Antimicrobial Resistance in Commensal Bacteria from Large-Scale Chicken Flocks in the Dél-Alföld Region of Hungary
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Origin of the Samples
2.2. Determination of Minimum Inhibitory Concentration (MIC)
2.3. Statistical Analysis
3. Results
3.1. Staphylococcus spp. Isolates
3.2. Enterococcus spp. Isolates
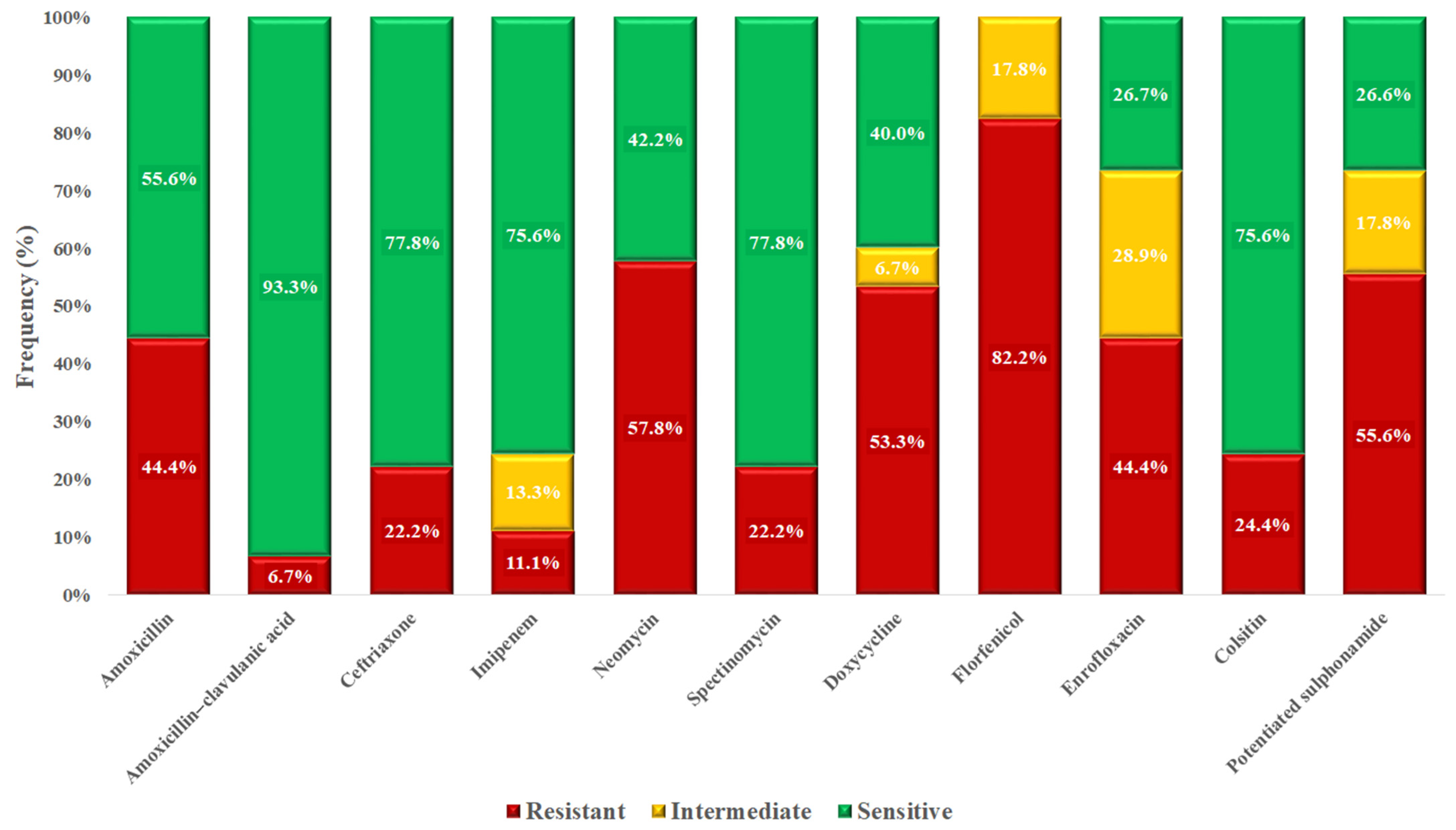
3.3. Escherichia coli Isolates
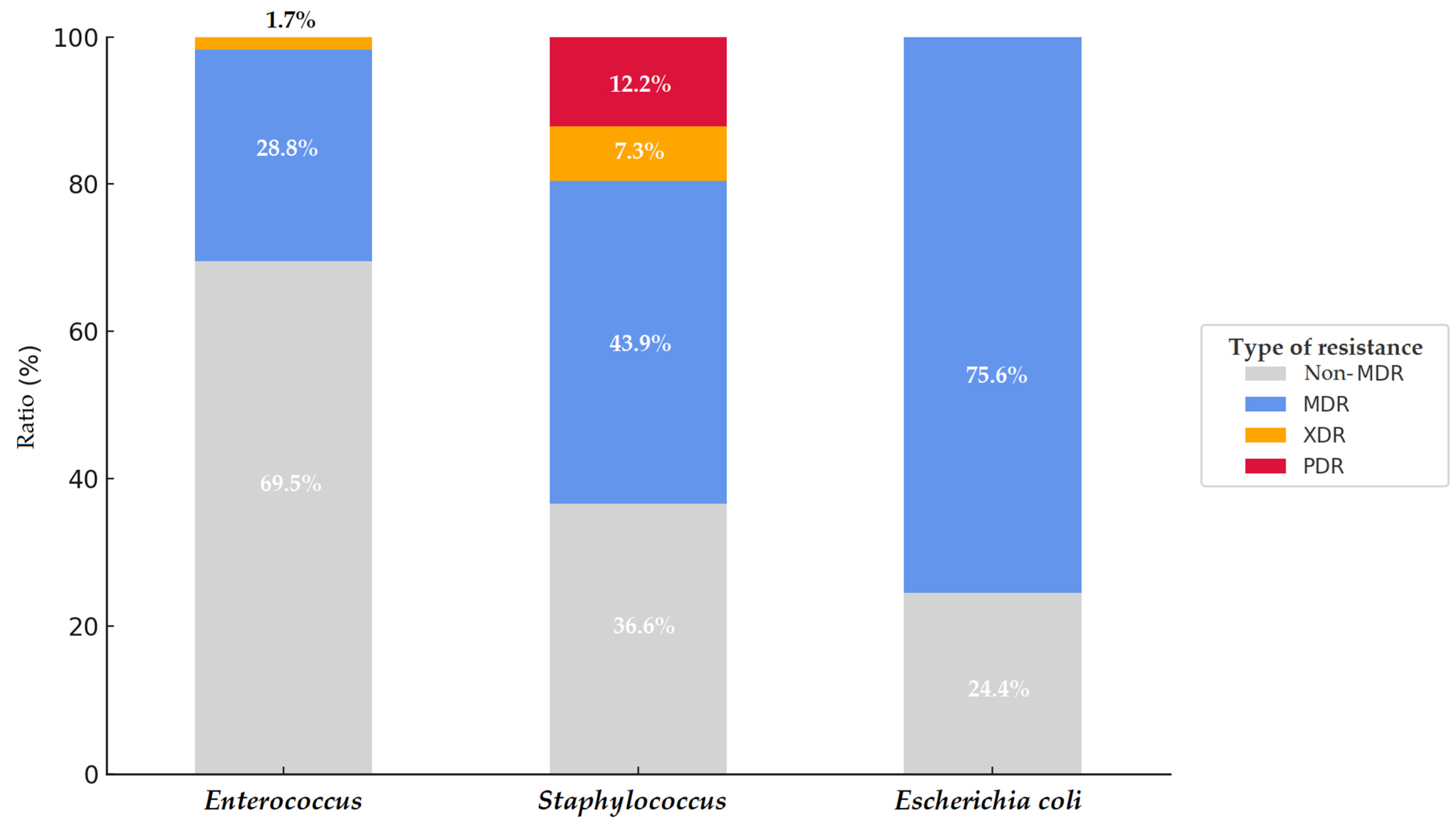
3.4. Multidrug-Resistant (MDR), Extensively Drug-Resistant (XDR), and Pan-Drug-Resistant (PDR) Isolates
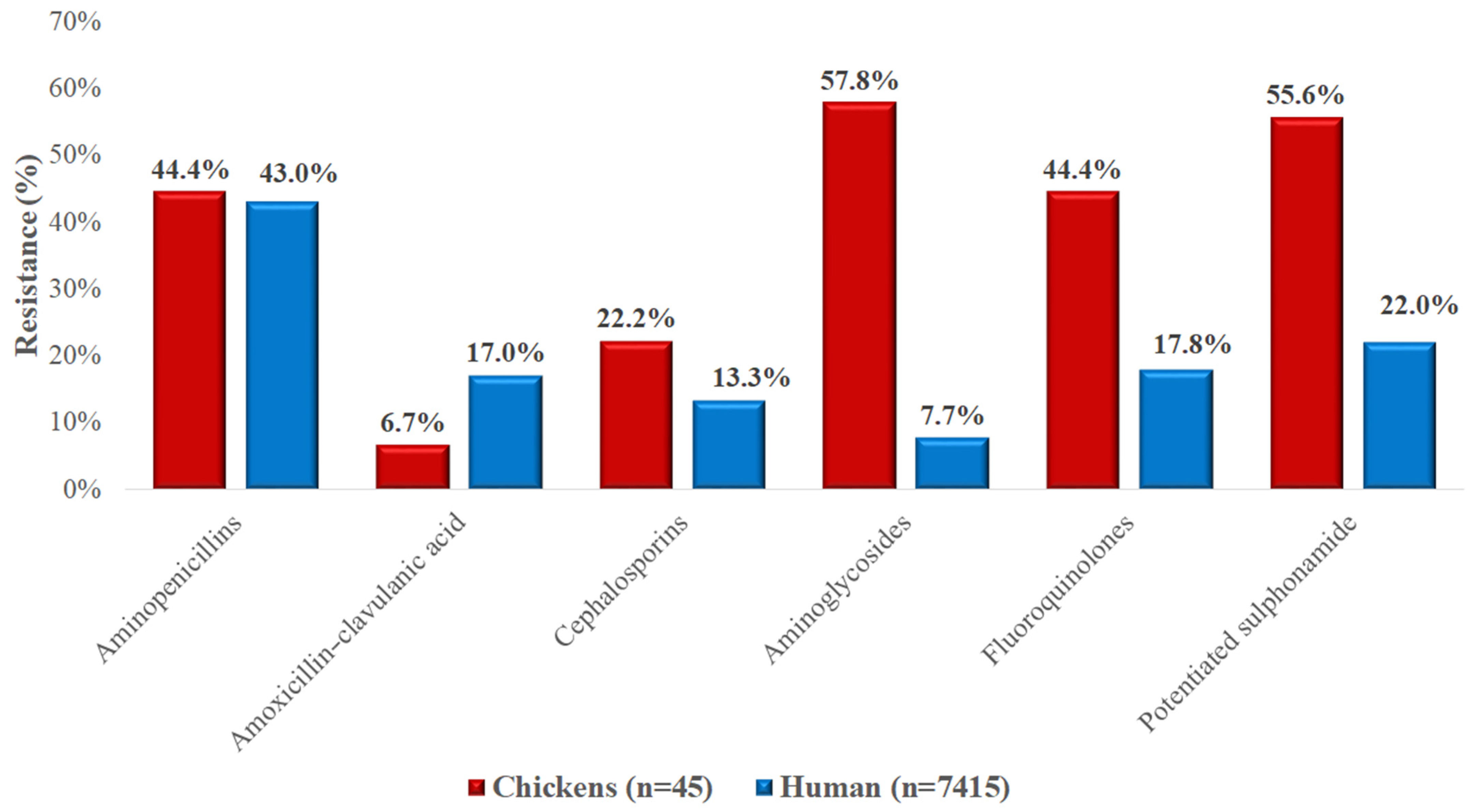
3.5. Comparison with Regional Human Resistance Data
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AMR | Antimicrobial resistance |
| CLSI | Clinical Laboratory Standards Institute |
| E. coli | Escherichia coli |
| MDR | Multidrug-resistant |
| PCA | Principal component analysis |
References
- Castanon, J.I.R. History of the Use of Antibiotic as Growth Promoters in European Poultry Feeds. Poult. Sci. 2007, 86, 2466–2471. [Google Scholar] [CrossRef]
- Centner, T.J. Efforts to Slacken Antibiotic Resistance: Labeling Meat Products from Animals Raised without Antibiotics in the United States. Sci. Total Environ. 2016, 563–564, 1088–1094. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.L.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Robles Aguilar, G.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Flynn, C.E.; Guarner, J. Emerging Antimicrobial Resistance. Mod. Pathol. 2023, 36, 100249. [Google Scholar] [CrossRef] [PubMed]
- Lebert, L.; Martz, S.-L.; Janecko, N.; Deckert, A.E.; Agunos, A.; Reid, A.; Rubin, J.E.; Reid-Smith, R.J.; McEwen, S.A. Prevalence and Antimicrobial Resistance among Escherichia coli and Salmonella in Ontario Smallholder Chicken Flocks. Zoonoses Public Health 2018, 65, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Simonsen, G.S. Antimicrobial Resistance Surveillance in Europe and Beyond. Eurosurveillance 2018, 23, 1800560. [Google Scholar] [CrossRef] [PubMed]
- Such, N.; Molnár, A.; Pál, L.; Farkas, V.; Menyhárt, L.; Husvéth, F.; Dublecz, K. The Effect of Pre- and Probiotic Treatment on the Gumboro-Titer Values of Broilers. Magy. Állatorvosok Lapja 2021, 143, 119–127. [Google Scholar]
- Kovács, L.; Nagy, D.; Könyves, L.; Jerzsele, Á.; Kerek, Á. Antimicrobial Properties of Essential Oils—Animal Health Aspects. Magy. Állatorvosok Lapja 2023, 145, 497–510. [Google Scholar] [CrossRef]
- Jerzsele, Á.; Somogyi, Z.; Szalai, M.; Kovács, D. Effects of Fermented Wheat Germ Extract on Artificial Salmonella Typhimurium Infection in Broiler Chickens. Magy. Állatorvosok Lapja 2020, 142, 77–85. [Google Scholar]
- Sebők, C.; Márton, R.A.; Meckei, M.; Neogrády, Z.; Mátis, G. Antimicrobial Peptides as New Tools to Combat Infectious Diseases. Magy. Állatorvosok Lapja 2024, 146, 181–191. [Google Scholar] [CrossRef]
- Olasz, Á.; Jerzsele, Á.; Balta, L.; Dobra, P.F.; Kerek, Á. In Vivo Efficacy of Different Extracts of Propolis in Broiler Salmonellosis. Magy. Állatorvosok Lapja 2023, 145, 461–475. [Google Scholar] [CrossRef]
- Kerek, Á.; Csanády, P.; Jerzsele, Á. Antibacterial Efficiency of Propolis—Part 1. Magy. Állatorvosok Lapja 2022, 144, 285–298. [Google Scholar]
- Kerek, Á.; Csanády, P.; Jerzsele, Á. Antiprotozoal and Antifungal Efficiency of Propolis—Part 2. Magy. Állatorvosok Lapja 2022, 144, 691–704. [Google Scholar]
- Hetényi, N.; Bersényi, A.; Hullár, I. Physiological Effects of Medium-Chain Fatty Acids and Triglycerides, and Their Potential Use in Poultry and Swine Nutrition: A Literature Review. Magy. Állatorvosok Lapja 2024, 146, 651–659. [Google Scholar] [CrossRef]
- Costa, M.C.; Bessegatto, J.A.; Alfieri, A.A.; Weese, J.S.; Filho, J.A.B.; Oba, A. Different Antibiotic Growth Promoters Induce Specific Changes in the Cecal Microbiota Membership of Broiler Chicken. PLoS ONE 2017, 12, e0171642. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Massow, A.; Stanley, M.; Papariella, M.; Chen, X.; Kraft, B.; Ebner, P. Contamination Rates and Antimicrobial Resistance in Enterococcus spp., Escherichia coli, and Salmonella Isolated from “No Antibiotics Added”—Labeled Chicken Products. Foodborne Pathog. Dis. 2011, 8, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Schwaiger, K.; Schmied, E.-M.V.; Bauer, J. Comparative Analysis on Antibiotic Resistance Characteristics of Listeria spp. and Enterococcus spp. Isolated from Laying Hens and Eggs in Conventional and Organic Keeping Systems in Bavaria, Germany. Zoonoses Public Health 2010, 57, 171–180. [Google Scholar] [CrossRef] [PubMed]
