 0.632 | 0.647 | 0.800 | 0.810 | 0.785 | 0.710 | 0.651 | 0.850 | 0.818 | 0.835 |
| CA425 | 0.816 | 0.765 | 0.850 | 1.000 | 0.763 | 0.730 | 0.756 | 0.722 | 0.817 | 0.767 |
| HMS1 | 0.605 | 0.588 | 0.700 | 0.571 | 0.634 | 0.568 | 0.586 | 0.633 | 0.638 | 0.708 |
| HMS2 | 0.684 | 0.588 | 0.800 | 0.619 | 0.828 | 0.581 | 0.757 | 0.811 | 0.796 | 0.817 |
| HMS3 | 0.763 | 0.882 | 0.950 | 0.667 | 0.613 | 0.620 | 0.822 | 0.826 | 0.768 | 0.783 |
| HMS6 | 0.553 | 0.647 | 0.750 | 0.857 | 0.828 | 0.618 | 0.706 | 0.745 | 0.806 | 0.779 |
| HMS7 | 0.868 | 0.412 | 0.650 | 0.857 | 0.613 | 0.767 | 0.438 | 0.818 | 0.835 | 0.606 |
| HTG4 | 0.658 | 0.647 | 0.550 | 0.333 | 0.613 | 0.565 | 0.684 | 0.701 | 0.376 | 0.642 |
| HTG10 | 0.763 | 0.882 | 0.850 | 0.762 | 0.839 | 0.751 | 0.781 | 0.825 | 0.842 | 0.866 |
| LEX3 | 0.842 | 0.529 | 0.900 | 0.762 | 0.871 | 0.805 | 0.781 | 0.867 | 0.862 | 0.850 |
| VHL20 | 0.605 | 0.941 | 0.850 | 0.905 | 0.850 | 0.721 | 0.748 | 0.855 | 0.862 | 0.849 |
| Mean | 0.735 | 0.706 | 0.763 | 0.740 | 0.776 | 0.695 | 0.716 | 0.784 | 0.773 | 0.791 |
| Marker | Fis | PIC | ||||||||
| TB | JH | QH | AM | MH | TB | JH | QH | AM | MH | |
| AHT4 | 0.009 | 0.030 | 0.242 | 0.003 | −0.006 | 0.685 | 0.802 | 0.601 | 0.708 | 0.821 |
| AHT5 | −0.259 | −0.093 | 0.097 | 0.065 | −0.038 | 0.636 | 0.702 | 0.714 | 0.715 | 0.751 |
| ASB2 | −0.033 | 0.145 | −0.059 | 0.178 | −0.016 | 0.748 | 0.559 | 0.808 | 0.758 | 0.810 |
| ASB17 | −0.072 | −0.098 | 0.156 | 0.027 | −0.004 | 0.711 | 0.753 | 0.781 | 0.788 | 0.874 |
| ASB23 | 0.111 | 0.006 | 0.059 | 0.010 | 0.060 | 0.667 | 0.594 | 0.803 | 0.770 | 0.811 |
| CA425 | −0.117 | −0.012 | −0.177 | −0.225 | 0.004 | 0.678 | 0.695 | 0.666 | 0.780 | 0.732 |
| HMS1 | −0.066 | −0.003 | −0.106 | 0.105 | 0.104 | 0.469 | 0.508 | 0.551 | 0.573 | 0.656 |
| HMS2 | −0.177 | 0.223 | 0.013 | 0.223 | −0.014 | 0.507 | 0.685 | 0.767 | 0.740 | 0.785 |
| HMS3 | −0.231 | −0.074 | −0.150 | 0.132 | 0.217 | 0.578 | 0.770 | 0.782 | 0.704 | 0.749 |
| HMS6 | 0.106 | 0.083 | −0.007 | −0.064 | −0.063 | 0.535 | 0.629 | 0.688 | 0.755 | 0.740 |
| HMS7 | −0.133 | 0.059 | 0.206 | −0.027 | −0.012 | 0.720 | 0.385 | 0.763 | 0.789 | 0.572 |
| HTG4 | −0.165 | 0.054 | 0.216 | 0.114 | 0.045 | 0.462 | 0.609 | 0.622 | 0.346 | 0.606 |
| HTG10 | −0.017 | −0.129 | −0.030 | 0.095 | 0.031 | 0.702 | 0.724 | 0.782 | 0.798 | 0.846 |
| LEX3 | −0.046 | 0.364 | −0.038 | 0.116 | −0.025 | 0.767 | 0.771 | 0.828 | 0.819 | 0.827 |
| VHL20 | 0.161 | −0.258 | −0.052 | −0.050 | −0.001 | 0.657 | 0.687 | 0.815 | 0.826 | 0.827 |
| Mean | −0.062 | 0.020 | 0.025 | 0.047 | 0.019 | 0.635 | 0.658 | 0.731 | 0.725 | 0.761 |
| Makers | Breeds | No. of Horses | No. of Markers | Ho | He | Fis | PIC |
|---|---|---|---|---|---|---|---|
| STRs * | TB ** | 38 | 15 | 0.735 | 0.695 | −0.058 | 0.635 |
| JH | 17 | 0.706 | 0.719 | 0.018 | 0.658 | ||
