Evaluation of PD-L1 and TIM-3 Pathways in T Cells During Experimental Bovine Leukemia Virus Infection in Sheep
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Blood Sample Collection and Experimental BLV Infection in Sheep
2.2. Cell Isolation
2.3. Nucleic Acid Extraction and Proviral Load Measurement
2.4. Flow Cytometric Analysis of PD-L1 and TIM-3 Expression
2.5. T-Cell Marker and Cytokine Production Analysis
2.6. Statistical Analysis
3. Results
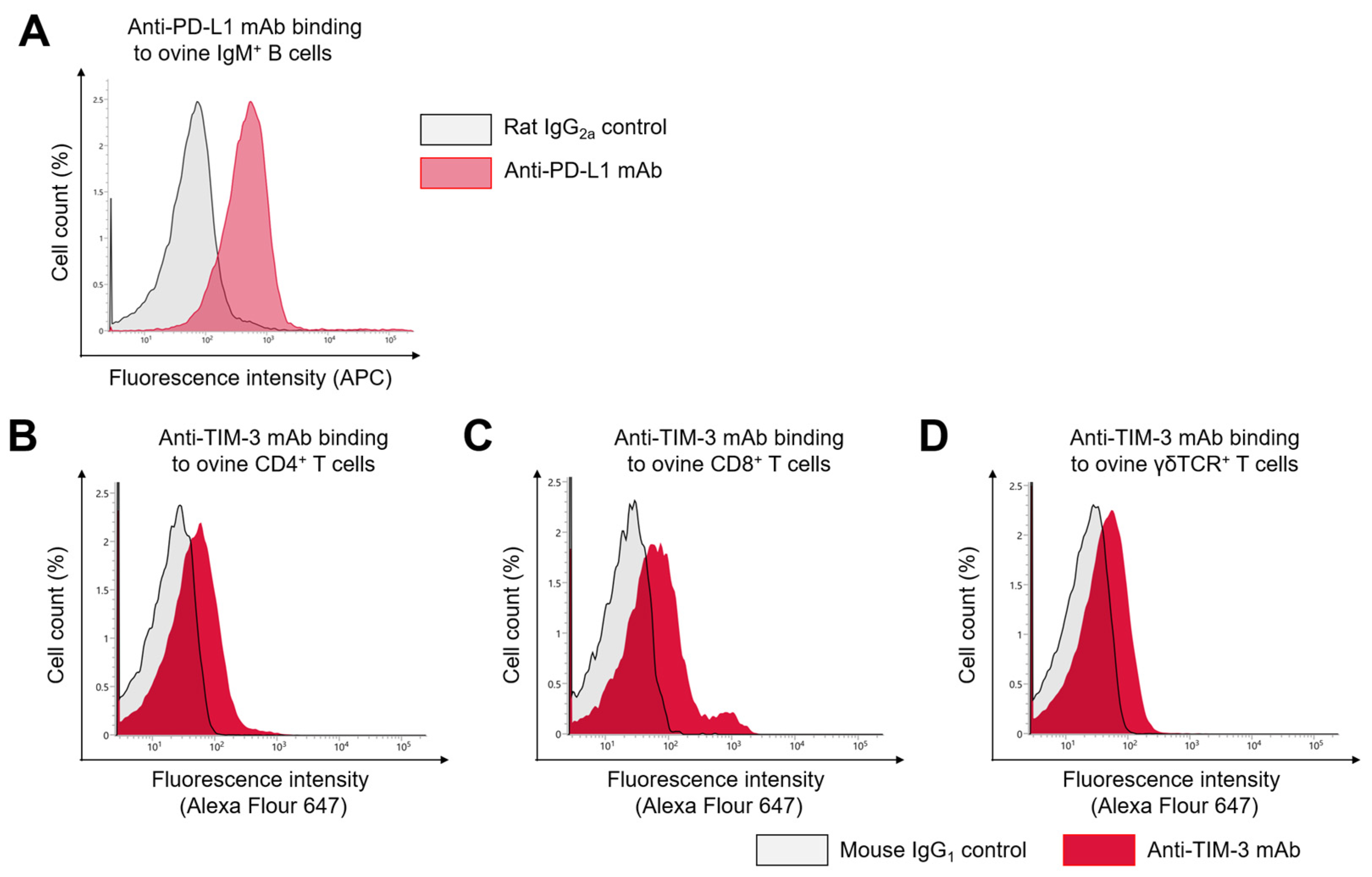
3.1. Cross-Reactivity of Anti-Bovine PD-L1 and TIM-3 MAbs with Ovine PD-L1 and TIM-3
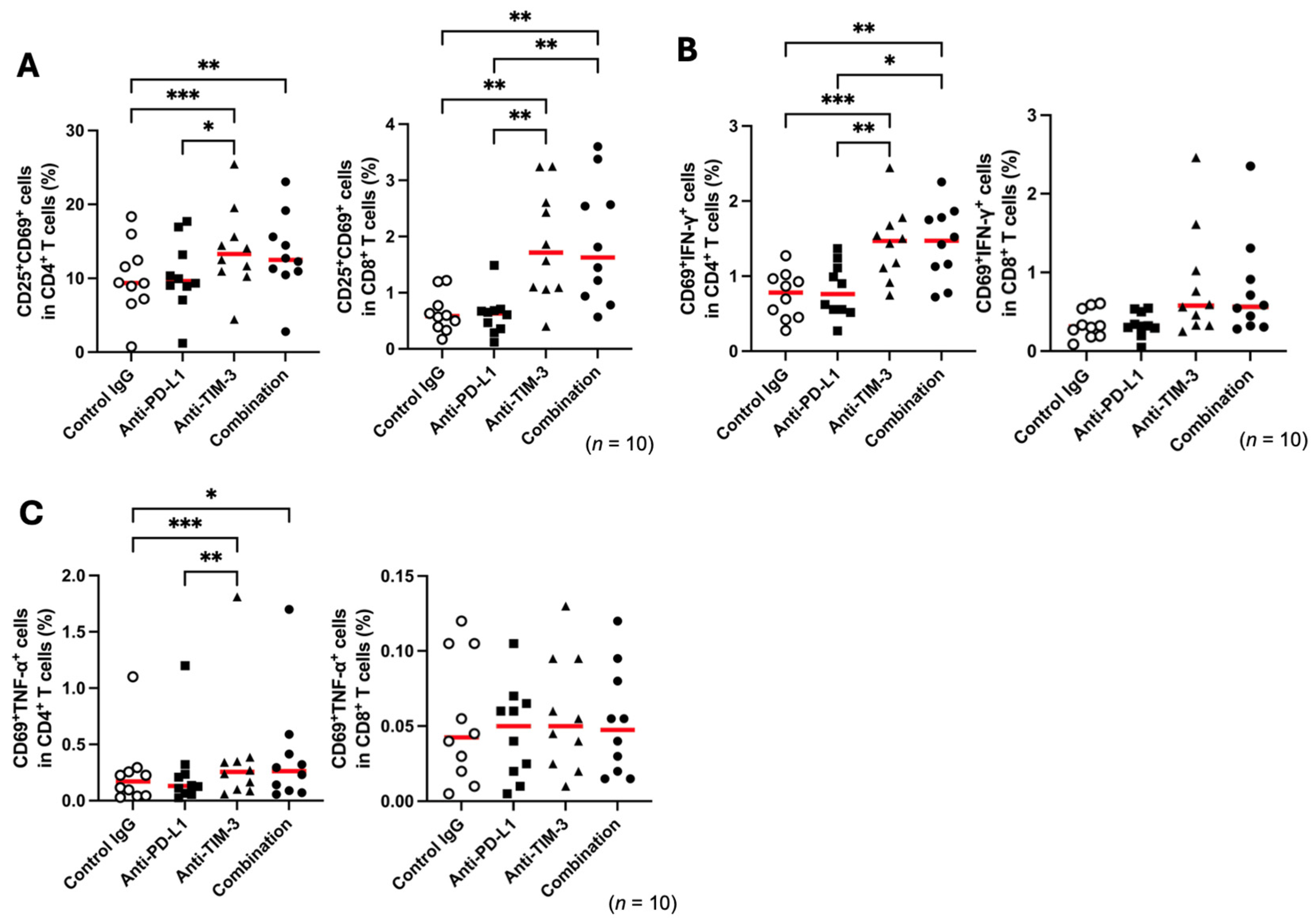
3.2. TIM-3 Blockade Enhances T-Cell Activation in Healthy Sheep
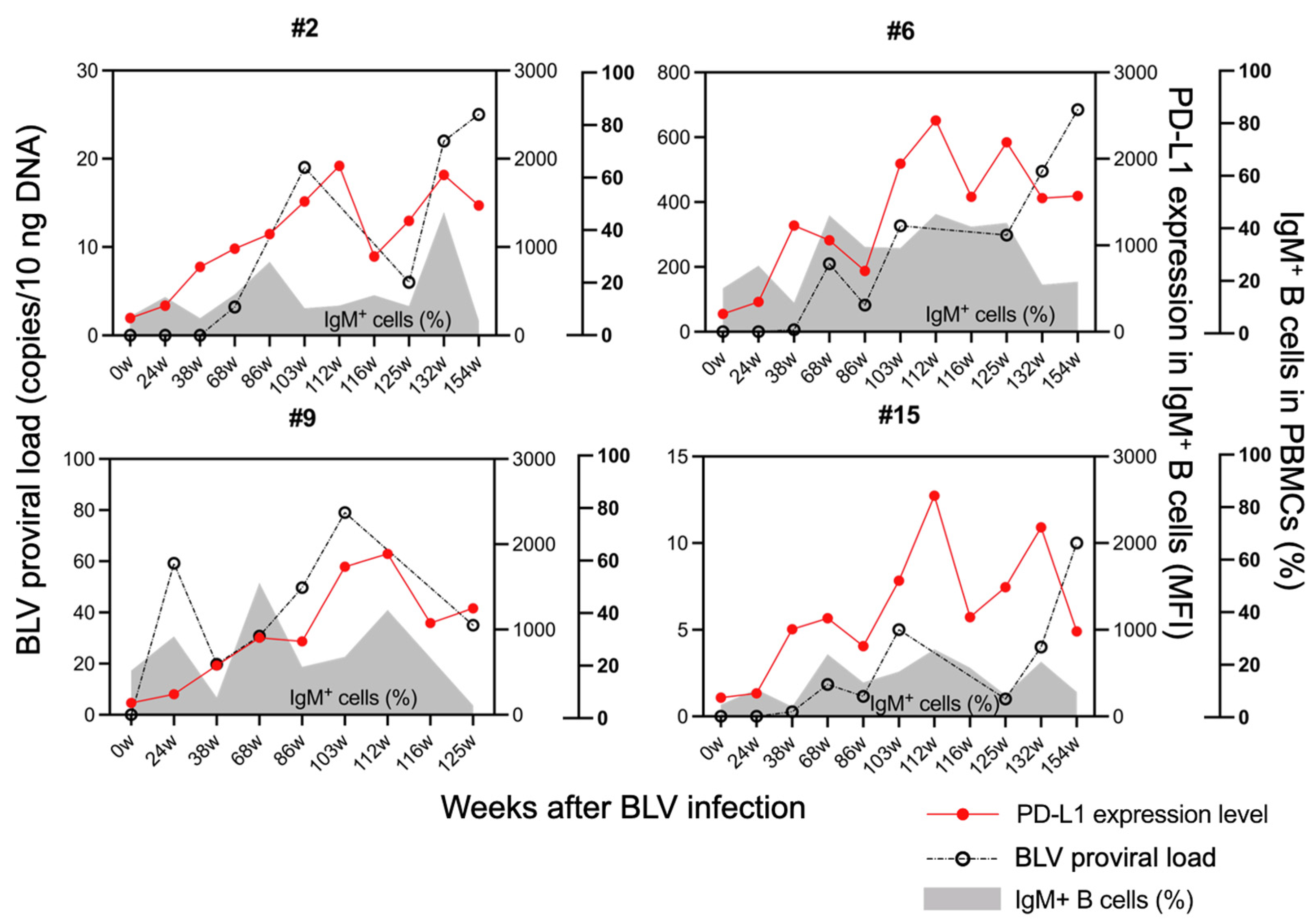
3.3. Upregulation of PD-L1 Expression and Proviral Load Following BLV Infection in Sheep
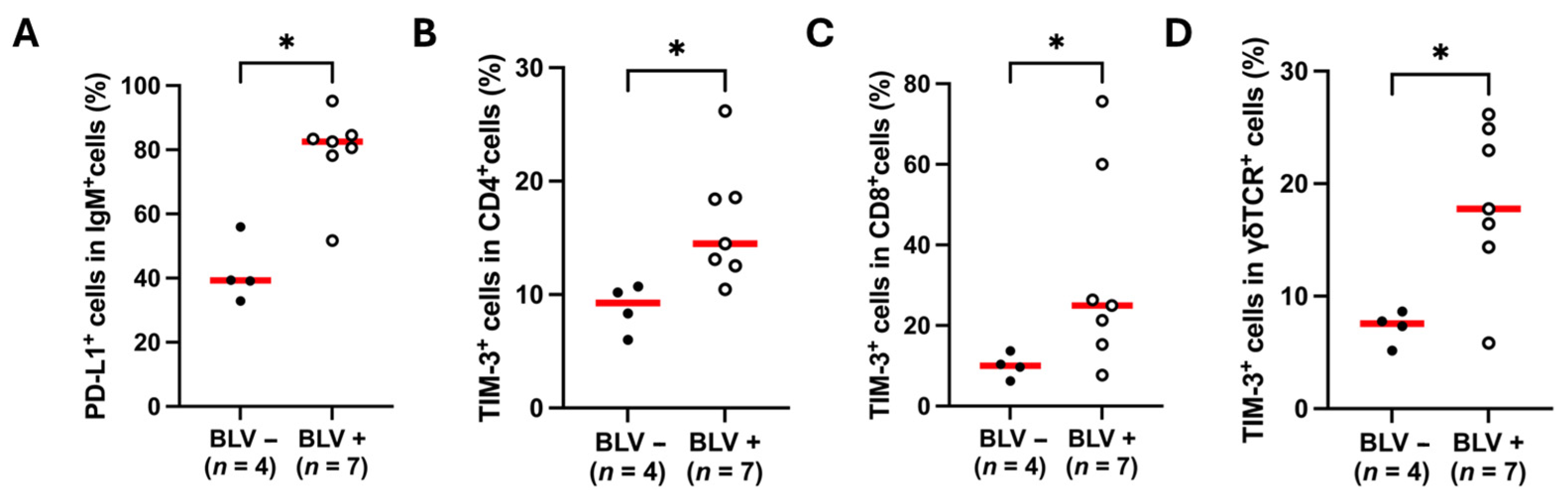
3.4. Upregulation of PD-L1 and TIM-3 in BLV-Infected Sheep
3.5. Blockade of TIM-3 Pathway Restores T-Cell Function in BLV-Infected Sheep
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AL | Aleukemic |
| APC | Allophycocyanin |
| BLV | Bovine leukemia virus |
| ConA | Concanavalin A |
| Cy5.5 | Cyanin5.5 |
| Cy7 | Cyanin7 |
| EBL | Enzootic bovine leukosis |
| EDTA | Ethylenediaminetetraacetic acid |
| ICI | Immune checkpoint inhibitors |
| IFN-γ | Interferon gamma |
| mAb | Monoclonal antibody |
| MFI | Median fluorescence intensity |
| PBMC | Peripheral blood mononuclear cells |
| PBS | Phosphate-buffered saline |
| PD-1 | Programmed death protein 1 |
| PD-L1 | Programmed death-ligand 1 |
| PE | Phycoerythrin |
| PerCP | Peridinin-chlorophyll-protein |
| PL | Persistent lymphocytosis |
| TIM-3 | T-cell immunoglobulin and mucin domain-3 |
| TNF-α | Tumor necrosis factor alpha |
References
- Zajac, A.J.; Blattman, J.N.; Murali-Krishna, K.; Sourdive, D.J.D.; Suresh, M.; Altman, J.D.; Ahmed, R. Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 1998, 188, 2205–2213. [Google Scholar] [CrossRef]
- Mueller, S.N.; Ahmed, R. High antigen levels are the cause of T cell exhaustion during chronic viral infection. Proc. Natl. Acad. Sci. USA 2009, 106, 8623–8628. [Google Scholar] [CrossRef] [PubMed]