- McDermott, P.F.; Cullen, P.; Hubert, S.K.; McDermott, S.D.; Bartholomew, M.; Simjee, S.; Wagner, D.D. Changes in Antimicrobial Susceptibility of Native Enterococcus faecium in Chickens Fed Virginiamycin. Appl. Environ. Microbiol. 2005, 71, 4986–4991. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.; Uwiera, R.R.E.; Kalmokoff, M.L.; Brooks, S.P.J.; Inglis, G.D. Antimicrobial Growth Promoter Use in Livestock: A Requirement to Understand Their Modes of Action to Develop Effective Alternatives. Int. J. Antimicrob. Agents 2017, 49, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Chauvin, C.; Gicquel-Bruneau, M.; Perrin-Guyomard, A.; Humbert, F.; Salvat, G.; Guillemot, D.; Sanders, P. Use of Avilamycin for Growth Promotion and Avilamycin-Resistance among Enterococcus faecium from Broilers in a Matched Case–Control Study in France. Prev. Vet. Med. 2005, 70, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Aarestrup, F.M.; McNicholas, P.M. Incidence of High-Level Evernimicin Resistance in Enterococcus faecium among Food Animals and Humans. Antimicrob. Agents Chemother. 2002, 46, 3088–3090. [Google Scholar] [CrossRef] [PubMed]
- Boucher, H.W.; Thauvin-Eliopoulos, C.; Loebenberg, D.; Eliopoulos, G.M. In Vivo Activity of Evernimicin (SCH 27899) against Methicillin-Resistant Staphylococcus aureus in Experimental Infective Endocarditis. Antimicrob. Agents Chemother. 2001, 45, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Becker, K.; Skov, R.L.; von Eiff, C. Staphylococcus, Micrococcus, and Other Catalase-Positive Cocci. In Manual of Clinical Microbiology; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2015; pp. 354–382. ISBN 978-1-68367-280-7. [Google Scholar]
- Park, S.; Ronholm, J. Staphylococcus aureus in Agriculture: Lessons in Evolution from a Multispecies Pathogen. Clin. Microbiol. Rev. 2021, 34, e00182-20. [Google Scholar] [CrossRef] [PubMed]
- Leonard, F.C.; Markey, B.K. Meticillin-Resistant Staphylococcus aureus in Animals: A Review. Vet. J. 2008, 175, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Cardona, C.J.; Bickford, A.A.; Charlton, B.R.; Cooper, G.L. Enterococcus durans Infection in Young Chickens Associated with Bacteremia and Encephalomalacia. Avian Dis. 1993, 37, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Borgen, K.; Sørum, M.; Wasteson, Y.; Kruse, H. VanA-Type Vancomycin-Resistant Enterococci (VRE) Remain Prevalent in Poultry Carcasses 3 Years after Avoparcin Was Banned. Int. J. Food Microbiol. 2001, 64, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Migura, L.; Pleydell, E.; Barnes, S.; Davies, R.H.; Liebana, E. Characterization of Vancomycin-Resistant Enterococcus faecium Isolates from Broiler Poultry and Pig Farms in England and Wales. J. Clin. Microbiol. 2005, 43, 3283–3289. [Google Scholar] [CrossRef] [PubMed]
- Kühn, I.; Iversen, A.; Möllby, R. The PhenePlateTM System for Studies of the Diversity of Enterococcal Populations from the Food Chain and the Environment. Int. J. Food Microbiol. 2003, 88, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Lauderdale, T.-L.; McDonald, L.C.; Shiau, Y.-R.; Chen, P.-C.; Wang, H.-Y.; Lai, J.-F.; Ho, M. Vancomycin-Resistant Enterococci from Humans and Retail Chickens in Taiwan with Unique VanB Phenotype-VanA Genotype Incongruence. Antimicrob. Agents Chemother. 2002, 46, 525–527. [Google Scholar] [CrossRef] [PubMed]
- van den Bogaard, A.E.; Willems, R.; London, N.; Top, J.; Stobberingh, E.E. Antibiotic Resistance of Faecal Enterococci in Poultry, Poultry Farmers and Poultry Slaughterers. J. Antimicrob. Chemother. 2002, 49, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, C.; Diarrassouba, F.; Brousseau, R.; Masson, L.; Topp, E.; Diarra, M.S. Pathotype and Antibiotic Resistance Gene Distributions of Escherichia coli Isolates from Broiler Chickens Raised on Antimicrobial-Supplemented Diets. Appl. Environ. Microbiol. 2009, 75, 6955–6962. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.; Manzanilla, E.G.; Ekhlas, D.; Leonard, F.C. Antimicrobial Resistance in Commensal Escherichia coli of the Porcine Gastrointestinal Tract. Antibiotics 2023, 12, 1616. [Google Scholar] [CrossRef] [PubMed]
