Genomic Adaptation, Environmental Challenges, and Sustainable Yak Husbandry in High-Altitude Pastoral Systems
Simple Summary
Abstract
1. Introduction
2. Genomic Insights into Hypoxia Tolerance in Yaks
3. Genetic Resources of Yaks
4. Available Breeds of Yaks
5. Genetic and Physiological Foundation of Production Traits of High-Altitude Adaptation in Yaks
6. Interspecies Hybridization Between Yaks and Cattle
7. Microbial Threats to Yak Health
8. Climate Change and Effects on Yak Productivity
| Country | Breeds and no of Individuals | Parameter | Gene | Associated Primers | Annealing Temperature | Base Pairs | Reference |
|---|---|---|---|---|---|---|---|
| China |
| Hemoglobin concentration | EPAS1 | EPAS1-S CGTGGTGACCCAAGATGGTG EPAS1-AGGTCACAGGGATGAGTGAAGTCAA GAPDH-S CCACGAGAAGTATAACAACACC GAPDH-A GTCATAAGTCCCTCCACGAT | 60 60 | 573–691 422–542 | [65] |
| China |
| Spermatogenic arrest | Dmrt7 | F: 5′-CCTCCAGATTGACTCTTAACTC-3′ R: 5′-GGACCCAAGGAAGGTAAGA-3′ | 64 | 1113 | [71] |
| China | Cattle–yak/350 | Milk fat | SORBS1 | F: CACTTGCTCTCCCCTTCCTG R: CAACGTTCAGCCTCTGGACT F: ATGCCCTGTGCTGTCAACTT R: TACAGTGGTCGCTGCCATAC F: GGACAGGAGAGTTCTGTGGC R: AAGGACAGAGCTGCTGGAAC F: AGAGTGCCTCACTGCATGTC R: ACAGACTGGTGAACAGCCAC F: ACCGGATTGAGCCACAGTTT R: GGCACCAAGATTTTCCCAGC F: ACTGAGGTCTCTCAGCCAGT R: TACAGTGGTCGCTGCCATAC F: CTGTCTGACCCTGCTCTGTG R: GCCGGTGAGAAACTCAGGAA F: TGCCATCTCCTCCCTACACA R: GTCCACACCATGGCCACTAA F: CCAAGATGAGCACGGAAGGT R: GGGATTGTGGTGGTACCCAG F: TCTCCAGACATCCCGTGTGA R: GGTCTTGTGGGCATCCACTT F: GTTGAACGGATCTCCCCCAA R: GCAACTGGAAACTGCCCTTC F: AAGCCCCTAACCTTGGTGTG R: AGAGCACGTGCAGGCTAAAT | 62 60 63 59 60 59 61 61 61 61 63 61 | 5791–6750 7741–8740 18,181–19,180 24,361–25,360 28,681–29,680 43,111–44,110 79,641–80,640 85,681–86,680 94,081–95,080 96,061–97,060 112,141–113,140 114,001–115,000 | [70] |
| Pakistan |
|
|
| F: TTGATCAGGTGGCAGAGAAC R: CAAGGCTGATGACCATCAAC F: CGAGCTGTTCCAGTACTTTC R: TGCTTGGGATTGTGATCCTG F: GTCCCAAGACAAGTATCAGG R: TAAGACTGCGCATGTGCTTG F: GCTATGGACAGACATACTGG R: ACAAGTGATGGACTCTCCAG F: TTGAGCTACAACGTTGTCCC R: GTAGGGCATATGCTGTGTAG F: TAGAGGAGGACAGAATCCAG R: CTCAGATTAAACACAGAAAAC F: CTGTGTTCC ATC GACGAAAC R: CTCATGGGTGCACTTCATAC F: ACCCCTGCATACTAACCTAC R: TCAGAGCACCTGGTAAATCG F: ACATCTTCAAAATGCGGCCC R: ATGGCCCATCGCTTTTTGTG F: CTGTCTTCTCTCCCAGAA R: GGACACACAGCAGAAACAAG F: TGGACAAACAGCAGTGCAGG R: TCCTTCCTGAAAGCAACACC F: GGCAGAGTTCATGTATCTCG R: TTG AAGCCATGCACCTTGAG | 52 | [67] | |
| Gansu, China | Gannan yak |
| FASN gene | F: CTGTCACCTTCCTCACTTGCCCT R: GAGGAGGAATCGGCCAGGATGTT F: CCCTCTAAAGCCGTCCTCACCA R: CCAGACCTTCATTTGCCAATCCTC F: ACAAGACAAGCCCGAGGAG R: TAGCAGGCAGTTCCGAGAG | 72 | 390 220 203 | [66] |
| China | Bos grunniens/81 | Fat content | DGAT1 K232A | F: 5′-GGCGGGGTGCGAACTAAG-3′ R: 5′-GCACAGCACTTTATTGACACATTC-3′. | 55 | 1760 | [68] |
| China | Yak (Bos grunniens)
|
| MSTN and CAST | MSTN: F: AAAGAGGGGCTGTGTAATGC R: ATGGTAATGACCGTTTCCGT CAST: F: CGTGCCTCGGACCTCTAT R: CGTCTTTATCCTTGGCTTCT | 52~54 | 260 254 | [72] |
| China | Bos Grunniens/387 | Growth | KLF6 | F: ATGCTCATGGGAAGGGTGTG R: CTTGGCACCAGTGTGCTTTC | 55–60 | 82 | [69] |
| China | Bos grunniens | MyHC I and MyHC IIB expression | ACTB | F: ATTGCCGATGGTGATGAC R: ACGGAGCGTGGCTACAG | 60 | 177 | [75] |
| GAPDH | F: TCACCAGGGCTGCTTTTA R: CTGTGCCGTTGAACTTGC | 126 | |||||
| UXT | F: AGGTGGATTTGGGCTGTAAC R: CTTGGTGAGGTTGTCGCTGA | 170 | |||||
| TBP | F: GTCCAATGATGCCTTACGG R: TGCTGCTCCTCCAGAATAGA | 82 | |||||
| YWHAZ | F: AATGTTGTAGGAGCCCGTAG R: CTGCTTGTGAAGCGTTGG | 190 | |||||
| RPL13A | F: CAAGCGGATGAACACCAA R: GCAGCAGGAACCACCATT | 192 | |||||
| SDHA | F: GGGAACATGGAGGAGGACA R: CCAAAGGCACGCTGGTAGA | 188 | |||||
| RPS15 | F: GACCTTCCGCAAGTTCACCT R: ACCACCTCGGGCTTCTCCAT | 198 | |||||
| HPRT1 | F: GTGATGAAGGAGATGGG R: ACAGGTCGGCAAAGAAC | 79 | |||||
| PPIA | F: TTTTGAAGCATACAGGTCC R: CCACTCAGTCTTGGCAGT | 98 | |||||
| HMBS | F: GAACAAAGGAGCCAAGAAC R: CAGAGGGCTGGGATGTAG | 121 | |||||
| MRPL39 | F: AAACCTTTGACCAAGTCCTGT R: TTCCTCTTTGAATGCCCTCTC | 135 | |||||
| PPP1R11 | F: CAGAAAAGACAGAAGGGTGC R: TTCCGAAGTTTGATGGTTAG | 164 | |||||
| B2M | F: CTGAGGAATGGGGAGAAG R: TGGGACAGCAGGTAGAAA | 80 | |||||
| China | Datong yak/55 |
| TLR2 | F: GGACAATGCCACGTGCTT R: GCACTGATCTCAAGCTCCTCAAG F: TGAGGAGCTTGAGATCAGTG R: ACTGTGTATCCTTGTGCTGG F: CCTAGGTAATGTGGAGACG R: AAGGAGGCATCTGGTAGAG F: CCAGCACAAGGATACACAGT R: CTTCATGTACCACAGTCCGT F: TTCCTGTTGCTCCTGCTCAC R: GACCACCACCAGACCAAGAC | 58 58 58 58 58 | 552 822 574 526 599 | [77] |
| China | Ashidan yaks/335 |
| AHR | F: TCATACCGGGCTCTTTGCAG R: GTACCCTGAACACCCGAAGG BTF3 F: AACCAGGAGAAACTCGCCAA R: TTCGGTGAAATGCCCTCTCG F: CACCCGTCTTCACCCATCAG R: TGCCTCCATGTGAACTTGCT F: CTTCCTGGGCATGGAATCCTG R: CAGCACCGTGTTGGCGTAG | 58 58 54 54 | 223 166 164 103 | [78] |
| China | Ashidan yaks/274 |
| HSF1 | F: TCCGGAGGTGGTCCACAT R: GAACTCGGTGTCATCCCTCTCT F: CCATCATCTCCGACATCACC R: CTCCTCCTTTACGCGAACC F: ATTGCCGATGGTGATGAC R: ACGGAGCGTGGCTACAG | 58 63.3 55 | 290 113 177 | [80] |
| China |
|
| CHKB | F: GCAGTCTCGGTTCCAGTTCT R: AATGCAAGGAGTCGGAGGTG F: AACCAGGAGAAACTCGCCAA R: TTCGGTGAAATGCCCTCTCG F: AGCTAATCGGTATGCCCTGG R: AACTGGAACCGAGACTGCG F: ATGAAAGGGCCATCACCATC R: GTGGTTCACGCCCATCACA | 60.57 59.32 55.40 57.45 60.18 60.11 55.85 60.00 | 90 166 118 204 | [81] |
| China | Ashidan yaks/336 |
| HPGDS | F: ATCCGGGCACTGTTAGAAGG R: GCCTGCAAAGTCTGTACTGT F: AACCAGGAGAAACTCGCCAA R: TTCGGTGAAATGCCCTCTCG F: ACCTGCCCATTTCTATCCTGAC R: ACTGTTTCTTAGCCCATCGCAT F: AATGAAAGGGCCATCACCATC R: GTGGTTCACGCCCATCACA | 170 166 187 204 | [79] | |
| China | Ashidan yaks/350 |
| CADM2 | F: GACTTCCCAGGATTGCCTGT R: CCCTGGGAGCACAGTTGTTT F: GGCTGTCACGTTCTTCTCTCA R: AGGGTTCATCCTGGAGGCTT F: AACCAGGAGAAACTCGCCAA R: TTCGGTGAAATGCCCTCTCG | 62 | 186 196 166 | [82] |
| China | Ashidan yaks/311 | Body weight; Withers height; Body length; Chest girth. | SOX6 | F: GCAACTACCACACCGTCACCTC R: TCCGCCGTCTGTCTTCATACCA F: AACCAGGAGAAACTCGCCAA R: TTCGGTGAAATGCCCTCTCG F: CGTTTGGGCAGGAGTTTGGA R: CGTTTGGTGGCTGTGGAGTT F: GCAGGTCATCACCATCGG R: CCGTGTTGGCGTAGAGGT | 59 59 60 60 | 114 166 148 158 | [97] |
| China | Ashidan yaks/315 | Body weight; Withers height; Body length; Chest girth. | MICALL2 MOGAT2 | F: CCGTCGTCTAATGCCAGTGA R: CATCTTTCCGCTGGACGGTA F: CGCTGGTCAAGACTGCCTAT R: ACAGTGAGGAAAACCCGGTG F: AACCAGGAGAAACTCGCCAA R: TTCGGTGAAATGCCCTCTCG F: CCTCATGGTGGACTGGTTCC R: CAATGATGTCGCTTCGGCTG F: CGCTGGTCAAGACTGCCTAT R: CATCATCAGATGTGGGCGGA F: AATGAAAGGGCCATCACCATC R: GTGGTTCACGCCCATCACA | 58.0 60.0 60.0 59.9 59.8 58.8 | 133 126 166 239 155 204 | [98] |