- Wherry, E.J. T cell exhaustion. Nat. Immunol. 2011, 12, 492–499. [Google Scholar] [CrossRef]
- Pauken, K.E.; Wherry, E.J. Overcoming T cell exhaustion in infection and cancer. Trends Immunol. 2015, 36, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Barber, D.L.; Wherry, E.J.; Masopust, D.; Zhu, B.; Allison, J.P.; Sharpe, A.H.; Freeman, G.J.; Ahmed, R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature 2006, 439, 682–687. [Google Scholar] [CrossRef]
- Han, Y.; Liu, D.; Li, L. PD-1/PD-L1 pathway: Current researches in cancer. Am. J. Cancer Res. 2020, 10, 727–742. [Google Scholar]
- Iwai, Y.; Ishida, M.; Tanaka, Y.; Okazaki, T.; Honjo, T.; Minato, N. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA 2002, 99, 12293–12297. [Google Scholar] [CrossRef]
- Shi, F.; Shi, M.; Zeng, Z.; Qi, R.; Liu, Z.; Zhang, J.; Yang, Y.; Tien, P.; Wang, F. PD-1 and PD-L1 upregulation promotes CD8 + T-cell apoptosis and postoperative recurrence in hepatocellular carcinoma patients. Int. J. Cancer 2011, 128, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.T.; Anderson, A.C.; Tan, W.G.; West, E.E.; Ha, S.J.; Araki, K.; Freeman, G.J.; Kuchroo, V.K.; Ahmed, R. Cooperation of Tim-3 and PD-1 in CD8 T-cell exhaustion during chronic viral infection. Proc. Natl. Acad. Sci. USA 2010, 107, 14733–14738. [Google Scholar] [CrossRef]
- Sakuishi, K.; Apetoh, L.; Sullivan, J.M.; Blazar, B.R.; Kuchroo, V.K.; Anderson, A.C. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J. Exp. Med. 2010, 207, 2187–2194. [Google Scholar] [CrossRef]
- Dong, H.; Strome, S.E.; Salomao, D.R.; Tamura, H.; Hirano, F.; Flies, D.B.; Roche, P.C.; Lu, J.; Zhu, G.; Tamada, K.; et al. Tumor-associated B7-H1 promotes T-Cell apoptosis: A potential mechanism of immune evasion. Nat. Med. 2002, 8, 793–800. [Google Scholar] [CrossRef]
- Monney, L.; Sabatos, C.A.; Gaglia, J.L.; Ryu, A.; Waldner, H.; Chernova, T.; Manning, S.; Greenfield, E.A.; Coyle, A.J.; Sobel, R.A.; et al. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature 2002, 415, 536–541. [Google Scholar] [CrossRef]