- Pinto, B.F.; Silva, S.A.M.; Rodrigues, I.C.; Lopes-Jorge, J.M.; Niza-Ribeiro, J.; Prata, J.C.; Costa, P.M. da Antimicrobial Resistance in Swine and Cattle Farms. Microbiol. Res. 2025, 16, 83. [Google Scholar] [CrossRef]
- Sevilla-Navarro, S.; Catalá-Gregori, P.; Torres-Boncompte, J.; Orenga, M.T.; Garcia-Llorens, J.; Cortés, V. Antimicrobial Resistance Trends of Escherichia coli Isolates: A Three-Year Prospective Study of Poultry Production in Spain. Antibiotics 2022, 11, 1064. [Google Scholar] [CrossRef] [PubMed]
- Watts, A.; Wigley, P. Avian Pathogenic Escherichia coli: An Overview of Infection Biology, Antimicrobial Resistance and Vaccination. Antibiotics 2024, 13, 809. [Google Scholar] [CrossRef] [PubMed]
- Nolan, L.K.; Vaillancourt, J.-P.; Barbieri, N.L.; Logue, C.M. Colibacillosis. In Diseases of Poultry; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2020; pp. 770–830. ISBN 978-1-119-37119-9. [Google Scholar]
- Jørgensen, S.L.; Stegger, M.; Kudirkiene, E.; Lilje, B.; Poulsen, L.L.; Ronco, T.; Pires Dos Santos, T.; Kiil, K.; Bisgaard, M.; Pedersen, K.; et al. Diversity and Population Overlap between Avian and Human Escherichia coli Belonging to Sequence Type 95. mSphere 2019, 4, e00333-18. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, Q.; Cheng, Y.; Liu, R.; Zhao, R.; Wang, J.; Wang, Y.; Yang, S.; Chen, A. Effect of Bacterial Resistance of Escherichia coli From Swine in Large-Scale Pig Farms in Beijing. Front. Microbiol. 2022, 13, 820833. [Google Scholar] [CrossRef] [PubMed]
- Morris, C.; Wickramasingha, D.; Abdelfattah, E.M.; Pereira, R.V.; Okello, E.; Maier, G. Prevalence of Antimicrobial Resistance in Fecal Escherichia coli and Enterococcus spp. Isolates from Beef Cow-Calf Operations in Northern California and Associations with Farm Practices. Front. Microbiol. 2023, 14, 1086203. [Google Scholar] [CrossRef] [PubMed]
- Shoaib, M.; Tang, M.; Aqib, A.I.; Zhang, X.; Wu, Z.; Wen, Y.; Hou, X.; Xu, J.; Hao, R.; Wang, S.; et al. Dairy Farm Waste: A Potential Reservoir of Diverse Antibiotic Resistance and Virulence Genes in Aminoglycoside- and Beta-Lactam-Resistant Escherichia coli in Gansu Province, China. Environ. Res. 2024, 263, 120190. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Rehman, M.A.; Yin, X.; Carrillo, C.D.; Wang, Q.; Yang, C.; Gong, J.; Diarra, M.S. Antimicrobial Resistance Phenotypes and Genotypes of Escherichia coli Isolates from Broiler Chickens Fed Encapsulated Cinnamaldehyde and Citral. J. Food Prot. 2021, 84, 1385–1399. [Google Scholar] [CrossRef] [PubMed]
- Poirel, L.; Madec, J.-Y.; Lupo, A.; Schink, A.-K.; Kieffer, N.; Nordmann, P.; Schwarz, S. Antimicrobial Resistance in Escherichia coli. Microbiol. Spectr. 2018, 6, 1–27. [Google Scholar] [CrossRef] [PubMed]
- van Hoek, A.H.A.M.; Mevius, D.; Guerra, B.; Mullany, P.; Roberts, A.P.; Aarts, H.J.M. Acquired Antibiotic Resistance Genes: An Overview. Front. Microbiol. 2011, 2, 203. [Google Scholar] [CrossRef] [PubMed]
- Kerek, Á.; Szabó, Á.; Jerzsele, Á. Antimicrobial Susceptibility Profiles of Commensal Enterococcus spp. Isolates from Chickens in Hungarian Poultry Farms Between 2022 and 2023. Antibiotics 2024, 13, 1194. [Google Scholar] [CrossRef] [PubMed]
- Kovács, L.; Szabó, Á.; Barnácz, F.; Csirmaz, B.; Jerzsele, Á.; Kerek, Á. Antimicrobial Susceptibility Profiles of Commensal Staphylococcus spp. Isolates from Turkeys in Hungarian Poultry Farms Between 2022 and 2023. Antibiotics 2025, 14, 200. [Google Scholar] [CrossRef] [PubMed]
- Lange, B.; Strathmann, M.; Oßmer, R. Performance Validation of Chromogenic Coliform Agar for the Enumeration of Escherichia coli and Coliform Bacteria. Lett. Appl. Microbiol. 2013, 57, 547–553. [Google Scholar] [CrossRef] [PubMed]
- Dingle, T.C.; Butler-Wu, S.M. Maldi-Tof Mass Spectrometry for Microorganism Identification. Clin. Lab. Med. 2013, 33, 589–609. [Google Scholar] [CrossRef] [PubMed]
- CLSI M07; Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically, 11th ed. Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2018.