| China | Ashidan yaks/326 | Body weight; Withers height; Body length; Chest girth. |
| F: AACCAGGAGAAACTCGCCAA R: TTCGGTGAAATGCCCTCTCG F: GCTTCCCAGTTCGCTTAG R: TTTCTGCCTTGGATGCTC F: CCTTGGGTCACTCGGGTTG R: GCGGCTCGGATTCTTTCG F: CCACGAGAAGTATAACAACACC R: GTCATAAGTCCCTCCACGAT F: AAGAAACTGGCTGCGTCTCA R: TAATGGCGGCAGTTGACCTT F: CCGCACATCAAGACGGAGCA R: TGACGGGTAGCCAGGGAACG | 63 55.6 63 56.1 56.1 64 | 166 104 141 120 168 213 | [85] |
| China | Yaks/354 | Body weight; Average daily gain. | MC4R | F: 5′-TGGGA CATTTATTCACAGCAG-3′ R: 5′CCTACACAG AAGAAAAAGCT-3′ | 55 | 1238 | [86] |
| Genes | Functions of Genes Associated with Environmental Stress | References |
|---|---|---|
| MMP3 | As a master regulator of the cellular response to hypoxia, hypoxia-inducible factor-1α is thought to have matrix metalloproteinases-3 (MMP3) as one of its primary target genes. | [99,100] |
| ATP8; ATP6. |
| [101] |
| HIF-1 | Because it regulates the localized tissue hypoxia that takes place in these settings, hypoxia-inducible factor 1 (HIF-1) has been found to have a significant role in the pathophysiology of tumor vascularization, myocardial ischemia, and stroke. | [102] |
| AQP4 | The production of brain edema is one of the several physiopathological processes in which aquaporin-4 (AQP4) is implicated. Additionally, it controls calcium signaling, waste removal, potassium buffering, and extracellular space volume. | [103] |
| EPAS1 | One essential transcription factor that controls the expression of genes involved in oxygen sensing is the endothelial PAS domain protein 1 gene (EPAS1). | [65] |
| VEGF-A | The vascular endothelial growth factor-A gene (VEGF-A), a crucial regulator of angiogenesis and endothelial cell mitogen, plays a major role in adaptation to high altitudes. | [104] |
| HIF-1α | HIF-1α is the oxygen-regulated subunit of HIF-1 that controls the transcription of genes related to oxygen homeostasis in response to hypoxia. | [105] |
| LDH | An essential component of anaerobic metabolism, lactate dehydrogenase (LDH) catalyzes the transformation of pyruvate to lactate during glycolysis in mammals. | [106] |
| HIFs | Oxygen-dependent transcriptional activators known as hypoxia-inducible factors (HIFs) are critical for mammalian development and tumor angiogenesis. In reaction to hypoxia, they control the transcription of genes related to oxygen homeostasis. | [107] |
| The primary roles of ALDH4A1, ALDH2, and ECI1 in bioenergy metabolism under hypoxic settings suggest that they might be engaged in hypoxia adaptation processes. | [108] |
| The crucial involvement of collagen-related pathways in high-altitude adaptation is highlighted by five collagen genes: COL1A2, COL3A1, COL5A2, COL14A1, and COL15A1. | [109] |
| In the heart-related modules of yaks, UQCRC1 and COX5A are frequently found to be differentially expressed hub genes linked to the energy source for cardiac contraction. The lung-related module also contains the common differential hub gene CAPS, which is connected to the contraction of the smooth muscle in the pulmonary arteries. Furthermore, the heart of yaks contains a unique hub gene called CHRM2, which is differentially expressed and essential for the independent control of cardiac function. | [110] |
| It is suggested that MAPKAPK3, ATP7A, PXN, NFATC2, DIAPH1, and F2R are new and intriguing options for controlling hypoxia adaptation in the heart. | [111] |
| Two mitochondrial genes, MT-ND1 and MT-ND2, encode subunits of NADH dehydrogenase, which are associated with high-altitude adaptation and are necessary for the electron transport chain in oxidative phosphorylation (OXPHOS). | [112] |
| CSF2 | By regulating the production of the heat shock protein 70 kDa 1A, colony-stimulating factor 2 (CSF2) is known to support the growth and survival of preimplantation embryos in rats and ruminants. | [113] |
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wiener, G.; Lin, H.J.; Jun, L.R. Origins, domestication and distribution of yak. In The Yak, 2nd ed.; Gerald, W., Han, J., Long, R., Eds.; FAO (Food and Agricultural Organization of the United Nations): Rome, Italy, 2003. [Google Scholar]
- Younas, M. Phenotypic, Productive, and Reproductive Traits of Domestic Yak (Bos grunniens grunniens) in District Chitral, Pakistan. J. Agric. Biol. 2025, 3, 344–349. [Google Scholar] [CrossRef]
- Khan, B.; Abbas, S.; Khan, Z.; Khan, G.; Anjum, M.; Baig, S.; Jamal, S. Indigenous practices of yak breeding in Gilgit-Baltistan: Current status and future prospects for transboundary yak husbandry in the Karakoram-Pamir mountain area. In Yak on the Move, Transboundary Challenges and Opportunities for Yak Raising in a Changing Hindu Kush Himalayan Region; Wu, N., Yi, S., Srijana, J., Neha, B., Eds.; ICIMOD: Kathmandu, Nepal, 2016; pp. 167–178. [Google Scholar]
- Jasra, A.; Hashmi, M.; Waqar, K.; Ali, M. Traditional yak herding in high-altitude areas of Gilgit-Baltistan, Pakistan: Transboundary and biodiversity conservation challenges. In Yak on the Move, Transboundary Challenges and Opportunities for Yak Raising in a Changing Hindu Kush Himalayan Region; Wu, N., Yi, S., Srijana, J., Neha, B., Eds.; ICIMOD: Kathmandu, Nepal, 2016. [Google Scholar]
- Joshi, S.; Shrestha, L.; Bisht, N.; Wu, N.; Ismail, M.; Dorji, T.; Dangol, G.; Long, R. Ethnic and Cultural Diversity amongst Yak Herding Communities in the Asian Highlands. Sustainability 2020, 12, 957. [Google Scholar] [CrossRef]
- Jing, X.; Ding, L.; Zhou, J.; Huang, X.; Degen, A.; Long, R. The adaptive strategies of yaks to live in the Asian highlands. Anim. Nutr. 2022, 9, 249–258. [Google Scholar] [CrossRef]
- Liu, X.; Liu, W.; Lenstra, J.A.; Zheng, Z.; Wu, X.; Yang, J.; Li, B.; Yang, Y.; Qiu, Q.; Liu, H. Evolutionary origin of genomic structural variations in domestic yaks. Nat. Commun. 2023, 14, 5617. [Google Scholar] [CrossRef]
- Qiu, Q.; Wang, L.; Wang, K.; Yang, Y.; Ma, T.; Wang, Z.; Zhang, X.; Ni, Z.; Hou, F.; Long, R.; et al. Yak whole-genome resequencing reveals domestication signatures and prehistoric population expansions. Nat. Commun. 2015, 6, 10283. [Google Scholar] [CrossRef]
- Kalwar, Q.; Ma, X.; Xi, B.; Korejo, R.A.; Bhuptani, D.K.; Chu, M.; Yan, P. Yak milk and its health benefits: A comprehensive review. Front. Vet. Sci. 2023, 10, 2023. [Google Scholar] [CrossRef] [PubMed]