| QH | 20 | 0.767 | 0.785 | 0.023 | 0.731 | ||
| AM | 21 | 0.740 | 0.773 | 0.043 | 0.725 | ||
| MH | 93 | 0.776 | 0.791 | 0.018 | 0.761 | ||
| SNPs | TB | 38 | 71 | 0.451 | 0.471 | 0.041 | 0.355 |
| JH | 17 | 0.470 | 0.491 | 0.043 | 0.362 | ||
| QH | 20 | 0.460 | 0.477 | 0.036 | 0.355 | ||
| AM | 18 | 0.415 | 0.468 | 0.113 | 0.349 | ||
| MH | 93 | 0.487 | 0.483 | −0.009 | 0.364 |
| Marker | Breeds | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AHT4 | AHT5 | ASB2 | HMS3 | HMS6 | HMS7 | HTG4 | HTG10 | VHL20 | ASB17 | ASB23 | HMS1 | LEX3 | CA425 | HMS2 | ||
| Sire | H/O | K/M | M/Q | I/O | P/P | M/N | K/M | I/I | L/M | N/O | K/K | J/M | M/- | N/O | K/L | TB ** |
| Dam | J/O | J/K | N/R | I/O | P/P | O/O | K/K | I/R | I/I | N/R | J/S | J/M | H/O | J/N | K/L | |
| Foal | O/O | M/N * | M/N | I/I | P/P | N/O | K/M | I/I | I/L | M/R | L/S | J/M | N/- | M/N | L/L | |
| Sire | I/O | J/N | K/M | I/N | L/P | N/O | P/P | L/S | I/M | M/Q | J/U | M/M | O/O | J/M | M/O | MH |
| Dam | H/O | N/N | K/K | I/I | L/O | O/O | K/P | R/S | I/Q | N/Q | L/U | J/M | L/O | J/N | J/O | |
| Foal | J/O | J/N | K/N | I/I | L/L | O/O | K/P | L/S | M/Q | M/M | U/U | M/M | O/O | J/N | J/N | |
| Marker | TB * | MH | ||||
|---|---|---|---|---|---|---|
| Sire | Dam | Foal | Sire | Dam | Foal | |
| MNEc_2_10_58909591_BIEC2_126732 | AB ** | BB | AA *** | AB | AB | AA |
| MNEc_2_6_31320852_BIEC2_946446 | AB | AA | AA | AB | BB | AB |
| MNEc_2_16_81464884_BIEC2_364741 | BB | BB | AB | AA | AA | AB |
| MNEc_2_10_43452669_BIEC2_119640 | AA | AB | AA | AB | AB | BB |
| MNEc_2_31_17012751_BIEC2_839012 | AB | BB | AB | AB | BB | AA |
| MNEc_2_26_29137373_BIEC2_692543 | AB | BB | AB | AB | AB | AB |
| MNEc_2_8_61558651_BIEC2_1057053 | BB | AB | AB | BB | BB | AB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.; Lee, S.; Oyungerel, B.; Cho, G. Evaluation of the Effectiveness of Single-Nucleotide Polymorphisms Versus Microsatellites for Parentage Verification in Horse Breeds. Vet. Sci. 2025, 12, 890. https://doi.org/10.3390/vetsci12090890
Kim D, Lee S, Oyungerel B, Cho G. Evaluation of the Effectiveness of Single-Nucleotide Polymorphisms Versus Microsatellites for Parentage Verification in Horse Breeds. Veterinary Sciences. 2025; 12(9):890. https://doi.org/10.3390/vetsci12090890
Chicago/Turabian StyleKim, Dongsoo, Sunyoung Lee, Baatartsogt Oyungerel, and Giljae Cho. 2025. "Evaluation of the Effectiveness of Single-Nucleotide Polymorphisms Versus Microsatellites for Parentage Verification in Horse Breeds" Veterinary Sciences 12, no. 9: 890. https://doi.org/10.3390/vetsci12090890
APA StyleKim, D., Lee, S., Oyungerel, B., & Cho, G. (2025). Evaluation of the Effectiveness of Single-Nucleotide Polymorphisms Versus Microsatellites for Parentage Verification in Horse Breeds. Veterinary Sciences, 12(9), 890. https://doi.org/10.3390/vetsci12090890