- Moawad, A.A.; Hotzel, H.; Awad, O.; Roesler, U.; Hafez, H.M.; Tomaso, H.; Neubauer, H.; El-Adawy, H. Evolution of Antibiotic Resistance of Coagulase-Negative Staphylococci Isolated from Healthy Turkeys in Egypt: First Report of Linezolid Resistance. Microorganisms 2019, 7, 476. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Yeh, K.-S.; Liu, H.-T.; Lin, J.-H. Staphylococcus aureus Isolated from Pork and Chicken Carcasses in Taiwan: Prevalence and Antimicrobial Susceptibility. J. Food Prot. 2009, 72, 608–611. [Google Scholar] [CrossRef] [PubMed]
- Nemeghaire, S.; Argudín, M.A.; Haesebrouck, F.; Butaye, P. Molecular Epidemiology of Methicillin-Resistant Staphylococcus sciuri in Healthy Chickens. Vet. Microbiol. 2014, 171, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Jahan, M.; Krause, D.O.; Holley, R.A. Antimicrobial Resistance of Enterococcus Species from Meat and Fermented Meat Products Isolated by a PCR-Based Rapid Screening Method. Int. J. Food Microbiol. 2013, 163, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Bywater, R.; McConville, M.; Phillips, I.; Shryock, T. The Susceptibility to Growth-Promoting Antibiotics of Enterococcus faecium Isolates from Pigs and Chickens in Europe. J. Antimicrob. Chemother. 2005, 56, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Lima-Filho, J.V.; Martins, L.V.; Nascimento, D.C.d.O.; Ventura, R.F.; Batista, J.E.C.; Silva, A.F.B.; Ralph, M.T.; Vaz, R.V.; Rabello, C.B.-V.; Silva, I.d.M.M.d.; et al. Zoonotic Potential of Multidrug-Resistant Extraintestinal Pathogenic Escherichia coli Obtained from Healthy Poultry Carcasses in Salvador, Brazil. Braz. J. Infect. Dis. 2013, 17, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Boulianne, M.; Arsenault, J.; Daignault, D.; Archambault, M.; Letellier, A.; Dutil, L. Drug Use and Antimicrobial Resistance among Escherichia coli and Enterococcus spp. Isolates from Chicken and Turkey Flocks Slaughtered in Quebec, Canada. Can. J. Vet. Res. 2016, 80, 49–59. [Google Scholar] [PubMed]
- Hesp, A.; van Schaik, G.; Wiegel, J.; Heuvelink, A.; Mevius, D.; Veldman, K. Antimicrobial Resistance Monitoring in Commensal and Clinical Escherichia coli from Broiler Chickens: Differences and Similarities. Prev. Vet. Med. 2022, 204, 105663. [Google Scholar] [CrossRef] [PubMed]
- Szabó, Á.; Jerzsele, Á.; Kovács, L.; Kerek, Á. Antimicrobial Susceptibility Profiles of Commensal Staphylococcus spp. Isolates from Chickens in Hungarian Poultry Farms Between 2022 and 2023. Antibiotics 2025, 14, 103. [Google Scholar] [CrossRef] [PubMed]
- Wittwer, M.; Keller, J.; Wassenaar, T.M.; Stephan, R.; Howald, D.; Regula, G.; Bissig-Choisat, B. Genetic Diversity and Antibiotic Resistance Patterns in a Campylobacter Population Isolated from Poultry Farms in Switzerland. Appl. Environ. Microbiol. 2005, 71, 2840–2847. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Linke, L.; Magnuson, R.; Jauch, L.; Hyatt, D.R. Antimicrobial Resistance and Genetic Diversity of Staphylococcus aureus Collected from Livestock, Poultry and Humans. One Health 2022, 15, 100407. [Google Scholar] [CrossRef] [PubMed]
- Syed, M.A.; Ullah, H.; Tabassum, S.; Fatima, B.; Woodley, T.A.; Ramadan, H.; Jackson, C.R. Staphylococci in Poultry Intestines: A Comparison between Farmed and Household Chickens. Poult. Sci. 2020, 99, 4549–4557. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.B.; Seo, K.W.; Jeon, H.Y.; Lim, S.-K.; Lee, Y.J. Characteristics of the Antimicrobial Resistance of Staphylococcus aureus Isolated from Chicken Meat Produced by Different Integrated Broiler Operations in Korea. Poult. Sci. 2018, 97, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Nhung, N.T.; Chansiripornchai, N.; Carrique-Mas, J.J. Antimicrobial Resistance in Bacterial Poultry Pathogens: A Review. Front. Vet. Sci. 2017, 4, 126. [Google Scholar] [CrossRef] [PubMed]
- Miranda, J.M.; Vázquez, B.I.; Fente, C.A.; Calo-Mata, P.; Cepeda, A.; Franco, C.M. Comparison of Antimicrobial Resistance in Escherichia coli, Staphylococcus aureus, and Listeria monocytogenes Strains Isolated from Organic and Conventional Poultry Meat. J. Food Prot. 2008, 71, 2537–2542. [Google Scholar] [CrossRef] [PubMed]