- Shah, A.M.; Bano, I.; Qazi, I.H.; Matra, M.; Wanapat, M. “The Yak”—A remarkable animal living in a harsh environment: An overview of its feeding, growth, production performance, and contribution to food security. Front. Vet. Sci. 2023, 10, 1086985. [Google Scholar] [CrossRef] [PubMed]
- Ning, W. Social, cultural and economic context of yak production. In The Yak, 2nd ed; FAO: Rome, Italy, 2003. [Google Scholar]
- Acharya, K.P.; Phuyal, S.; Saied, A.A. A one health approach to improve the safety of traditional yak blood drinking in Nepal. Commun. Med. 2025, 5, 84. [Google Scholar] [CrossRef]
- He, L.; Li, X.; Liu, G.; Cidan, Y.-J.; Ullah, Q.; Mushtaq, R.; Xiu, Y.; Sun, G.; Luosang, D.; Jiang, N. Yak’s Resilience and Adaptation to High-Altitude Stress. Pak. J. Zool. 2024, 1–13. [Google Scholar] [CrossRef]
- Ayalew, W.; Chu, M.; Liang, C.; Wu, X.; Yan, P. Adaptation Mechanisms of Yak (Bos grunniens) to High-Altitude Environmental Stress. Animals 2021, 11, 2344. [Google Scholar] [CrossRef]
- Mishra, K.P.; Ganju, L. Influence of High Altitude Exposure on the Immune System: A Review. Immunol. Investig. 2010, 39, 219–234. [Google Scholar] [CrossRef]
- Brent, M.B. A review of the skeletal effects of exposure to high altitude and potential mechanisms for hypobaric hypoxia-induced bone loss. Bone 2022, 154, 116258. [Google Scholar] [CrossRef]
- Ibrahim, E.; Sohail, S.K.; Ihunwo, A.; Eid, R.A.; Al-Shahrani, Y.; Rezigalla, A.A. Effect of high-altitude hypoxia on function and cytoarchitecture of rats’ liver. Sci. Rep. 2025, 15, 12771. [Google Scholar] [CrossRef]
- Tong, W.; Allison, B.J.; Brain, K.L.; Patey, O.V.; Niu, Y.; Botting, K.J.; Ford, S.G.; Garrud, T.A.; Wooding, P.F.B.; Shaw, C.J.; et al. Chronic Hypoxia in Ovine Pregnancy Recapitulates Physiological and Molecular Markers of Preeclampsia in the Mother, Placenta, and Offspring. Hypertension 2022, 79, 1525–1535. [Google Scholar] [CrossRef]
- Parraguez, V.H.; Urquieta, B.; Pérez, L.; Castellaro, G.; De los Reyes, M.; Torres-Rovira, L.; Aguado-Martínez, A.; Astiz, S.; González-Bulnes, A. Fertility in a high-altitude environment is compromised by luteal dysfunction: The relative roles of hypoxia and oxidative stress. Reprod. Biol. Endocrinol. 2013, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Sales, F.; Peralta, O.A.; Narbona, E.; McCoard, S.; De Los Reyes, M.; González-Bulnes, A.; Parraguez, V.H. Hypoxia and Oxidative Stress Are Associated with Reduced Fetal Growth in Twin and Undernourished Sheep Pregnancies. Animals 2018, 8, 217. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Li, S.; He, Z.; Ma, X. Physiological and Genetic Basis of High-Altitude Indigenous Animals’ Adaptation to Hypoxic Environments. Animals 2024, 14, 3031. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Long, K.; Ma, J.; Zhang, J.; He, D.; Jin, L.; Tang, Q.; Jiang, A.; Wang, X.; Hu, Y. Comparative analysis of the microRNA transcriptome between yak and cattle provides insight into high-altitude adaptation. PeerJ 2017, 5, e3959. [Google Scholar] [CrossRef]
- Durmowicz, A.G.; Hofmeister, S.; Kadyraliev, T.; Aldashev, A.A.; Stenmark, K.R. Functional and structural adaptation of the yak pulmonary circulation to residence at high altitude. J. Appl. Physiol. 1993, 74, 2276–2285. [Google Scholar] [CrossRef]
- Gera, R.; Mishra, A.K.; Arora, R.; Parsad, R.; Bagiyal, M.; Chhabra, P.; Sharma, U.; Ahlawat, S.; Sharma, R.; Kumar, R. High Altitude Adaptation in Livestock and Poultry: Exploring Morphological, Physiological, Biochemical and Omic Insights: A Review. Indian J. Anim. Res. 2025, 59, 357–364. [Google Scholar] [CrossRef]
- Das, P.J.; Bam, J.; Paul, V.; Medhi, D.; Roy, A.N.; Deb, S.M. The Yak Wool; ICAR-National Research Centre on Yak: Dirang, India, 2018; pp. 1–108. [Google Scholar]
- Shao, B.; Long, R.; Ding, Y.; Wang, J.; Ding, L.; Wang, H. Morphological adaptations of yak (Bos grunniens) tongue to the foraging environment of the Qinghai-Tibetan Plateau. J. Anim. Sci. 2010, 88, 2594–2603. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Zhao, F.; Hu, J.; Zhao, Z.; Shi, B.; Li, S. A conjoint analysis of renal structure and omics characteristics reveal new insight to yak high-altitude hypoxia adaptation. Genomics 2024, 116, 110857. [Google Scholar] [CrossRef]
- Beall, C.M.; Cavalleri, G.L.; Deng, L.; Elston, R.C.; Gao, Y.; Knight, J.; Li, C.; Li, J.C.; Liang, Y.; McCormack, M.; et al. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. USA 2010, 107, 11459–11464. [Google Scholar] [CrossRef]
- Hanaoka, M.; Droma, Y.; Basnyat, B.; Ito, M.; Kobayashi, N.; Katsuyama, Y.; Kubo, K.; Ota, M. Genetic Variants in EPAS1 Contribute to Adaptation to High-Altitude Hypoxia in Sherpas. PLoS ONE 2012, 7, e50566. [Google Scholar] [CrossRef]
- Heath, D.; Williams, D.; Dickinson, J. The pulmonary arteries of the yak. Cardiovasc. Res. 1984, 18, 133–139. [Google Scholar] [CrossRef]
- Ding, X.Z.; Liang, C.N.; Guo, X.; Wu, X.Y.; Wang, H.B.; Johnson, K.A.; Yan, P. Physiological insight into the high-altitude adaptations in domesticated yaks (Bos grunniens) along the Qinghai-Tibetan Plateau altitudinal gradient. Livest. Sci. 2014, 162, 233–239. [Google Scholar] [CrossRef]
- Kim, J.J.; Park, H.-M.; Kyoung, A.Y.; Lim, S.-K.; Lee, J.E.; Park, B.C. Redefining copy number variation and single-nucleotide polymorphism counting via novel concepts based on recent PCR enhancements. Biochem. Biophys. Res. Commun. 2024, 740, 150988. [Google Scholar] [CrossRef]
- Zhang, M.; Huang, C.; Bao, Q.; Wang, T.; Ma, X.; Meng, G.; Zhou, X.; Zheng, Q.; Chu, M.; Liang, C.; et al. Whole-genome resequencing revealed the genetic diversity and body weight-related genes of Gannan yak. Front. Vet. Sci. 2025, 12, 2025. [Google Scholar] [CrossRef]
- Yu, X.; Li, C.; Zhao, Z.; Zhang, Y. Effects of Slaughter Age on the Quality of Gannan Yak Meat: Analysis of Edible Quality, Nutritional Value, and GC × GC-ToF-MS of the Longissimus Dorsi Muscle. Food Sci. Nutr. 2025, 13, e70381. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Guo, S.; Wang, X.; Cao, M.; Pei, J.; Li, R.; Bao, P.; Wang, J.; Lamao, J.; Gongbao, D.; et al. Whole-Genome Resequencing Highlights the Unique Characteristics of Kecai Yaks. Animals 2022, 12, 2682. [Google Scholar] [CrossRef] [PubMed]
- Grant, P.R.; Grant, B.R.; Huey, R.B.; Johnson, M.T.J.; Knoll, A.H.; Schmitt, J. Evolution caused by extreme events. Philos. Trans. R. Soc. B: Biol. Sci. 2017, 372, 20160146. [Google Scholar] [CrossRef]