- Anderson, A.C.; Anderson, D.E.; Bregoli, L.; Hastings, W.D.; Kassam, N.; Lei, C.; Chandwaskar, R.; Karman, J.; Su, E.W.; Hirashima, M.; et al. Promotion of tissue inflammation by the immune receptor Tim-3 expressed on innate immune cells. Science 2007, 318, 1141–1143. [Google Scholar] [CrossRef]
- Ndhlovu, L.C.; Lopez-Vergè, S.; Barbour, J.D.; Jones, R.B.; Jha, A.R.; Long, B.R.; Schoeffler, E.C.; Fujita, T.; Nixon, F.; Lanier, L. Tim-3 marks human natural killer cell maturation and suppresses cell-mediated cytotoxicity. Blood 2012, 119, 3732–3743. [Google Scholar] [CrossRef]
- Phong, B.L.; Avery, L.; Sumpter, T.L.; Gorman, J.V.; Watkins, S.C.; Colgan, J.D.; Kane, L.P. Tim-3 enhances FcεRI-proximal signaling to modulate mast cell activation. J. Exp. Med. 2015, 212, 2289–2304. [Google Scholar] [CrossRef]
- Edwards, S.C.; Hedley, A.; Hoevenaar, W.H.M.; Wiesheu, R.; Glauner, T.; Kilbey, A.; Shaw, R.; Boufea, K.; Batada, N.; Hatano, S.; et al. PD-1 and TIM-3 differentially regulate subsets of mouse IL-17A–producing γδ Tcells. J. Exp. Med. 2023, 220, e20211431. [Google Scholar] [CrossRef]
- Zhu, C.; Anderson, A.C.; Schubart, A.; Xiong, H.; Imitola, J.; Khoury, S.J.; Zheng, X.X.; Strom, T.B.; Kuchroo, V.K. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat. Immunol. 2005, 6, 1245–1252. [Google Scholar] [CrossRef]
- Tang, D.; Lotze, M.T. Tumor immunity times out: TIM-3 and HMGB1. Nat. Immunol. 2012, 13, 808–810. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.H.; Zhu, C.; Kondo, Y.; Anderson, A.C.; Gandhi, A.; Russell, A.; Dougan, S.K.; Petersen, B.S.; Melum, E.; Pertel, T.; et al. CEACAM1 regulates TIM-3-mediated tolerance and exhaustion. Nature 2015, 517, 386–390. [Google Scholar] [CrossRef] [PubMed]
- Curiel, T.J.; Wei, S.; Dong, H.; Alvarez, X.; Cheng, P.; Mottram, P.; Krzysiek, R.; Knutson, K.L.; Daniel, B.; Zimmermann, M.C.; et al. Blockade of B7-H1 improves myeloid dendritic cell–mediated antitumor immunity. Nat. Med. 2003, 9, 562–567. [Google Scholar] [CrossRef] [PubMed]
- Akinleye, A.; Rasool, Z. Immune checkpoint inhibitors of PD-L1 as cancer therapeutics. J. Hematol. Oncol. 2019, 12, 92. [Google Scholar] [CrossRef]