- Mkize, N.; Zishiri, O.T.; Mukaratirwa, S. Genetic Characterisation of Antimicrobial Resistance and Virulence Genes in Staphylococcus aureus Isolated from Commercial Broiler Chickens in the Durban Metropolitan Area, South Africa. J. S. Afr. Vet. Assoc. 2017, 88, e1–e7. [Google Scholar] [CrossRef] [PubMed]
- Bekele, B.; Ashenafi, M. Distribution of Drug Resistance among Enterococci and Salmonella from Poultry and Cattle in Ethiopia. Trop. Anim. Health Prod. 2010, 42, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Semedo-Lemsaddek, T.; Bettencourt Cota, J.; Ribeiro, T.; Pimentel, A.; Tavares, L.; Bernando, F.; Oliveira, M. Resistance and Virulence Distribution in Enterococci Isolated from Broilers Reared in Two Farming Systems. Ir. Vet. J. 2021, 74, 22. [Google Scholar] [CrossRef] [PubMed]
- Szoke, Z.; Fauszt, P.; Mikolas, M.; David, P.; Szilagyi-Tolnai, E.; Pesti-Asboth, G.; Homoki, J.R.; Kovacs-Forgacs, I.; Gal, F.; Stundl, L.; et al. Comprehensive Analysis of Antimicrobial Resistance Dynamics among Broiler and Duck Intensive Production Systems. Sci. Rep. 2025, 15, 4673. [Google Scholar] [CrossRef] [PubMed]
- Parkhi, C.M.; Liverpool-Tasie, L.S.O.; Reardon, T. Do Smaller Chicken Farms Use More Antibiotics? Evidence of Antibiotic Diffusion from Nigeria. Agribusiness 2023, 39, 242–262. [Google Scholar] [CrossRef]
- Pesavento, G.; Calonico, C.; Ducci, B.; Magnanini, A.; Lo Nostro, A. Prevalence and Antibiotic Resistance of Enterococcus spp. Isolated from Retail Cheese, Ready-to-Eat Salads, Ham, and Raw Meat. Food Microbiol. 2014, 41, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ayeni, F.A.; Odumosu, B.T.; Oluseyi, A.E.; Ruppitsch, W. Identification and Prevalence of Tetracycline Resistance in Enterococci Isolated from Poultry in Ilishan, Ogun State, Nigeria. J. Pharm. Bioallied Sci. 2016, 8, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Lanza, I.P.; Silva, G.R.; Menezes, L.D.M.; Assis, D.C.S.; Figueiredo, H.C.P.; Lana, A.M.Q.; Lara, L.J.C.; Figueiredo, T.C.; Souza, M.R.; Cançado, S.V. Research Note: Antimicrobial Resistance Profile of Enterococcus spp. Isolated from the Eggshell of Laying Hens Submitted to Pharmacological Treatment. Poult. Sci. 2022, 101, 101606. [Google Scholar] [CrossRef] [PubMed]
- Noh, E.B.; Kim, Y.B.; Seo, K.W.; Son, S.H.; Ha, J.S.; Lee, Y.J. Antimicrobial Resistance Monitoring of Commensal Enterococcus faecalis in Broiler Breeders. Poult. Sci. 2020, 99, 2675–2683. [Google Scholar] [CrossRef] [PubMed]
- Karunarathna, R.; Ahmed, K.A.; Liu, M.; Yu, C.; Popowich, S.; Goonewardene, K.; Gunawardana, T.; Kurukulasuriya, S.; Gupta, A.; Ayalew, L.E.; et al. Non-Viable Chicken Embryos: An Overlooked Niche Harbouring a Significant Source of Multidrug Resistant Bacteria in the Poultry Production. Int. J. Vet. Sci. Med. 2020, 8, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Moon, D.C.; Kim, S.-J.; Mechesso, A.F.; Song, H.-J.; Kang, H.Y.; Choi, J.-H.; Yoon, S.-S.; Lim, S.-K. Nationwide Surveillance on Antimicrobial Resistance Profiles of Enterococcus faecium and Enterococcus faecalis Isolated from Healthy Food Animals in South Korea, 2010 to 2019. Microorganisms 2021, 9, 925. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, M.; Santos, V.; Fernandes, A.; Bernardo, F.; Vilela, C.L. Antimicrobial Resistance and in Vitro Biofilm-Forming Ability of Enterococci from Intensive and Extensive Farming Broilers. Poult. Sci. 2010, 89, 1065–1069. [Google Scholar] [CrossRef] [PubMed]
- Makarov, D.A.; Ivanova, O.E.; Pomazkova, A.V.; Egoreva, M.A.; Prasolova, O.V.; Lenev, S.V.; Gergel, M.A.; Bukova, N.K.; Karabanov, S.Y. Antimicrobial Resistance of Commensal Enterococcus Faecalis and Enterococcus faecium from Food-Producing Animals in Russia. Vet. World 2022, 15, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Roy, K.; Islam, M.S.; Paul, A.; Ievy, S.; Talukder, M.; Sobur, M.A.; Ballah, F.M.; Khan, M.S.R.; Rahman, M.T. Molecular Detection and Antibiotyping of Multi-Drug Resistant Enterococcus faecium from Healthy Broiler Chickens in Bangladesh. Vet. Med. Sci. 2022, 8, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Sørum, M.; Holstad, G.; Lillehaug, A.; Kruse, H. Prevalence of Vancomycin Resistant Enterococci on Poultry Farms Established after the Ban of Avoparcin. Avian Dis. 2004, 48, 823–828. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.O.; Baptiste, K.E. Vancomycin-Resistant Enterococci: A Review of Antimicrobial Resistance Mechanisms and Perspectives of Human and Animal Health. Microb. Drug Resist. 2018, 24, 590–606. [Google Scholar] [CrossRef] [PubMed]