- Ahmad, H.I.; Mahmood, S.; Hassan, M.; Sajid, M.; Ahmed, I.; Shokrollahi, B.; Shahzad, A.H.; Abbas, S.; Raza, S.; Khan, K.; et al. Genomic insights into Yak (Bos grunniens) adaptations for nutrient assimilation in high-altitudes. Sci. Rep. 2024, 14, 5650. [Google Scholar] [CrossRef]
- Cai, B.; Wu, X.; Shi, Y.; Kang, Y.; Ding, Z.; Guo, S.; Cao, M.; Hu, L.; Zhang, B.; Wang, X.; et al. Whole-Genome Sequencing Unveils the Uniqueness of Yushu Yaks (Bos grunniens). Int. J. Mol. Sci. 2025, 26, 3879. [Google Scholar] [CrossRef] [PubMed]
- Ørsted, M.; Hoffmann, A.A.; Sverrisdóttir, E.; Nielsen, K.L.; Kristensen, T.N. Genomic variation predicts adaptive evolutionary responses better than population bottleneck history. PLoS Genet. 2019, 15, e1008205. [Google Scholar] [CrossRef] [PubMed]
- Barghi, N.; Schlötterer, C. Distinct Patterns of Selective Sweep and Polygenic Adaptation in Evolve and Resequence Studies. Genome Biol. Evol. 2020, 12, 890–904. [Google Scholar] [CrossRef]
- Mahar, K.; Gurao, A.; Kumar, A.; Chitkara, M.; Gowane, G.R.; Ahlawat, S.; Niranjan, S.K.; Pundir, R.K.; Arora, R.; Kataria, R.S.; et al. Genomic insights into high-altitude adaptation and evolutionary dynamics of Indian yaks in the Trans-Himalayan region. Conserv. Genet. 2025, 26, 49–62. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, N.a.; Cheng, H.; Wang, Y.; Liu, X.; Qiao, B.; Zhao, L. Mapping conservation priorities for wild yak (Bos mutus) habitats on the Tibetan Plateau, China. Sci. Total Environ. 2024, 914, 169803. [Google Scholar] [CrossRef]
- Wiener, G. The yak, an essential element of the high altitude regions of Central Asia. Études Mongoles Sibériennes Centrasiatiques Tibétaines 2013, 43, 43–44. [Google Scholar]
- Hameed, A.; Schlecht, E.; Tariq, M.; Buerkert, A.; Scheper, C.; König, S.; Roessler, R. Phenotypic and genetic diversity of domestic yak (Bos grunniens) in high-altitude rangelands of Gilgit-Baltistan, Pakistan. J. Anim. Breed. Genet. 2022, 139, 723–737. [Google Scholar] [CrossRef]
- Behl, R.; Vij, P.; Niranjan, S.; Jayakumar, S.; Behl, J.; Vijh, R. Yak genetic resources of India: Distribution, types and characteristics. Indian J. Anim. Sci. 2020, 90, 831–836. [Google Scholar] [CrossRef]
- Wang, X.; Pei, J.; Xiong, L.; Bao, P.; Chu, M.; Ma, X.; La, Y.; Liang, C.; Yan, P.; Guo, X. Genetic diversity, phylogeography, and maternal origin of yak (Bos grunniens). BMC Genom. 2024, 25, 481. [Google Scholar] [CrossRef]
- Zhong, J.; Chen, Z.; Zhao, S.; Xiao, Y. Classification of ecological types of the Chinese yak. Acta Ecol. Sin. 2006, 26, 2068–2072. [Google Scholar] [CrossRef]
- Porter, V.; Alderson, L.; Hall, S.J.; Sponenberg, D.P. Mason’s World Encyclopedia of Livestock Breeds and Breeding, 2 Volume Pack; CABI: Wallingford, UK, 2016. [Google Scholar]
- Dong, S.; Long, R.; Kang, M. Milking performance of China yak (Bos grunniens): A preliminary report. Afr. J. Agric. Res. 2007, 2, 052–057. [Google Scholar] [CrossRef]
- Huang, C.; Fu, D.; Wu, X.; Chu, M.; Ma, X.; Jia, C.; Guo, X.; Bao, P.; Yan, P.; Chunnian, L. Characterization of the complete mitochondrial genome of the Bazhou yak (Bos grunniens). Mitochondrial DNA Part B 2019, 4, 3234–3235. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Li, G.; Li, R.; Yang, Y.; He, D.; Liu, W.; Yosri, M.; Ma, Z. Mitogenome characterization and phylogeny of Huzhu white yak (Bos grunniens) in China. Mitochondrial DNA Part B 2022, 7, 901–904. [Google Scholar] [CrossRef]
- Mipam, T.D.; Wen, Y.; Fu, C.; Li, S.; Zhao, H.; Ai, Y.; Li, L.; Zhang, L.; Zou, D. Maternal phylogeny of a newly-found yak population in China. Int. J. Mol. Sci. 2012, 13, 11455–11470. [Google Scholar] [CrossRef]
- Das, P.J.; Kour, A.; Deori, S.; Begum, S.S.; Pukhrambam, M.; Maiti, S.; Sivalingam, J.; Paul, V.; Sarkar, M. Characterization of Arunachali Yak: A Roadmap for Pastoral Sustainability of Yaks in India. Sustainability 2022, 14, 12655. [Google Scholar] [CrossRef]
- Krofel, M.; Groff, C.; Oberosler, V.; Augugliaro, C.; Rovero, F. Snow leopard (Panthera uncia) predation and consumption of an adult yak in the Mongolian Altai. Ethol. Ecol. 2021, 33, 636–643. [Google Scholar] [CrossRef]
- Indra, R.; Magash, A. Composition, quality and consumption of yak milk in Mongolia. In Yak Production in Central Asian Highlands; Jianlin, H., Richard, C., Hanotte, O., McVeigh, C., Rege, J.E.O., Eds.; ILRI (International Livestock Research Institute): Nairobi, Kenya, 2002; p. 493. [Google Scholar]
- Qi, X.B.; Jianlin, H.; Wang, G.; Rege, J.E.; Hanotte, O. Assessment of cattle genetic introgression into domestic yak populations using mitochondrial and microsatellite DNA markers. Anim. Genet. 2010, 41, 242–252. [Google Scholar] [CrossRef]
- Rasool, G.; Khan, B.; Jasra, A. Yak pastoralism in Pakistan. In Yak Production in Central Asian Highlands; Jianlin, H., Richard, C., Hanotte, O., McVeigh, C., Rege, J.E.O., Eds.; ILRI (International Livestock Research Institute): Nairobi, Kenya, 2002; p. 95. [Google Scholar]
- Hussain, T.; Wajid, A.; Soail, M.; Ali, A.; Abbas, K.; Marikar, F.M.; Musthafa, M.M.; Babar, E. Molecular Phylogeny and Genetic Diversity of Domestic Yaks (Bos grunniens) in Pakistan based on Mitochondrial and Microsatellite Markers. Vet. Stanica 2021, 52, 671–684. [Google Scholar] [CrossRef]
- Zhu, Y.-B.; Basang, W.-D.; Pingcuo, Z.-D.; Cidan, Y.-J.; Luo, S.; Luosang, D.-Z.; Dawa, Y.-L. Genetic diversity and population structure of seven Tibet Yak ecotype populations using microsatellite markers. Pak. J. Zool. 2019, 51, 1979–1982. [Google Scholar] [CrossRef]
- Li, G.; Wei, X.; Chen, S.; Li, R.; Zhou, S.; Li, W.; Lin, Y.; Yosri, M.; Hanif, Q.; Ma, Z. Characterization of whole mitogenome sequence of the Tongde yak (Bos grunniens). Mitochondrial DNA B Resour. 2021, 6, 2498–2500. [Google Scholar] [CrossRef]
- Zia, U.-e.; Mahmood, T.; Ali, M. Dairy development in Pakistan; FAO: Rome, Italy, 2011. [Google Scholar]
- Baig, J.; Bilal, G.; Ulhaq, M.; Waheed, H.; Khan, M.; Qayyum, A.; Ahad, F.; Farooq, K.; Anwar, M.; Israr, T. Estimates of Heritabilities of Milk Fat and Milk Protein and Their Correlations with Milk Yield in Sahiwal Cattle of Punjab Pakistan. Trop. Anim. Sci. J. 2023, 46, 403–409. [Google Scholar] [CrossRef]
- Tariq, M.; Younas, M.; Khan, A.B.; Schlecht, E. Body Measurements and Body Condition Scoring as Basis for Estimation of Live Weight in Nili-Ravi Buffaloes. Pak. Vet. J. 2013, 33, 2074–7764. [Google Scholar]