- Larkin, J.; Lao, C.D.; Urba, W.J.; McDermott, D.F.; Horak, C.; Jiang, J.; Wolchok, J.D. Efficacy and safety of nivolumab in patients with BRAF V600 mutant and BRAF wild-type advanced melanoma. JAMA Oncol. 2015, 1, 433. [Google Scholar] [CrossRef]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.-J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017, 18, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Koyama, S.; Akbay, E.A.; Li, Y.Y.; Herter-Sprie, G.S.; Buczkowski, K.A.; Richards, W.G.; Gandhi, L.; Redig, A.J.; Rodig, S.J.; Asahina, H.; et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat. Commun. 2016, 7, 10501. [Google Scholar] [CrossRef]
- Postow, M.A.; Chesney, J.; Pavlick, A.C.; Robert, C.; Grossmann, K.; McDermott, D.; Linette, G.P.; Meyer, N.; Giguere, J.K.; Agarwala, S.S.; et al. Nivolumab and Ipilimumab versus Ipilimumab in untreated melanoma. N. Engl. J. Med. 2015, 372, 2006–2017. [Google Scholar] [CrossRef] [PubMed]
- do Nascimento, A.M.M.; de Souza, C.M.S.; Oliveira, A.C.D.; Blagitz, M.G.; Ramos Sanchez, E.M.; Della Libera, A.M.M.P.; Leite, R.d.M.H.; Fernandes, A.C.d.C.; Souza, F.N. The bovine leukemia virus infection prolongs immunosuppression in dairy cows during the periparturient period by sustaining higher expression of immunological checkpoints in T cells. Vet. Immunol. Immunopathol. 2023, 263, 110636. [Google Scholar] [CrossRef]
- Schwartz, I.; Lévy, D. Pathobiology of bovine leukemia virus. Vet. Res. 1994, 25, 521–536. [Google Scholar]
- Chi, J.; VanLeeuwen, J.A.; Weersink, A.; Keefe, G.P. Direct production losses and treatment costs from bovine viral diarrhoea virus, bovine leukosis virus, Mycobacterium avium subspecies paratuberculosis, and Neospora caninum. Prev. Vet. Med. 2002, 55, 137–153. [Google Scholar] [CrossRef]
- Ott, S.L.; Johnson, R.; Wells, S.J. Association between bovine-leukosis virus seroprevalence and herd-level productivity on US dairy farms. Prev. Vet. Med. 2003, 61, 249–262. [Google Scholar] [CrossRef]