- Hassan, I.Z.; Wandrag, B.; Gouws, J.J.; Qekwana, D.N.; Naidoo, V. Antimicrobial Resistance and Mcr-1 Gene in Escherichia coli Isolated from Poultry Samples Submitted to a Bacteriology Laboratory in South Africa. Vet. World 2021, 14, 2662–2669. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, P.; Anjay, A.; Kumari, S.; Dayal, S.; Kumar, S. Antimicrobial Resistance and Molecular Characterisation of E. coli from Poultry in Eastern India. Vet. Ital. 2018, 54, 197–204. [Google Scholar] [CrossRef] [PubMed]
- van den Bogaard, A.E.; London, N.; Driessen, C.; Stobberingh, E.E. Antibiotic Resistance of Faecal Escherichia coli in Poultry, Poultry Farmers and Poultry Slaughterers. J. Antimicrob. Chemother. 2001, 47, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Majewski, M.; Józefiak, A.; Kimsa-Furdzik, M.; Dziubdziela, L.; Hudak-Nowak, M.; Wilczyński, J.; Anusz, K. Antimicrobial Resistance of Escherichia coli and Klebsiella spp. Conventionally Sampled from Factory-Farmed Chickens—Clinical Submissions. Ann. Agric. Environ. Med. 2021, 28, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Mandal, A.K.; Talukder, S.; Hasan, M.M.; Tasmim, S.T.; Parvin, M.S.; Ali, M.Y.; Islam, M.T. Epidemiology and Antimicrobial Resistance of Escherichia coli in Broiler Chickens, Farmworkers, and Farm Sewage in Bangladesh. Vet. Med. Sci. 2021, 8, 187–199. [Google Scholar] [CrossRef] [PubMed]
- Shaib, H.; Aoun, P.; Ghaddar, A.; Al Labadi, H.; Obeid, Y. Multidrug Resistance and Plasmid Profiles of Escherichia coli Isolated from Lebanese Broiler Farms. Int. J. Microbiol. 2023, 2023, 8811675. [Google Scholar] [CrossRef] [PubMed]
- Moffo, F.; Mouiche, M.M.M.; Djomgang, H.K.; Tombe, P.; Wade, A.; Kochivi, F.L.; Dongmo, J.B.; Mbah, C.K.; Mapiefou, N.P.; Mingoas, J.-P.K.; et al. Associations between Antimicrobial Use and Antimicrobial Resistance of Escherichia coli Isolated from Poultry Litter under Field Conditions in Cameroon. Prev. Vet. Med. 2022, 204, 105668. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, S.-Y.; Zhang, J.-X.; He, X.-X.; Xiong, W.-G.; Sun, Y.-X. Variations of Antibiotic Resistance Profiles in Chickens during Administration of Amoxicillin, Chlortetracycline and Florfenicol. J. Appl. Microbiol. 2018, 125, 1692–1701. [Google Scholar] [CrossRef] [PubMed]
- Much, P.; Sun, H.; Lassnig, H.; Koeberl-Jelovcan, S.; Schliessnig, H.; Stueger, H.P. Differences in Antimicrobial Resistance of Commensal Escherichia coli Isolated from Caecal Contents of Organically and Conventionally Raised Broilers in Austria, 2010–2014 and 2016. Prev. Vet. Med. 2019, 171, 104755. [Google Scholar] [CrossRef] [PubMed]
- Kempf, I.; Jouy, E.; Chauvin, C. Colistin Use and Colistin Resistance in Bacteria from Animals. Int. J. Antimicrob. Agents 2016, 48, 598–606. [Google Scholar] [CrossRef] [PubMed]
- Kimera, Z.I.; Mshana, S.E.; Rweyemamu, M.M.; Mboera, L.E.G.; Matee, M.I.N. Antimicrobial Use and Resistance in Food-Producing Animals and the Environment: An African Perspective. Antimicrob. Resist. Infect. Control 2020, 9, 37. [Google Scholar] [CrossRef] [PubMed]
- Founou, L.L.; Founou, R.C.; Essack, S.Y. Antimicrobial Resistance in the Farm-to-Plate Continuum: More Than a Food Safety Issue. Future Sci. OA 2021, 7, FSO692. [Google Scholar] [CrossRef] [PubMed]
- Hedman, H.D.; Vasco, K.A.; Zhang, L. A Review of Antimicrobial Resistance in Poultry Farming within Low-Resource Settings. Animals 2020, 10, 1264. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.; Salvatierra, G.; Dávila-Barclay, A.; Ayzanoa, B.; Castillo-Vilcahuaman, C.; Huang, M.; Pajuelo, M.J.; Lescano, A.G.; Cabrera, L.; Calderón, M.; et al. Market Chickens as a Source of Antibiotic-Resistant Escherichia coli in a Peri-Urban Community in Lima, Peru. Front. Microbiol. 2021, 12, 635871. [Google Scholar] [CrossRef] [PubMed]
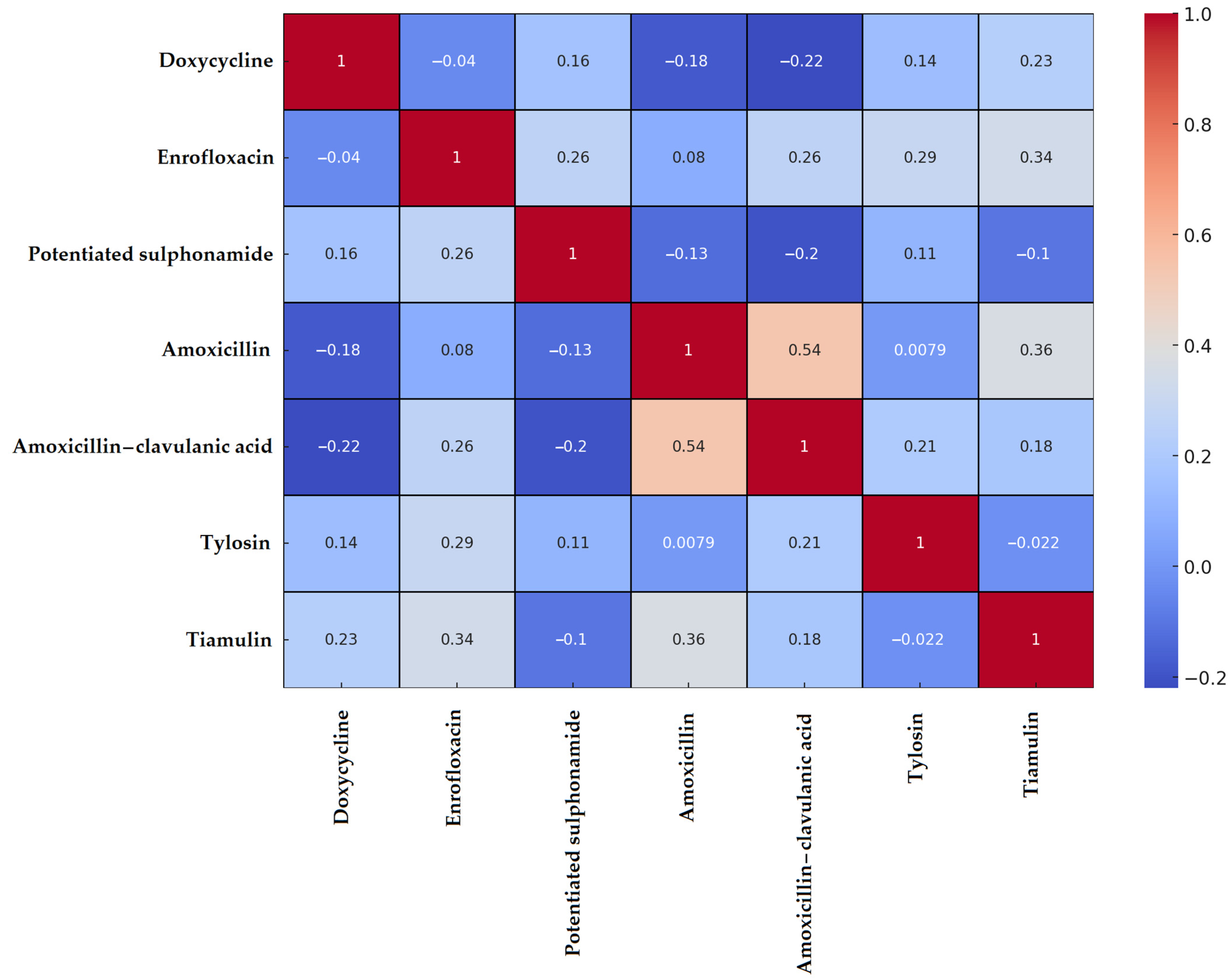
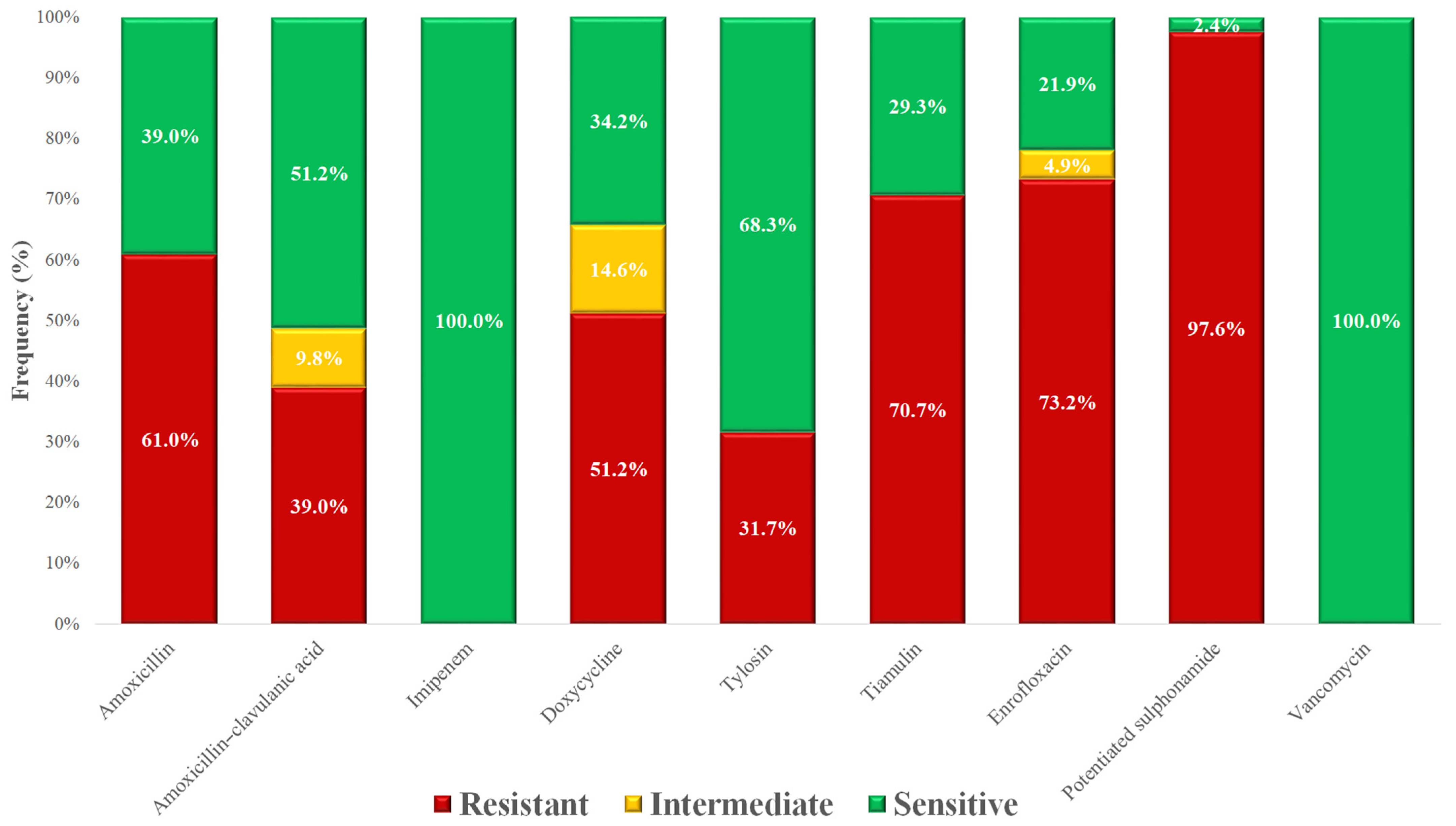