- Jia, C.; Wang, H.; Li, C.; Wu, X.; Zan, L.; Ding, X.; Guo, X.; Bao, P.; Pei, J.; Chu, M.; et al. Genome-wide detection of copy number variations in polled yak using the Illumina BovineHD BeadChip. BMC Genom. 2019, 20, 376. [Google Scholar] [CrossRef]
- Wu, X.; Ding, X.; Chu, M.; Guo, X.; Bao, P.; Liang, C.; Yan, P. Novel SNP of EPAS1 gene associated with higher hemoglobin concentration revealed the hypoxia adaptation of yak (Bos grunniens). J. Integr. Agric. 2015, 14, 741–748. [Google Scholar] [CrossRef]
- Shi, B.; Jiang, Y.; Chen, Y.; Zhao, Z.; Zhou, H.; Luo, Y.; Hu, J.; Hickford, J.G. Variation in the fatty acid synthase gene (FASN) and its association with milk traits in Gannan yaks. Animals 2019, 9, 613. [Google Scholar] [CrossRef]
- Zhang, Q.; Cidan, Y.; Luosang, D.; Pingcuo, Z.; Dawa, Y.; Chen, X.; Basang, W. Dominant-genotype Frequency Analysis of Economic Traits Related to SNP Candidate Markers in Three Yak Populations. Indian J. Anim. Res. 2022, 1, 7. [Google Scholar] [CrossRef]
- Liu, W.; Yue, Y.; Lin, Y.; Liu, Z.; Jin, S.; Xu, Y.; Zheng, Y. Yak DGAT1 gene: Cloning, tissue expression profile, splicing and polymorphism analysis. Livest. Sci. 2011, 142, 264–269. [Google Scholar] [CrossRef]
- Goshu, H.A.; Wu, X.; Chu, M.; Bao, P.; Ding, X.; Yan, P. Copy number variations of KLF6 modulate gene transcription and growth traits in Chinese Datong yak (Bos grunniens). Animals 2018, 8, 145. [Google Scholar] [CrossRef]
- Yang, L.; Min, X.; Zhu, Y.; Hu, Y.; Yang, M.; Yu, H.; Li, J.; Xiong, X. Polymorphisms of SORBS 1 Gene and Their Correlation with Milk Fat Traits of Cattleyak. Animals 2021, 11, 3461. [Google Scholar] [CrossRef]
- Yan, P.; Xiang, L.; Guo, X.; Bao, P.-J.; Jin, S.; Wu, X.-Y. The low expression of Dmrt7 is associated with spermatogenic arrest in cattle-yak. Mol. Biol. Rep. 2014, 41, 7255–7263. [Google Scholar] [CrossRef]
- Zheng, Y.; Lin, Y.; Yue, Y.; Xu, Y.; Jin, S. Expression profiles of myostatin and calpastatin genes and analysis of shear force and intramuscular fat content of yak longissimus muscle. Czech J. Anim. Sci. 2011, 56, 544–550. [Google Scholar] [CrossRef]
- Bhuvaneshwari, R.; Padmanaban, K.; Babu Rajendran, R. Histopathological alterations in muscle, liver and gill tissues of zebra fish Danio rerio due to environmentally relevant concentrations of organochlorine pesticides (OCPs) and heavy metals. Int. J. Environ. Res. 2015, 9, 1365–1372. [Google Scholar] [CrossRef]
- Jiang, H.; Chai, Z.-X.; Cao, H.-W.; Zhang, C.-F.; Zhu, Y.; Zhang, Q.; Xin, J.-W. Genome-wide identification of SNPs associated with body weight in yak. BMC Genom. 2022, 23, 833. [Google Scholar] [CrossRef]
- Wu, X.; Zhou, X.; Ding, X.; Chu, M.; Liang, C.; Pei, J.; Xiong, L.; Bao, P.; Guo, X.; Yan, P. Reference gene selection and myosin heavy chain (MyHC) isoform expression in muscle tissues of domestic yak (Bos grunniens). PLoS ONE 2020, 15, e0228493. [Google Scholar] [CrossRef]
- Wang, J.; Li, X.; Peng, W.; Zhong, J.; Jiang, M. Genome-Wide Association Study of Body Weight Trait in Yaks. Animals 2022, 12, 1855. [Google Scholar] [CrossRef]
- Peng, S.; Chen, L.; Zheng, T.-Y.; Zhang, L.; Li, Z.; Xiao-Jing, T.; Zhong-Ren, M.; Liu, L.-X. Molecular characterization of the TLR2 gene in Datong yak. Indian J. Anim. Res. 2020, 54, 957–961. [Google Scholar] [CrossRef]
- Dai, R.; Huang, C.; Wu, X.; Ma, X.; Chu, M.; Bao, P.; Pei, J.; Guo, X.; Yan, P.; Liang, C. Copy number variation (CNV) of the AHR gene in the Ashidan yak and its association with growth traits. Gene 2022, 826, 146454. [Google Scholar] [CrossRef]
- Huang, C.; Ge, F.; Ren, W.; Zhang, Y.; Wu, X.; Zhang, Q.; Ma, X.; Bao, P.; Guo, X.; Chu, M. Copy number variation of the HPGDS gene in the Ashidan yak and its associations with growth traits. Gene 2021, 772, 145382. [Google Scholar] [CrossRef]
- Ren, W.; Huang, C.; Ma, X.; La, Y.; Chu, M.; Guo, X.; Wu, X.; Yan, P.; Liang, C. Association of HSF1 gene copy number variation with growth traits in the Ashidan yak. Gene 2022, 842, 146798. [Google Scholar] [CrossRef]
- Goshu, H.A.; Chu, M.; Xiaoyun, W.; Pengjia, B.; Zhi, D.X.; Yan, P. Genomic copy number variation of the CHKB gene alters gene expression and affects growth traits of Chinese domestic yak (Bos grunniens) breeds. Mol. Genet. Genom. 2019, 294, 549–561. [Google Scholar] [CrossRef]
- Ge, F.; Jia, C.; Chu, M.; Liang, C.; Yan, P. Copy number variation of the CADM2 gene and its association with growth traits in yak. Animals 2019, 9, 1008. [Google Scholar] [CrossRef]
- Zhang, X.; Bao, P.; Ye, N.; Zhou, X.; Zhang, Y.; Liang, C.; Guo, X.; Chu, M.; Pei, J.; Yan, P. Identification of the key genes associated with the yak hair follicle cycle. Genes 2021, 13, 32. [Google Scholar] [CrossRef]
- Goshu, H.A.; Chu, M.; Wu, X.; Pengjia, B.; Ding, X.Z.; Yan, P. Association study and expression analysis of GPC1 gene copy number variation in Chinese Datong yak (Bos grunniens) breed. Ital. J. Anim. Sci. 2019, 18, 820–832. [Google Scholar] [CrossRef]
- Zhang, Z.; Chu, M.; Bao, Q.; Bao, P.; Guo, X.; Liang, C.; Yan, P. Two Different Copy Number Variations of the SOX5 and SOX8 Genes in Yak and Their Association with Growth Traits. Animals 2022, 12, 1587. [Google Scholar] [CrossRef]
- Cai, X.; Mipam, T.; Zhao, F.; Sun, L. SNPs detected in the yak MC4R gene and their association with growth traits. Animals 2015, 9, 1097–1103. [Google Scholar] [CrossRef]
- Gui, L.; Raza, S.H.A.; Sun, Y.; Sabek, A.; Abbas, S.Q.; Shah, M.A.; Khan, R.; Abdelnour, S. Molecular characterization and analysis of the association of growth hormone 1 gene with growth traits in Chinese indigenous yak (Bos grunniens). Trop. Anim. Health Prod. 2021, 53, 221. [Google Scholar] [CrossRef]
- Ding, X.; Liang, C.; Guo, X.; Xing, C.; Bao, P.; Chu, M.; Pei, J.; Zhu, X.; Yan, P. A novel single nucleotide polymorphism in exon 7 of LPL gene and its association with carcass traits and visceral fat deposition in yak (Bos grunniens) steers. Mol. Biol. Rep. 2012, 39, 669–673. [Google Scholar] [CrossRef]
- Presgraves, D.C.; Meiklejohn, C.D. Hybrid Sterility, Genetic Conflict and Complex Speciation: Lessons From the Drosophila simulans Clade Species. Front. Genet. 2021, 12, 669045. [Google Scholar] [CrossRef]
- Zhang, R. Interspecies Hybridization between Yak, Bos taurus and Bos indicus and Reproduction of the Hybrids. In Recent Advances in Yak Reproduction; Zhao, X., Zhang, R., Eds.; 2000; Available online: https://www.ivis.org/library/recent-advances-yak-reproduction/interspecies-hybridization-between-yak-bos-taurus-and-bos (accessed on 20 July 2025).