- Blagitz, M.G.; Souza, F.N.; Batista, C.F.; Azevedo, L.F.F.; Sanchez, E.M.R.; Diniz, S.A.; Silva, M.X.; Haddad, J.P.; Della Libera, A.M.M.P. Immunological implications of bovine leukemia virus infection. Res. Vet. Sci. 2017, 114, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Benitez, O.J.; LaDronka, R.M.; Norby, B.; Grooms, D.L.; Bartlett, P.C. The effect of bovine leukemia virus on dairy cow longevity. JDS Commun. 2022, 3, 185–188. [Google Scholar] [CrossRef]
- Lv, G.; Wang, J.; Lian, S.; Wang, H.; Wu, R. The global epidemiology of bovine leukemia virus: Current trends and future implications. Animals 2024, 14, 297. [Google Scholar] [CrossRef]
- Ikebuchi, R.; Konnai, S.; Shirai, T.; Sunden, Y.; Murata, S.; Onuma, M.; Ohashi, K. Increase of cells expressing PD-L1 in bovine leukemia virus infection and enhancement of anti-viral immune responses in vitro via PD-L1 blockade. Vet. Res. 2011, 42, 103. [Google Scholar] [CrossRef]
- Ikebuchi, R.; Konnai, S.; Okagawa, T.; Yokoyama, K.; Nakajima, C.; Suzuki, Y.; Murata, S.; Ohashi, K. Blockade of bovine PD-1 increases T cell function and inhibits bovine leukemia virus expression in B cells in vitro. Vet. Res. 2013, 44, 59. [Google Scholar] [CrossRef]
- Nakamura, H.; Konnai, S.; Okagawa, T.; Maekawa, N.; Sajiki, Y.; Watari, K.; Kamitani, K.; Saito, M.; Kato, Y.; Suzuki, Y.; et al. Combined immune checkpoint blockade enhances antiviral immunity against bovine leukemia virus. J. Virol. 2023, 97, e0143022. [Google Scholar] [CrossRef]
- Begg, D.J.; O’Brien, R.; Mackintosh, C.G.; Griffin, J.F.T. Experimental infection model for Johne’s disease in sheep. Infect. Immun. 2005, 73, 5603–5611. [Google Scholar] [CrossRef] [PubMed]
- Balseiro, A.; Altuzarra, R.; Vidal, E.; Moll, X.; Espada, Y.; Sevilla, I.A.; Domingo, M.; Garrido, J.M.; Juste, R.A.; Prieto, M.; et al. Assessment of BCG and inactivated Mycobacterium bovis vaccines in an experimental tuberculosis infection model in sheep. PLoS ONE 2017, 12, e0180546. [Google Scholar] [CrossRef] [PubMed]
- Wilson-Welder, J.H.; Nally, J.E.; Alt, D.P.; Palmer, M.V.; Coatney, J.; Plummer, P. Experimental transmission of bovine digital dermatitis to sheep: Development of an infection model. Vet. Pathol. 2018, 55, 245–257. [Google Scholar] [CrossRef]