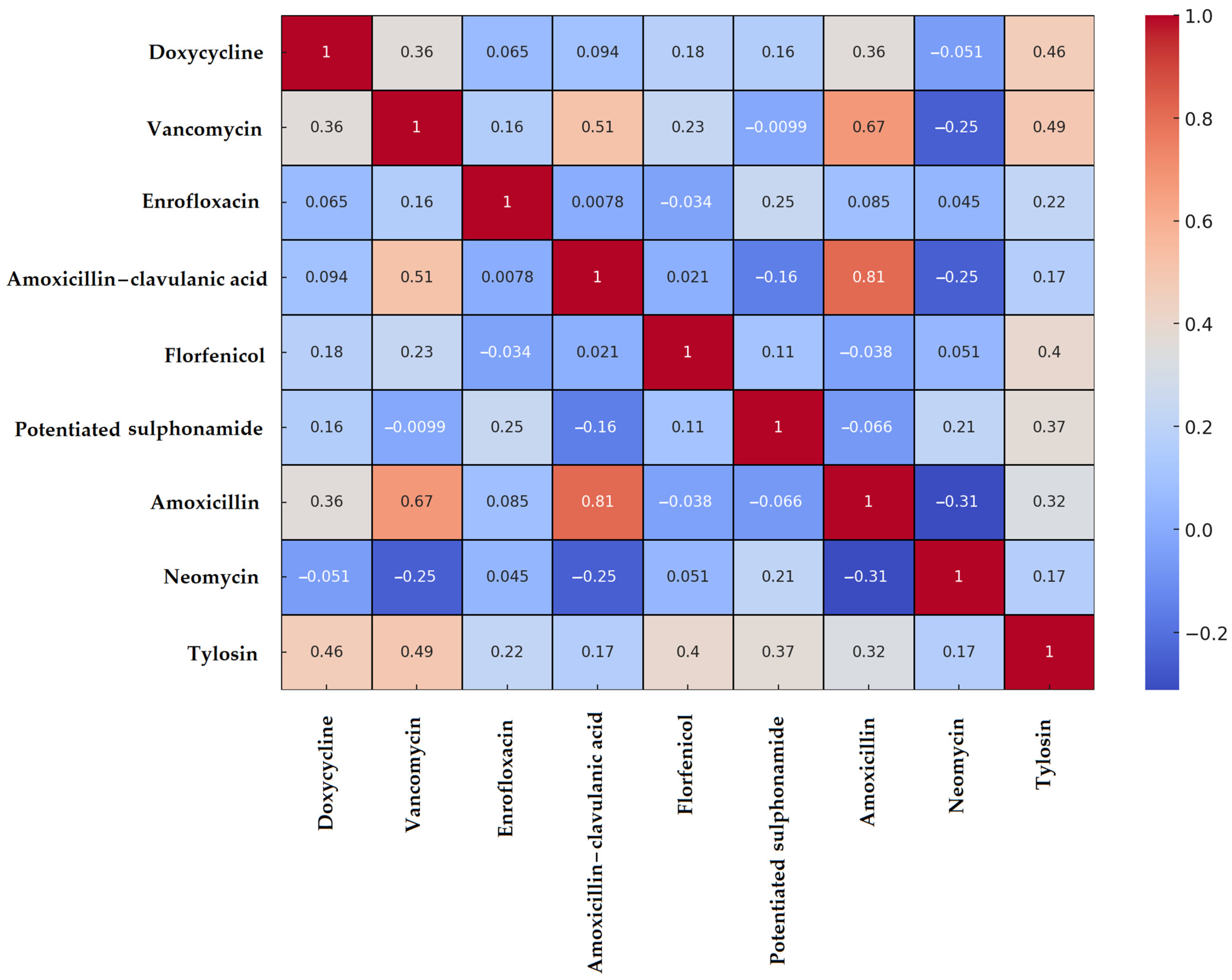

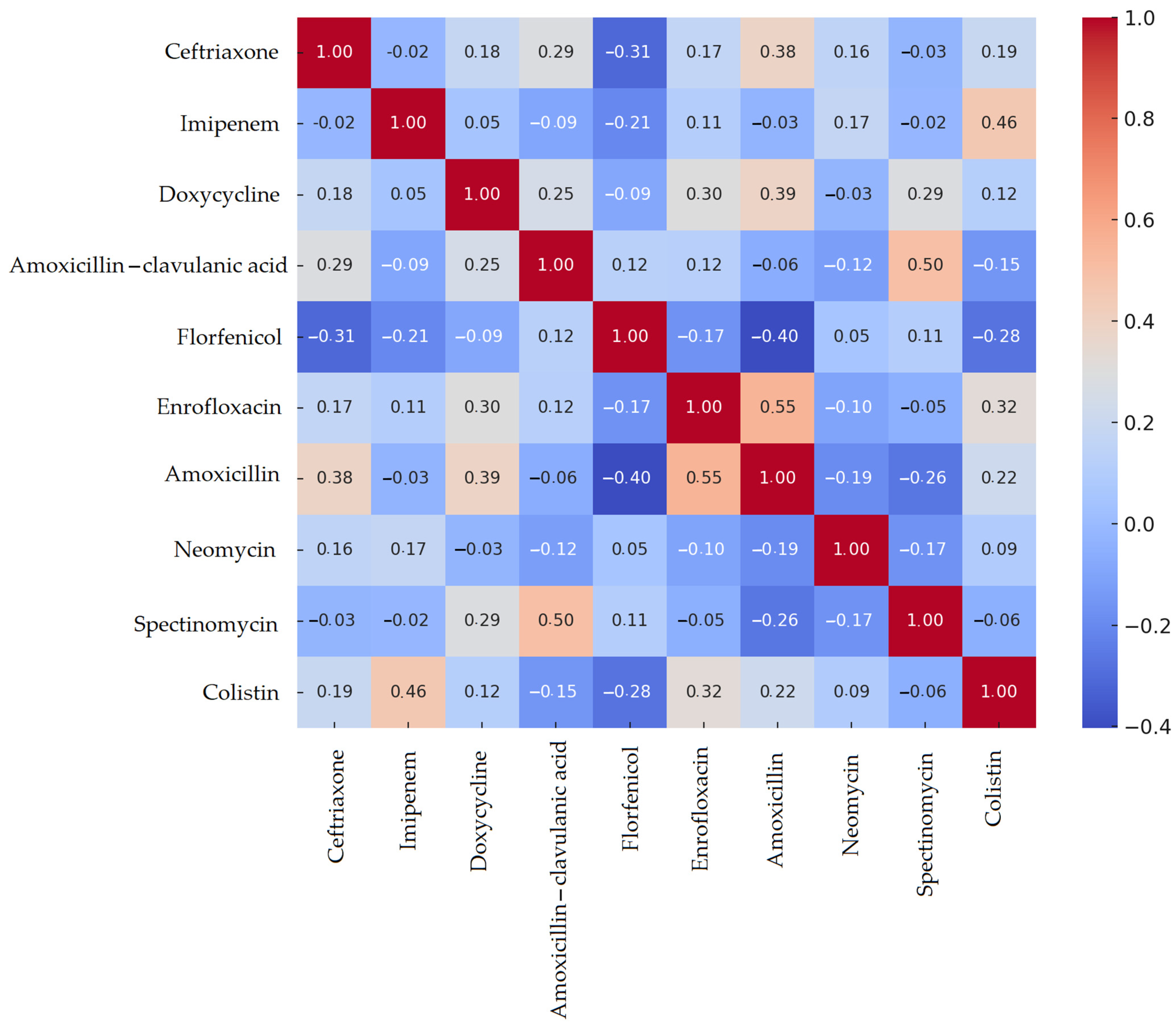


| Species | Number of Isolates |
|---|---|
| Staphylococcus spp. (n = 41) | |
| Staphylococcus aureus ssp. aureus | 32 |
| Staphylococcus delphini | 5 |
| Staphylococcus gallinarum | 4 |
| Enterococcus spp. (n = 59) | |
| Enterococcus faecalis | 26 |
| Enterococcus faecium | 21 |
| Enterococcus durans | 6 |
| Enterococcus gallinarum | 3 |
| Enterococcus hirae | 2 |
| Enterococcus mundtii | 1 |
| Escherichia coli (n = 45) | |
| Antibiotics | 1 Breakpoint | 0.001 | 0.002 | 0.004 | 0.008 | 0.016 | 0.031 | 0.063 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | 1024 | MIC50 | MIC90 | 2 ECOFF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| µg/mL | |||||||||||||||||||||||||
| Amoxicillin | 0.5 | 1 | 4 | 11 | 8 | 11 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0.5 | 2 | 0.5 | |||||||
| 2.4% | 9.8% | 26.8% | 19.5% | 26.8% | 4.9% | 2.4% | 2.4% | 2.4% | 0.0% | 0.0% | 0.0% | 0.0% | 2.4% | ||||||||||||
| 3 Amoxicillin–clavulanic acid | 1 | 2 | 1 | 5 | 13 | 4 | 13 | 0 | 3 | 0.25 | 1 | 0.5 | |||||||||||||
| 4.9% | 2.4% | 12.2% | 31.7% | 9.8% | 31.7% | 31.7% | 7.3% | ||||||||||||||||||
| Doxycycline | 0.5 | 2 | 1 | 2 | 0 | 5 | 0 | 2 | 2 | 6 | 5 | 0 | 1 | 4 | 1 | 2 | 0 | 3 | 5 | 0.5 | 128 | 0.5 | |||
| 4.9% | 2.4% | 4.9% | 0.0% | 12.2% | 0.0% | 4.9% | 4.9% | 14.6% | 12.2% | 0.0% | 2.4% | 9.8% | 2.4% | 4.9% | 0.0% | 7.3% | 12.2% | ||||||||
| Enrofloxacin | 4 | 1 | 2 | 3 | 2 | 1 | 2 | 2 | 9 | 2 | 9 | 2 | 5 | 1 | 1 | 1 | 8 | 64 | 0.5 | ||||||
| 2.4% | 4.9% | 7.3% | 4.9% | 2.4% | 4.9% | 4.9% | 22.0% | 4.9% | 22.0% | 4.9% | 12.2% | 2.4% | 2.4% | 2.4% | |||||||||||
| Imipenem | 8 | 5 | 7 | 9 | 12 | 7 | 1 | 0.25 | 1 | 0.125 | |||||||||||||||
| 12.2% | 17.1% | 22.0% | 29.3% | 17.1% | 2.4% | ||||||||||||||||||||
| 4 Potentiated sulphonamide | 4 | 1 | 0 | 0 | 8 | 6 | 10 | 0 | 2 | 3 | 11 | 64 | 1024 | 0.25 | |||||||||||
| 2.4% | 0.0% | 0.0% | 19.5% | 14.6% | 24.4% | 0.0% | 4.9% | 7.3% | 26.8% | ||||||||||||||||
| Tylosin | 64 | 1 | 0 | 0 | 1 | 5 | 0 | 2 | 5 | 4 | 1 | 3 | 5 | 0 | 1 | 0 | 1 | 0 | 0 | 12 | 4 | 1024 | 2 | ||
| 2.4% | 0.0% | 0.0% | 2.4% | 12.2% | 0.0% | 4.9% | 12.2% | 9.8% | 2.4% | 7.3% | 12.2% | 0.0% | 2.4% | 0.0% | 2.4% | 0.0% | 0.0% | 29.3% | |||||||
| Tiamulin | 4 | 2 | 3 | 0 | 3 | 4 | 6 | 2 | 4 | 1 | 15 | 0 | 0 | 0 | 1 | 16 | 64 | 2 | |||||||
| 4.9% | 7.3% | 0.0% | 7.3% | 9.8% | 14.6% | 4.9% | 9.8% | 2.4% | 36.6% | 0.0% | 0.0% | 0.0% | 2.4% | ||||||||||||
| Vancomycin | 32 | 1 | 17 | 10 | 8 | 5 | 0.5 | 2 | 2 | ||||||||||||||||
| 2.4% | 41.5% | 24.4% | 19.5% | 12.2% | |||||||||||||||||||||
| Antibiotics | 1 Breakpoint | 0.001 | 0.002 | 0.004 | 0.008 | 0.016 | 0.031 | 0.063 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | 1024 | MIC50 | MIC90 | 2 ECOFF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| µg/mL | |||||||||||||||||||||||||
| Amoxicillin | 16 | 1 | 13 | 16 | 17 | 2 | 0 | 1 | 3 | 0 | 2 | 1 | 1 | 2 | 1 | 128 | - | ||||||||
| 1.7% | 22.0% | 27.1% | 28.8% | 3.4% | 0.0% | 1.7% | 5.1% | 0.0% | 3.4% | 1.7% | 1.7% | 3.4% | |||||||||||||
| 3 Amoxicillin–clavulanic acid | 16 | 1 | 6 | 20 | 19 | 4 | 2 | 0 | 2 | 1 | 0 | 4 | 1 | 16 | - | ||||||||||
| 1.7% | 10.2% | 33.9% | 32.2% | 6.8% | 3.4% | 0.0% | 3.4% | 1.7% | 0.0% | 6.8% | |||||||||||||||
| Doxycycline | 16 | 3 | 1 | 3 | 0 | 0 | 0 | 3 | 0 | 3 | 6 | 21 | 8 | 8 | 1 | 1 | 1 | 4 | 16 | 1 | |||||
| 5.1% | 1.7% | 5.1% | 0.0% | 0.0% | 0.0% | 5.1% | 0.0% | 5.1% | 10.2% | 35.6% | 13.6% | 13.6% | 1.7% | 1.7% | 1.7% | ||||||||||
| Enrofloxacin | 4 | 3 | 0 | 9 | 29 | 8 | 2 | 4 | 0 | 1 | 0 | 0 | 2 | 0 | 1 | 0.5 | 4 | - | |||||||
| 5.1% | 0.0% | 15.3% | 49.2% | 13.6% | 3.4% | 6.8% | 0.0% | 1.7% | 0.0% | 0.0% | 3.4% | 0.0% | 1.7% | ||||||||||||
| Florfenicol | 8 | 6 | 21 | 24 | 5 | 2 | 1 | 8 | 16 | 8 | |||||||||||||||
| 10.2% | 35.6% | 40.7% | 8.5% | 3.4% | 1.7% | ||||||||||||||||||||
| Imipenem | 16 | 1 | 1 | 0 | 1 | 0 | 3 | 14 | 28 | 9 | 2 | 2 | 4 | 4 | |||||||||||
| 1.7% | 1.7% | 0.0% | 1.7% | 0.0% | 5.1% | 23.7% | 47.5% | 15.3% | 3.4% | ||||||||||||||||
| Neomycin | 1024 | 2 | 2 | 0 | 2 | 1 | 1 | 3 | 0 | 6 | 11 | 12 | 19 | 512 | 1024 | 256 | |||||||||