- Chen, S.-Y.; Li, C.; Luo, Z.; Li, X.; Jia, X.; Lai, S.-J. Favoring Expression of Yak Alleles in Interspecies F1 Hybrids of Cattle and Yak Under High-Altitude Environments. Front. Vet. Sci. 2022, 9, 2022. [Google Scholar] [CrossRef]
- Diao, N.-C.; Gong, Q.-L.; Li, J.-M.; Zhao, D.; Li, D.; Zhao, B.; Ge, G.-Y.; Li, D.-L.; Shi, K.; Du, R. Prevalence of bovine viral diarrhea virus (BVDV) in yaks between 1987 and 2019 in mainland China: A systematic review and meta-analysis. Microb. Pathog. 2020, 144, 104185. [Google Scholar] [CrossRef]
- Cui, P.; Feng, L.; Zhang, L.; He, J.; An, T.; Fu, X.; Li, C.; Zhao, X.; Zhai, Y.; Li, H.; et al. Antimicrobial Resistance, Virulence Genes, and Biofilm Formation Capacity Among Enterococcus species From Yaks in Aba Tibetan Autonomous Prefecture, China. Front. Microbiol. 2020, 11, 2020. [Google Scholar] [CrossRef]
- Rehman, M.U.; Zhang, H.; Iqbal, M.K.; Mehmood, K.; Huang, S.; Nabi, F.; Luo, H.; Lan, Y.; Li, J. Antibiotic resistance, serogroups, virulence genes, and phylogenetic groups of Escherichia coli isolated from yaks with diarrhea in Qinghai Plateau, China. Gut Pathog. 2017, 9, 24. [Google Scholar] [CrossRef]
- Krishnan, G.; Paul, V.; Hanah, S.; Bam, J.; Das, P. Effects of climate change on yak production at high altitude. Indian J. Anim. Sci. 2016, 86, 621–626. [Google Scholar] [CrossRef]
- Sapkota, S.; Acharya, K.P.; Laven, R.; Acharya, N. Possible Consequences of Climate Change on Survival, Productivity and Reproductive Performance, and Welfare of Himalayan Yak (Bos grunniens). Vet. Sci. 2022, 9, 449. [Google Scholar] [CrossRef]
- Li, X.; Huang, C.; Liu, M.; Dai, R.; Wu, X.; Ma, X.; Chu, M.; Bao, P.; Pei, J.; Guo, X. Copy Number Variation of the SOX6 Gene and Its Associations with Growth Traits in Ashidan Yak. Animals 2022, 12, 3074. [Google Scholar] [CrossRef]
- Liu, M.; Huang, C.; Dai, R.; Ren, W.; Li, X.; Wu, X.; Ma, X.; Chu, M.; Bao, P.; Guo, X. Copy Number Variations in the MICALL2 and MOGAT2 Genes Are Associated with Ashidan Yak Growth Traits. Animals 2022, 12, 2779. [Google Scholar] [CrossRef]
- Ding, X.; Yang, C.; Bao, P.; Wu, X.; Pei, J.; Yan, P.; Guo, X. Population genetic variations of the MMP3 gene revealed hypoxia adaptation in domesticated yaks (Bos grunniens). Asian-Australas. J. Anim. Sci. 2019, 32, 1801–1808. [Google Scholar] [CrossRef]
- Jiang, J.; Wu, Q.; Rajasekaran, S.; Wu, R. MMP3 at the crossroads: Linking molecular pathways to disease diagnosis and therapy. Pharmacol. Res. 2025, 216, 107750. [Google Scholar] [CrossRef]
- Wang, J.; Shi, Y.; Elzo, M.A.; Dang, S.; Jia, X.; Lai, S. Genetic diversity of ATP8 and ATP6 genes is associated with high-altitude adaptation in yak. Mitochondrial DNA Part A 2018, 29, 385–393. [Google Scholar] [CrossRef]
- Wang, D.; Li, H.; Li, Y.; Guo, S.; Yang, J.; Qi, D.; Jin, C.; Zhao, X. Hypoxia-inducible factor 1α cDNA cloning and its mRNA and protein tissue specific expression in domestic yak (Bos grunniens) from Qinghai-Tibetan plateau. Biochem. Biophys. Res. Commun. 2006, 348, 310–319. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Liu, J.; Xu, Y.; Dong, X.; Shao, B. Evolutionary adaptation of aquaporin-4 in yak (Bos grunniens) brain to high-altitude hypoxia of Qinghai-Tibetan Plateau. High. Alt. Med. Biol. 2020, 21, 167–175. [Google Scholar] [CrossRef]
- Wu, X.; Liang, C.; Ding, X.; Guo, X.; Bao, P.; Chu, M.; Liu, W.; Yan, P.J.G.M.R. Association of novel single-nucleotide polymorphisms of the vascular endothelial growth factor-A gene with high-altitude adaptation in yak (Bos grunniens). Genet. Mol. Res. 2013, 12, 5506–5515. [Google Scholar] [CrossRef]
- Dolt, K.S.; Mishra, M.K.; Karar, J.; Baig, M.A.; Ahmed, Z.; Pasha, M.Q. cDNA cloning, gene organization and variant specific expression of HIF-1α in high altitude yak (Bos grunniens). Gene 2007, 386, 73–80. [Google Scholar] [CrossRef]
- Kuang, L.; Zheng, Y.; Lin, Y.; Xu, Y.; Jin, S.; Li, Y.; Dong, F.; Jiang, Z. High-altitude adaptation of yak based on genetic variants and activity of lactate dehydrogenase-1. Biochem. Genet. 2010, 48, 418–427. [Google Scholar] [CrossRef]
- Xiong, X.; Fu, M.; Lan, D.; Li, J.; Zi, X.; Zhong, J. Yak response to high-altitude hypoxic stress by altering mRNA expression and DNA methylation of hypoxia-inducible factors. Anim. Biotechnol. 2015, 26, 222–229. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; La, Y.; Fu, D.; Jiang, H.; Bao, P.; Wu, X.; Chu, M.; Guo, X.; Yan, P. Differential abundance of brain mitochondrial proteins in yak and cattle: A proteomics-based study. Front. Vet. Sci. 2021, 8, 663031. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Zhang, Q.; He, Y.; Yang, L.; Zhang, X.; Shi, P.; Yang, L.; Liu, Z.; Zhang, F.; Liu, F.; et al. The transcriptomic landscape of yaks reveals molecular pathways for high altitude adaptation. Genome Biol. 2019, 11, 72–85. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, T.; Wang, W.; Chen, Y.; Cai, W.; Zhu, B.; Xu, L.; Gao, H.; Zhang, L.; Li, J. Comparative transcriptome analysis of gayal (Bos frontalis), yak (Bos grunniens), and cattle (Bos taurus) reveal the high-altitude adaptation. Front. Genet. 2022, 12, 778788. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhong, J.; Wang, J.; Chai, Z.; Zhang, C.; Xin, J.; Wang, J.; Cai, X.; Wu, Z.; Ji, Q. Whole-transcriptome analysis of yak and cattle heart tissues reveals regulatory pathways associated with high-altitude adaptation. Front. Genet. 2021, 12, 579800. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Hu, Y.; Wang, J.; Elzo, M.A.; Yang, X.; Lai, S. Genetic diversities of MT-ND1 and MT-ND2 genes are associated with high-altitude adaptation in yak. Mitochondrial DNA Part A 2018, 29, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Pan, Y.; Cui, Y.; Peng, X.; Chen, P.; Fan, J.; Li, G.; Zhao, T.; Zhang, J.; Qin, S. Colony-stimulating factor 2 enhances the developmental competence of yak (Poephagus grunniens) preimplantation embryos by modulating the expression of heat shock protein 70 kDa 1A. Theriogenology 2017, 93, 16–23. [Google Scholar] [CrossRef] [PubMed]
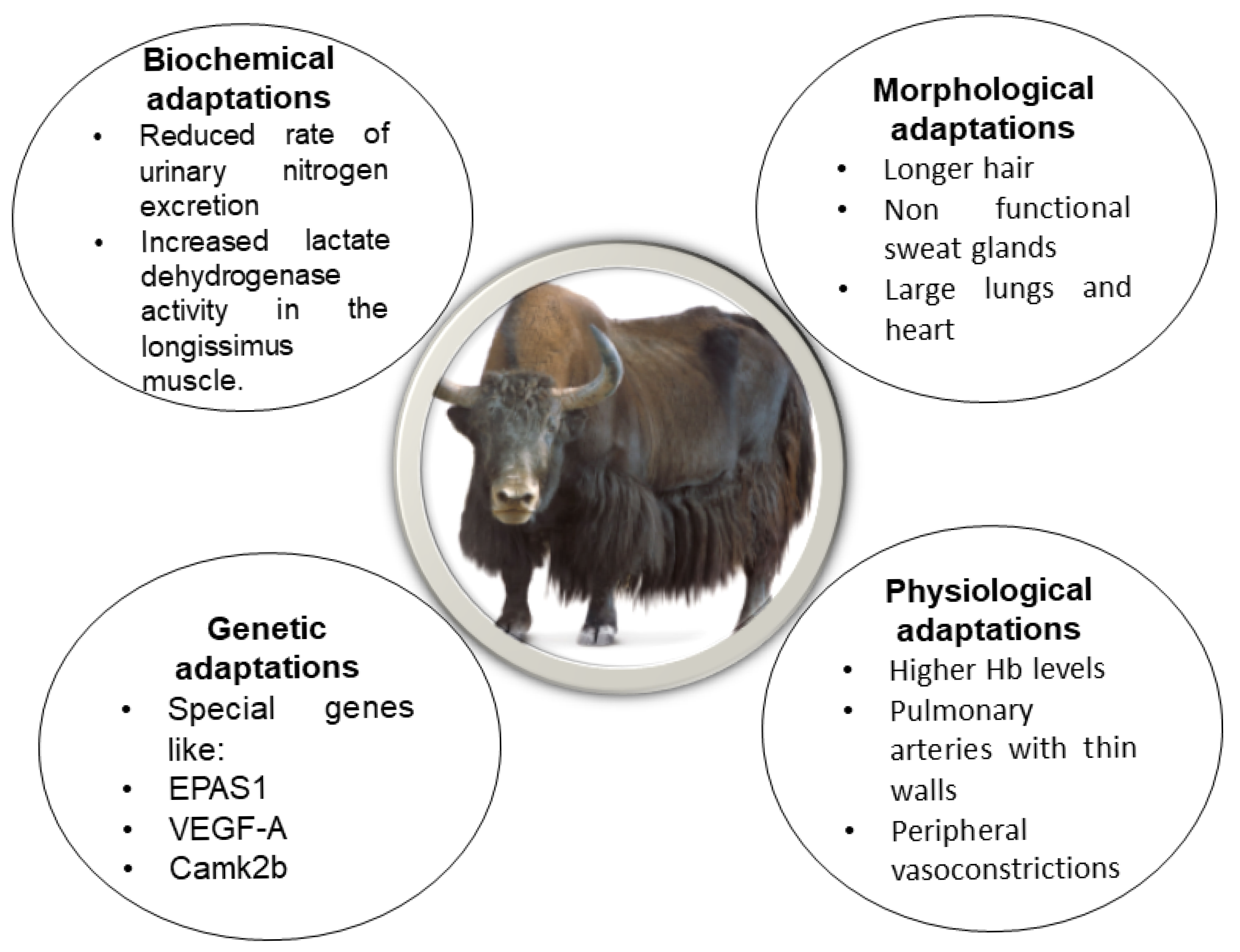
| Sr No. | Breed Name | Country | Location | Characteristics | Reference |
|---|---|---|---|---|---|
| 1. | Afghanistan yak | Afghanistan | West Asia |
| [47] |
| 2. | Merakpa yak | Bhutan | Eastern Bhutan, Tibet |
| [48] |
| 3. | Haapa yak | Western and central Bhutan, Tibet |
| ||
| 4. | Datong yak | China | East Asia |
| [1] |
| 5. | Huanhu yak | Qinghai Lake |
| [1] | |
| 6. | Guoluo yak | East Asia |
| [49] | |
| 7. | Batang yak | Batang area, Qinghai Province |
| ||
| 8. | Gannan yak | Gannan Tibetan, Gansu |
| ||
| 9. | Heihe yak | East Asia |
| ||
| 10. | Bazhou yak |
| [50] | ||
| 11. | Jiulong yak | Sichuan Province, Jiulong |
| [51] | |
| 12. | Maiwa yak | East Asia |
| ||
| 13. | Niangya yak, Liangya |
| |||
| 14. | Jinchuan yak | Maori and Akeli Village, Sichuan Province, Jinchuan |
| [52] | |
| 15. | Sarlag yak | East Asia |
| [48] | |
| 16. | Muli yak |
| [47] | ||
| 17. | Sibu yak, Tibetan high-mountain yak |
| |||
| 18. | Kyrgyz yak |
| |||
| 19. | Plateau yak of Qinghai |
| |||
| 20. | Xingjiang yak |
| |||
| 21. | Shandang yak |
| |||
| 22. | Tianzhu white yak |
| |||
| 23. | Zhongdian yak |
| |||
| 24. | Jiali/Alpine yak |
| |||
| 25. | Pali yak | Pali town, Yadong, Rigeze |
| [1] | |
| 26. | Arunachali yak | India | Northeastern states, India |
| [53] |
| 27. | Chour-gau yak | Ladakh |
| [48] | |
| 28. | Indian yak | South Asian |
| [47] | |
| 29. | Altai yak | Mongolia | East Asia |
| [54] |
| 30. | Hangai yak |
| |||
| 31. | Khainag yak |
| [55] | ||
| 32. | Nepalese ak | Nepal | South Asia |
| [56] |