- Porta, N.G.; Alvarez, I.; Suarez Archilla, G.; Ruiz, V.; Abdala, A.; Trono, K. Experimental infection of sheep with bovine leukemia virus (BLV): Minimum dose of BLV-FLK cells and cell-free BLV and neutralization activity of natural antibodies. Rev. Argent. Microbiol. 2019, 51, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Tiyamanee, W.; Konnai, S.; Okagawa, T.; Nojima, Y.; Ganbaatar, O.; Maekawa, N.; Hasebe, R.; Kagawa, Y.; Kato, Y.; Suzuki, Y.; et al. Molecular characterization of immunoinhibitory factors PD-1/PD-L1 in sheep. Vet. Immunol. Immunopathol. 2023, 261, 110609. [Google Scholar] [CrossRef]
- Kilkenny, C.; Browne, W.J.; Cuthill, I.C.; Emerson, M.; Altman, D.G. Improving bioscience research reporting: The arrive guidelines for reporting animal research. PLoS Biol. 2010, 8, e1000412. [Google Scholar] [CrossRef]
- Sajiki, Y.; Konnai, S.; Okagawa, T.; Nishimori, A.; Maekawa, N.; Goto, S.; Watari, K.; Minato, E.; Kobayashi, A.; Kohara, J.; et al. Prostaglandin E2–induced immune exhaustion and enhancement of antiviral effects by anti–PD-L1 antibody combined with COX-2 inhibitor in bovine leukemia virus infection. J. Immunol. 2019, 203, 1313–1324. [Google Scholar] [CrossRef]
- Nakamura, H.; Konnai, S.; Okagawa, T.; Maekawa, N.; Tiyamanee, W.; Ikehata, M.; Matsubara, K.; Watari, K.; Kamitani, K.; Saito, M.; et al. Comprehensive analysis of immune checkpoint molecules profiles phenotype and function of exhausted T cells in enzootic bovine leukosis. J. Immunol. 2025, in press. [Google Scholar] [CrossRef]
- Ikebuchi, R.; Konnai, S.; Okagawa, T.; Yokoyama, K.; Nakajima, C.; Suzuki, Y.; Murata, S.; Ohashi, K. Influence of PD-L1 cross-linking on cell death in PD-L1-expressing cell lines and bovine lymphocytes. Immunology 2014, 142, 551–561. [Google Scholar] [CrossRef]
- Corsale, A.M.; Shekarkar Azgomi, M.; Plano, F.; Di Simone, M.; Perez, C.; Picone, C.; Gigliotta, E.; Vullo, C.; Camarda, G.; Rotolo, C.; et al. High TIM-3 expression may contribute to the functional impairment of bone marrow and circulating gamma delta T lymphocytes during the progression of multiple myeloma. Blood 2022, 140 (Suppl. S1), 9943–9944. [Google Scholar] [CrossRef]