| 3.4% | 3.4% | 0.0% | 3.4% | 1.7% | 1.7% | 5.1% | 0.0% | 10.2% | 18.6% | 20.3% | 32.2% | ||||||||||||||
| 4 Potentiated sulphonamide | 64 | 1 | 6 | 6 | 8 | 15 | 8 | 6 | 3 | 1 | 1 | 0 | 4 | 8 | 128 | - | |||||||||
| 1.7% | 10.2% | 10.2% | 13.6% | 25.4% | 13.6% | 10.2% | 5.1% | 1.7% | 1.7% | 0.0% | 6.8% | ||||||||||||||
| Tylosin | 8 | 5 | 15 | 12 | 6 | 0 | 0 | 4 | 0 | 0 | 0 | 4 | 13 | 2 | 1024 | - | |||||||||
| 8.5% | 25.4% | 20.3% | 10.2% | 0.0% | 0.0% | 6.8% | 0.0% | 0.0% | 0.0% | 6.8% | 22.0% | ||||||||||||||
| Vancomycin | 32 | 1 | 1 | 8 | 12 | 16 | 8 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 7 | 2 | 1024 | 4 | |||||||
| 1.7% | 1.7% | 13.6% | 20.3% | 27.1% | 13.6% | 6.8% | 3.4% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | 11.9% | ||||||||||||
| Antibiotics | 1 Breakpoint | 0.001 | 0.002 | 0.004 | 0.008 | 0.016 | 0.031 | 0.063 | 0.125 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | 1024 | MIC50 | MIC90 | 2 ECOFF |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| µg/mL | |||||||||||||||||||||||||
| Amoxicillin | 32 | 1 | 1 | 1 | 14 | 7 | 1 | 0 | 1 | 1 | 5 | 9 | 4 | 8 | 512 | 8 | |||||||||
| 2.2% | 2.2% | 2.2% | 31.1% | 15.6% | 2.2% | 0.0% | 2.2% | 2.2% | 11.1% | 20.0% | 8.9% | ||||||||||||||
| 3 Amoxicillin-clavulanic acid | 32 | 4 | 16 | 9 | 13 | 3 | 0 | 0 | 0 | 0 | 0 | 8 | 16 | 8 | |||||||||||
| 8.9% | 35.6% | 20.0% | 28.9% | 6.7% | 0.0% | 0.0% | 0.0% | 0.0% | 0.0% | ||||||||||||||||
| Ceftriaxone | 4 | 1 | 1 | 5 | 6 | 12 | 6 | 2 | 1 | 1 | 0 | 0 | 1 | 2 | 1 | 2 | 1 | 2 | 1 | 0.063 | 64 | 0.125 | |||
| 2.2% | 2.2% | 11.1% | 13.3% | 26.7% | 13.3% | 4.4% | 2.2% | 2.2% | 0.0% | 0.0% | 2.2% | 4.4% | 2.2% | 4.4% | 2.2% | 4.4% | 2.2% | ||||||||
| Doxycycline | 16 | 8 | 10 | 3 | 5 | 11 | 8 | 16 | 64 | 8 | |||||||||||||||
| 17.8% | 22.2% | 6.7% | 11.1% | 24.4% | 17.8% | ||||||||||||||||||||
| Enrofloxacin | 2 | 3 | 3 | 5 | 0 | 1 | 5 | 8 | 4 | 2 | 5 | 7 | 1 | 1 | 1 | 16 | 0.125 | ||||||||
| 0.0% | 6.7% | 11.1% | 0.0% | 2.2% | 11.1% | 17.8% | 8.9% | 4.4% | 11.1% | 15.6% | 2.2% | 2.2% | |||||||||||||
| Florfenicol | 16 | 8 | 23 | 12 | 2 | 16 | 32 | 16 | |||||||||||||||||
| 17.8% | 51.1% | 26.7% | 4.4% | ||||||||||||||||||||||
| Imipenem | 4 | 1 | 0 | 11 | 0 | 3 | 2 | 9 | 8 | 6 | 3 | 2 | 0.5 | 4 | 0.5 | ||||||||||
| 2.2% | 0.0% | 24.4% | 0.0% | 6.7% | 4.4% | 20.0% | 17.8% | 13.3% | 6.7% | 4.4% | |||||||||||||||
| Colistin | 2 | 2 | 0 | 5 | 1 | 11 | 11 | 4 | 1 | 0 | 1 | 0 | 1 | 0 | 2 | 0 | 3 | 3 | 0.5 | 512 | 2 | ||||
| 4.4% | 0.0% | 11.1% | 2.2% | 24.4% | 24.4% | 8.9% | 2.2% | 0.0% | 2.2% | 0.0% | 2.2% | 0.0% | 4.4% | 0.0% | 6.7% | 6.7% | |||||||||
| Neomycin | 32 | 5 | 14 | 11 | 12 | 0 | 2 | 1 | 32 | 64 | 8 | ||||||||||||||
| 11.1% | 31.1% | 24.4% | 26.7% | 0.0% | 4.4% | 2.2% | |||||||||||||||||||
| 4 Potentiated sulphonamide | 4 | 1 | 7 | 4 | 7 | 1 | 0 | 8 | 0 | 7 | 10 | 128 | 1024 | 0.5 | |||||||||||
| 2.2% | 15.6% | 8.9% | 15.6% | 2.2% | 0.0% | 17.8% | 0.0% | 15.6% | 22.2% | ||||||||||||||||
| Spectinomycin | 128 | 4 | 31 | 10 | 64 | 128 | 64 | ||||||||||||||||||
| 8.9% | 68.9% | 22.2% | |||||||||||||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kerek, Á.; Szabó, Á.; Barnácz, F.; Csirmaz, B.; Kovács, L.; Jerzsele, Á. Antimicrobial Resistance in Commensal Bacteria from Large-Scale Chicken Flocks in the Dél-Alföld Region of Hungary. Vet. Sci. 2025, 12, 691. https://doi.org/10.3390/vetsci12080691
Kerek Á, Szabó Á, Barnácz F, Csirmaz B, Kovács L, Jerzsele Á. Antimicrobial Resistance in Commensal Bacteria from Large-Scale Chicken Flocks in the Dél-Alföld Region of Hungary. Veterinary Sciences. 2025; 12(8):691. https://doi.org/10.3390/vetsci12080691
Chicago/Turabian StyleKerek, Ádám, Ábel Szabó, Franciska Barnácz, Bence Csirmaz, László Kovács, and Ákos Jerzsele. 2025. "Antimicrobial Resistance in Commensal Bacteria from Large-Scale Chicken Flocks in the Dél-Alföld Region of Hungary" Veterinary Sciences 12, no. 8: 691. https://doi.org/10.3390/vetsci12080691
APA StyleKerek, Á., Szabó, Á., Barnácz, F., Csirmaz, B., Kovács, L., & Jerzsele, Á. (2025). Antimicrobial Resistance in Commensal Bacteria from Large-Scale Chicken Flocks in the Dél-Alföld Region of Hungary. Veterinary Sciences, 12(8), 691. https://doi.org/10.3390/vetsci12080691