| 33. | Siru yak | Pakistan |
| [57] | |
| 34. | Nagor yak |
| [44] | ||
| 35. | Balti yak |
| [56] | ||
| 36. | Himalayan yak |
| [58] | ||
| 37. | Nubra yak |
| [3] | ||
| 38. | Mishmi yak |
| [10] | ||
| 39. | Gaddi yak |
| |||
| 40. | Baltistani yak |
| [3] | ||
| 41. | Kharmangi yak |
| [58] | ||
| 42. | Gilgit yak |
| [59] | ||
| 43. | Skardu yak |
| |||
| 44. | Pakistani yak |
| [48] | ||
| 45. | Russian Federation yak | Russia | Northern Asia |
| [47] |
| 46. | Tajikistan yak | Tajikistan | Altai Territory, Tyan Shan |
| [5] |
| 47. | Tongde yak | Tongde County of Qinghai Province | East Asia |
| [60] |
| Location | Species | Trait Studied | Gene/SNP | Function of Associated Gene | Tissues | Method | Biological Impact | Origin | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Gannan–Tibetan Autonomous Prefecture, Gansu | Gannan yak/290 | Total solid content, Milk fat, Milk protein, Non-fat solids, and Milk lactose. | FASN | An enzyme called fatty acid synthase aids in the production of fatty acids (FAs) and is essential for mammalian de novo lipogenesis. |
| PCR-SSCP | The mammary gland and subcutaneous fat had the highest expression levels of fatty acid synthase, whereas the heart, small intestine, lung, kidney, abomasum, rumen, large intestine, longissimus dorsi muscle, and liver had the lowest expression levels. | China | [66] |
| Nagqu Jiali County, Tibet; Muzugongga County, Tibet; Neirong County, Tibet | Jiali Sibu Cawula/238 | Growth meat quality and lactation | ACSL1_A2079T ACSL1_G2409A CAPN4_G-1222A CYP4A11_G4806A GHSR_T1387C Hesx1_G618C Hesx1_T226C MyoD1_C1710T OXGR1_A347G TMEM-18_C1267T TMEM-18_C4447T UCP_ T1499C | Venous blood | Snapshot technology | The genetic diversity of the three yak populations was well preserved, and none of the populations had undergone artificial selection for economic traits. | Pakistan | [67] | |
Sichuan Province | Jiulong yak/32 | Milk fat content | DGAT1 K232A polymorphism | A key player in cellular triglyceride metabolism, diacylglycerol O-acyltransferase 1 (DGAT 1; EC 2.3.1.20) catalyzes the last stage of triglyceride production. It has a role in lactation, adipose tissue development, and intestinal fat absorption. Furthermore, it has the ability to catalyze the creation of diacylglycerols, waxes, and retinyl esters in vitro. | Longissimus muscle | Identification of DGAT1 gene splicing and assay of isoform proportion | There was no discernible difference between the liver and biceps femoris in yaks and cattle. | China | [68] |
| Analysis of DGAT1 mRNA levels was conducted using quantitative real-time RT-PCR. | The liver and adipose tissue of adult yaks had noticeably greater levels of DGAT1 mRNA than skeletal muscles, such the biceps femoris and longissimus dorsi. | ||||||||
| Jiulong yaks/58 Zhongdian yak/23 |
| PCR-SSCP; | PCR-SSCP analysis and direct sequencing of the PCR products revealed three genotypes of DGAT1: AA, GC, and AA/GC. Among the 81 samples, only one yak displayed the AA/GC genotype, while the others exhibited the AA genotype. | ||||||
| Datong Yak Farm, Qinghai Province Gansu Province, Tianzhu Tibetan Autonomous County Qinghai Province Datong Yak Farm, Qinghai Province Gansu Province, bordering Sichuan and Qinghai | Bos Grunniens/387 Polled Tianzhu white plateau Datong Gannan | Growth | KLF6 | A zinc finger transcription factor that is expressed in a variety of tissues, KLF6 belongs to the Kruppel-like factor family. It is essential for cell division, proliferation, development, and growth-related signaling pathways. |
| RT-qPCR | It is evident that KLF6 CNVRs play a major role in regulating the gene’s mRNA expression levels in the skeletal muscles of Bos grunniens. Additionally, there is a negative association between DNA copy numbers and gene expression, indicating that the expression of this gene influences quantitative growth features in yak populations. | China | [69] |
| Ngawa Tibetan and Qiang Autonomous Prefecture, Hongyuan County, Sichuan Province | Cattle yak/350 | Milk fat | SORBS1 | The Cbl-associated protein encoded by the sorbin and SH3 domain-containing 1 (SORBS1) gene is crucial for insulin signaling and stimulation. It is a member of the SORBS family. |
| PCR | The SORBS1 gene is a possible genetic marker for selecting milk fat qualities in cattle and yaks, since polymorphisms in this gene are highly correlated with these features. All nine of the SNPs that were found showed a strong association with the cattle yak’s milk fat traits. | China | [70] |
| Nil | Pali/56 Gannan/187 Tianzhu White/288 | Hemoglobin concentration | EPAS1 | The endothelial PAS domain protein 1 gene (EPAS1) is a key transcription factor that controls the expression of genes associated with oxygen levels. |
| RT-PCR | The lungs, kidneys, liver, heart, ovaries, spleen, muscles, and pancreas contain the highest quantities of EPAS1 mRNA in yaks. | China | [65] |
| Gannan Autonomous Prefecture, Gansu Province | Cattle Yaks Cattle-yaks | Spermatogenic arrest | Dmrt7 | Dmrt7 appears to be exclusive to mammals and is expressed only in the adult testes and embryonic gonads. It has no bearing on female gametogenesis but is essential for male gametogenesis. |
| RT-PCR | Although there was no discernible difference in the amounts of the Dmrt7 protein between the testes of cattle and yak, the expression of the Dmrt7 protein in cattle–yak was much lower than that in cattle and yak. In cattle and yak, male sterility is associated with this decreased expression of Dmrt7. | China | [71] |
| Nil | Bos grunniens Yak male calves/6 Adult male yaks/10 Chinese yellow cattle/8 |
| MSTN
| As negative regulators of skeletal muscle development, myostatin (MSTN) and calpastatin (CAST) are potential genes associated with muscle growth and tenderness. | Longissimus muscles | RT-PCR | Despite being smaller in body size than yellow cattle, adult yaks exhibited lower levels of MSTN and similar levels of CAST mRNA in the longissimus muscle compared to yellow cattle. | China | [72] |
| Bos grunniens/480 Pali yak (YD) Sibu yak (SB) Jiali yak (JL) | Body weight Parathyroid hormone (PTH) Adrenomedullin | G protein-coupled receptor kinase 4 (GRK4) males:
| Numerous studies have linked muscular dystrophy and obesity to G protein-coupled receptor kinase 4 (GRK4). | Venous blood | Enzyme linked immunosorbent assay (ELISA) | Yaks’ PTH and ADM levels were measured, and the results show that PTH levels and body weight were positively correlated, whereas ADM levels and body weight were negatively correlated. Additionally, there were differences in the AX-174555047 mutation. By modifying GRK4 expression, the SNP AX-174555047 may have an impact on body weight, which in turn impacts PTH and ADM function. | China | [73,74] |
| Qinghai Datong Yak Farm | Bos grunniens |
| MyHC | About 35% of muscles’ protein content is made up of myosin heavy chain (MyHC), the main structural protein. The skeletal muscle of several mammalian species contains four adult MyHC isoforms: MyHC I, IIA, IIX, and IIB. | Skeletal muscles:
| RT-qPCR | While GAPDH, the most often used reference gene, displayed the greatest fluctuation in expression across various muscle tissues, UXT and PRL13A were shown to be the most stable reference genes. The muscles with the highest concentration of type I muscle fibers and the lowest concentration of type IIB muscle fibers were the psoas major (Chapman), trapezius pars thoracica (TPT), and extensor digitorum lateralis (EDL). Conversely, the largest percentage of type IIB muscular fibers was found in the gluteobiceps (GB) muscle. | China | [75] |
| Longri Breeding Farm of Sichuan Province | Maiwa yaks/406 | Body weight |