- Yu, L.; Wang, Z.; Hu, Y.; Wang, Y.; Lu, N.; Zhang, C. Tumor-infiltrating gamma delta T-cells reveal exhausted subsets with remarkable heterogeneity in colorectal cancer. Int. J. Cancer 2023, 153, 1684–1697. [Google Scholar] [CrossRef] [PubMed]
- O’malley, D.M.; Neffa, M.; Monk, B.J.; Melkadze, T.; Huang, M.; Kryzhanivska, A.; Bulat, I.; Meniawy, T.M.; Bagameri, A.; Wang, E.W.; et al. Dual PD-1 and CTLA-4 checkpoint blockade using Balstilimab and Zalifrelimab combination as second-line treatment for advanced cervical cancer: An open-label phase II study. J. Clin. Oncol. 2021, 40, 762–771. [Google Scholar] [CrossRef]
- Livingstone, E.; Zimmer, L.; Hassel, J.C.; Fluck, M.; Eigentler, T.K.; Loquai, C.; Haferkamp, S.; Gutzmer, R.; Meier, F.; Mohr, P.; et al. Adjuvant Nivolumab plus Ipilimumab or Nivolumab alone versus placebo in patients with resected stage IV melanoma with no evidence of disease (IMMUNED): Final results of a randomised, double-blind, phase 2 trial. Lancet 2022, 400, 1117–1129. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, N.; Konnai, S.; Hosoya, K.; Kim, S.; Kinoshita, R.; Deguchi, T.; Owaki, R.; Tachibana, Y.; Yokokawa, M.; Takeuchi, H.; et al. Safety and clinical efficacy of an anti-PD-L1 antibody (C4G12) in dogs with advanced malignant tumours. PLoS ONE 2023, 18, e0291727. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.B.; Ndhlovu, L.C.; Barbour, J.D.; Sheth, P.M.; Jha, A.R.; Long, B.R.; Wong, J.C.; Satkunarajah, M.; Schweneker, M.; Chapman, J.M.; et al. Tim-3 expression defines a novel population of dysfunctional T cells with highly elevated frequencies in progressive HIV-1 infection. J. Exp. Med. 2008, 205, 2763–2779. [Google Scholar] [CrossRef]
- Sada-Ovalle, I.; Ocaña-Guzman, R.; Pérez-Patrigeón, S.; Chávez-Galán, L.; Sierra-Madero, J.; Torre-Bouscoulet, L.; Addo, M.M. Tim-3 blocking rescue macrophage and T cell function against Mycobacterium tuberculosis infection in HIV+ patients. J. Int. AIDS Soc. 2015, 18, 20078. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xu, C.; Yang, F.; Zong, L.; Qin, Y.; Gao, Y.; Su, Q.; Li, T.; Li, Y.; Xu, Y.; et al. Natural liller cells induce CD8+ T cell dysfunction via galectin-9/TIM-3 in chronic hepatitis B virus infection. Front. Immunol. 2022, 13, 884290. [Google Scholar] [CrossRef]
- Diupotex, M.; Zamora-Chimal, J.; Gajón, J.A.; Bonifaz, L.C.; Becker, I. CXCR5 and TIM-3 expressions define distinct exhausted T cell subsets in experimental cutaneous infection with Leishmania mexicana. Front. Immunol. 2023, 14, 1231836. [Google Scholar] [CrossRef]
- Banstola, A.; Reynolds, J.N.J. The sheep as a large animal model for the investigation and treatment of human disorders. Biology 2022, 11, 1251. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tiyamanee, W.; Okagawa, T.; Yamada, S.; Ikehata, M.; Nakamura, H.; Inoue, M.; Maekawa, N.; Kato, Y.; Murata, S.; Ohashi, K.; et al. Evaluation of PD-L1 and TIM-3 Pathways in T Cells During Experimental Bovine Leukemia Virus Infection in Sheep. Vet. Sci. 2025, 12, 810. https://doi.org/10.3390/vetsci12090810
Tiyamanee W, Okagawa T, Yamada S, Ikehata M, Nakamura H, Inoue M, Maekawa N, Kato Y, Murata S, Ohashi K, et al. Evaluation of PD-L1 and TIM-3 Pathways in T Cells During Experimental Bovine Leukemia Virus Infection in Sheep. Veterinary Sciences. 2025; 12(9):810. https://doi.org/10.3390/vetsci12090810
Chicago/Turabian StyleTiyamanee, Wisa, Tomohiro Okagawa, Shinji Yamada, Mari Ikehata, Hayato Nakamura, Maho Inoue, Naoya Maekawa, Yukinari Kato, Shiro Murata, Kazuhiko Ohashi, and et al. 2025. "Evaluation of PD-L1 and TIM-3 Pathways in T Cells During Experimental Bovine Leukemia Virus Infection in Sheep" Veterinary Sciences 12, no. 9: 810. https://doi.org/10.3390/vetsci12090810
APA StyleTiyamanee, W., Okagawa, T., Yamada, S., Ikehata, M., Nakamura, H., Inoue, M., Maekawa, N., Kato, Y., Murata, S., Ohashi, K., Murakami, K., & Konnai, S. (2025). Evaluation of PD-L1 and TIM-3 Pathways in T Cells During Experimental Bovine Leukemia Virus Infection in Sheep. Veterinary Sciences, 12(9), 810. https://doi.org/10.3390/vetsci12090810