| MFSD4 has consistently shown a significant impact on the main intake effects in skeletal muscle. LRRC37B has been reliably associated with body size in pigs, while neural cell adhesion molecule 2 (NCAM2) has been demonstrated to correlate with body weight in Simmental cattle. | Blood | GWAS | Seven markers were found to be significantly associated with the body weight trait. Among these, several candidate genes, including MFSD4, LRRC37B, and NCAM2, were identified. | China | [76] |
| Datong Yak Breeding Farm in Qinghai Province | Datong yak/55 |
| TLR2 | Toll-like receptors (TLRs) are important pattern recognition receptors and are widely expressed on the surfaces of innate immune system cells, such as monocytes and macrophages. | Whole blood jugular vein | PCR | The protein plays an important role in the body’s immune regulation mechanism. | China | [77] |
| Datong Yak Farm, Qinghai Province | Ashidan yaks/335 |
| AHR | The basic helix–loop–helix PAS family includes the ligand-dependent transcription factor known as the aromatic hydrocarbon receptor (AHR). It serves as an environmental sensor that is conserved throughout a variety of biological evolutionary processes. |
| qPCR | The liver, heart, adipose tissue, kidneys, spleen, and lungs had the highest levels of AHR expression. | China | [78] |
| Datong County, Qinghai Province | Ashidan yaks/336 |
| HPGDS | In male reproduction, HPGDS contributes to the negative control of cell proliferation through its involvement in PGD2 formation. As a potential gene, the HPGDS gene is also linked to characteristics of chicken meat quality. |
| PCR | In general, the 30-month-old yak had a higher level of HPGDS gene expression than the 6-month-old yak. | China | [79] |
| Datong Yak Farm, Qinghai Province | Ashidan yaks/274 |
| HSF1 | HSF1 is expressed in the cardiomyocytes, tissues, and organs. It exerts an irreplaceable effect in anti-apoptosis, anti-inflammatory, and anti-ischemia-reperfusion injury of cardiomyocytes. Furthermore, HSF1 is significant for the normal development of the body. |
| qPCR | HSF1 relative expression in muscles, followed by heart, liver, kidney, adipose tissue, lung, and spleen | China | [80] |
Datong Yak Farm in Qinghai Province;
| Datong yaks/222 Polled yaks/165 Tianzhu yaks/30 ; Gannan yaks/30 |
| CHKB | The CHKB gene is essential for maintaining normal mitochondrial function and plays a key role in the biosynthesis of phosphatidylcholine. It also regulates osteoclast and osteoblast functions, contributes to meat production and quality, supports growth and muscle development, and maintains bone homeostasis. Additionally, CHKB is involved in eye movement and the regulation of wakefulness. |
| qPCR | In 90-day-old fetuses, the CHKB gene was highly expressed in the lungs, brain, spleen, and kidneys; moderately expressed in the liver and muscle tissues; and showed low expression levels in the heart. In contrast, at the adult stage, CHKB expression was significantly higher in adipose, spleen, and lung tissues compared to other tissues. Moderate expression was observed in muscle and brain tissues, while the remaining tissues exhibited only low expression levels. | China | [81] |
| Datong Yak Farm, Qinghai Province | Ashidan yaks/350 |
| CADM2 | Variants of the CADM2 gene have been previously recognized as playing a vital role in influencing human body mass index (BMI) values via the central nervous system. Additionally, analyses in mice have revealed that CADM2 is closely associated with body weight and energy homeostasis through brain activity. | Blood | qPCR | The CNV2 mutation significantly influenced body weight in yaks at six months of age. | China | [82] |
| Tianzhu white yak propagation bases of Wuwei City, Gansu Province | Yak | Hair follicles’ (HFs) cycle |
| ------------------------- |
| RT-qPCR | Hub genes, including FER, ELMO1, PCOLCE, and HOXC13, were identified through screening in various modules. | China | [83] |
| Qinghai Province, Datong Yak Farm in Qinghai | Bos grunniens/536 | Growth traits Gene expression | GPC1 | The GPC1 gene plays a crucial role among proteoglycans in differentially regulating muscle cell proliferation, differentiation, and cellular responsiveness to FGF2. Notably, the copy number variations (CNVs) of the GPC1 gene are associated with meat production and quality, which are economically important traits that have been thoroughly considered for artificial selection in yak breeding. |
| qPCR | GPC1 exhibited significantly high expression levels in muscle and spleen tissues; moderate expression in the brain and lungs; and weak expression in the liver, kidneys, and heart. | China | [84] |
| Datong Farm, Qinghai Province | Ashidan yaks/326 |
|
| Normal development and bone formation depend on the SOX5 and SOX8 genes. |
| qPCR | Compared to the heart, spleen, kidney, and muscles, the expression of SOX5 was substantially higher in the lung. In a similar vein, SOX8 expression in the lung was noticeably greater than that in the muscles and liver. | China | [85] |
| Maiwa yak in Hongyuan County, Sichuan Province | Yaks/354 |
|
| MC4R (melanocortin 4 receptor) is expressed in the appetite-regulating areas of the brain and is involved in leptin signaling pathways. |
| PCR | SNP4 was associated with significant changes in the seventh transmembrane domain of the MC4R protein, leading to functional deterioration or even loss of function of MC4R. This may contribute to increased feed intake, body weight, and average daily gain in yaks with CC genotypes. | China | [86] |
| Qilian County, Qinghai Province | Bos grunniens/423 |
|
| It regulates essential cellular and physiological processes by binding to various hormones of the somatotropic axis, influencing muscle accretion, bone development, and fat catabolism. |
| PCR | A significant association was observed between this SNP and several growth traits in which the genotype GG exhibited the best values. | China | [87] |
| Gansu | Tianzhu white yak/111 Qinghai Plateau yak/70 Xinjiang yak/50 Gannan yak/95 Datong yak/72 |
| LPL | Lipoprotein lipase (LPL) is considered as a key enzyme in lipid deposition and metabolism in tissues. It is assumed to be a major candidate gene for genetic markers in lipid deposition. | Blood | PCR–SSCP analysis and DNA sequencing | The results indicate that the LPL gene is a strong candidate gene that affects carcass traits and fat deposition in yaks. | China | [88] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naz, S.; Chatha, A.M.M.; Ullah, Q.; Farooq, M.; Jamil, T.; Muner, R.D.; Kiran, A. Genomic Adaptation, Environmental Challenges, and Sustainable Yak Husbandry in High-Altitude Pastoral Systems. Vet. Sci. 2025, 12, 714. https://doi.org/10.3390/vetsci12080714
Naz S, Chatha AMM, Ullah Q, Farooq M, Jamil T, Muner RD, Kiran A. Genomic Adaptation, Environmental Challenges, and Sustainable Yak Husbandry in High-Altitude Pastoral Systems. Veterinary Sciences. 2025; 12(8):714. https://doi.org/10.3390/vetsci12080714
Chicago/Turabian StyleNaz, Saima, Ahmad Manan Mustafa Chatha, Qudrat Ullah, Muhammad Farooq, Tariq Jamil, Raja Danish Muner, and Azka Kiran. 2025. "Genomic Adaptation, Environmental Challenges, and Sustainable Yak Husbandry in High-Altitude Pastoral Systems" Veterinary Sciences 12, no. 8: 714. https://doi.org/10.3390/vetsci12080714
APA StyleNaz, S., Chatha, A. M. M., Ullah, Q., Farooq, M., Jamil, T., Muner, R. D., & Kiran, A. (2025). Genomic Adaptation, Environmental Challenges, and Sustainable Yak Husbandry in High-Altitude Pastoral Systems. Veterinary Sciences, 12(8), 714. https://doi.org/10.3390/vetsci12080714






