Investigation of Infection of Enterocytozoon bieneusi and Giardia duodenalis in Beef Cattle in Yunnan, China
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Sampling Area
2.2. DNA Extraction and PCR
2.3. PCR Amplification
2.4. Sequence Analysis
2.5. Statistical Analysis
3. Results
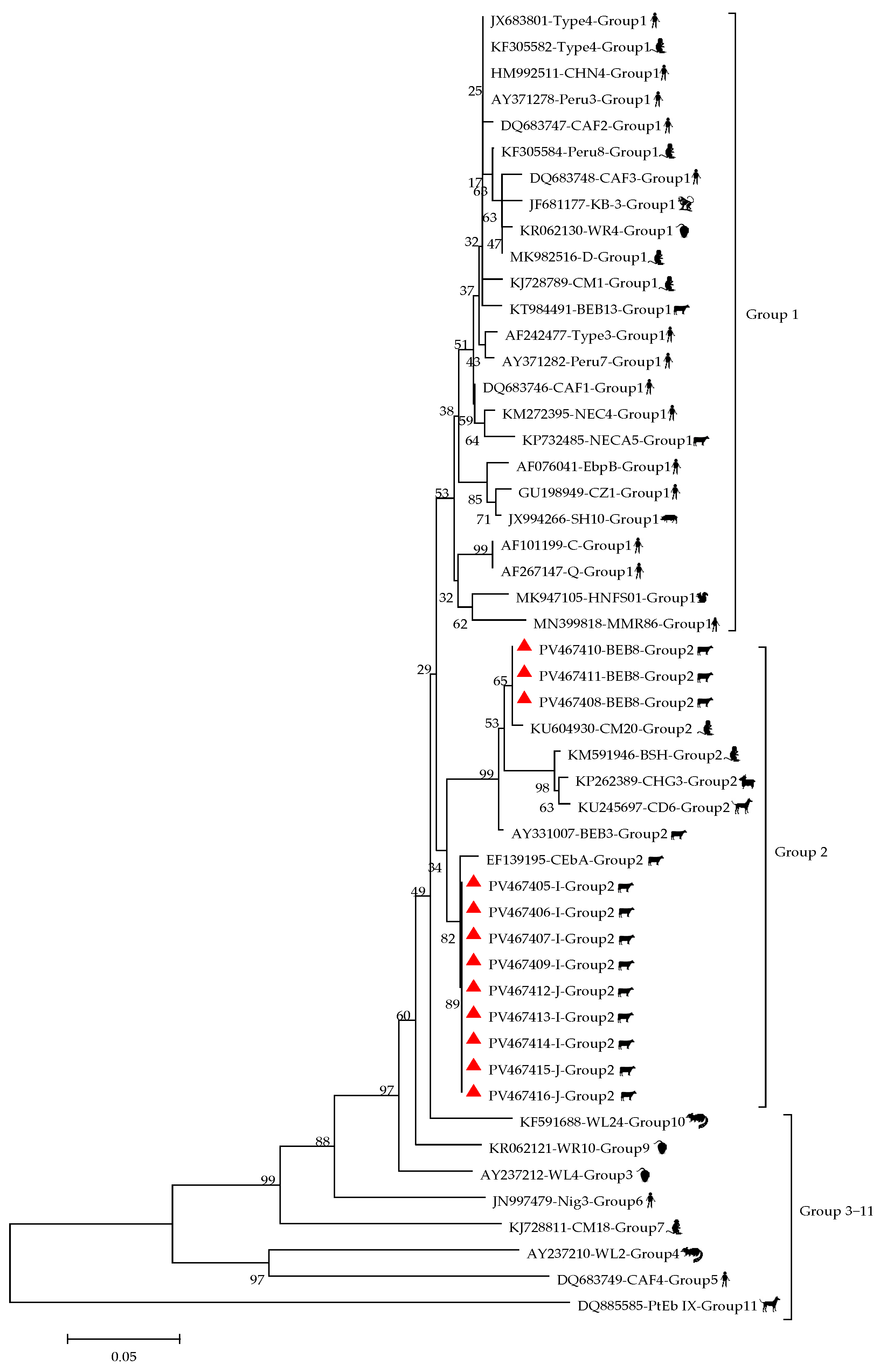
3.1. Prevalence and Genotypes of E. bieneusi
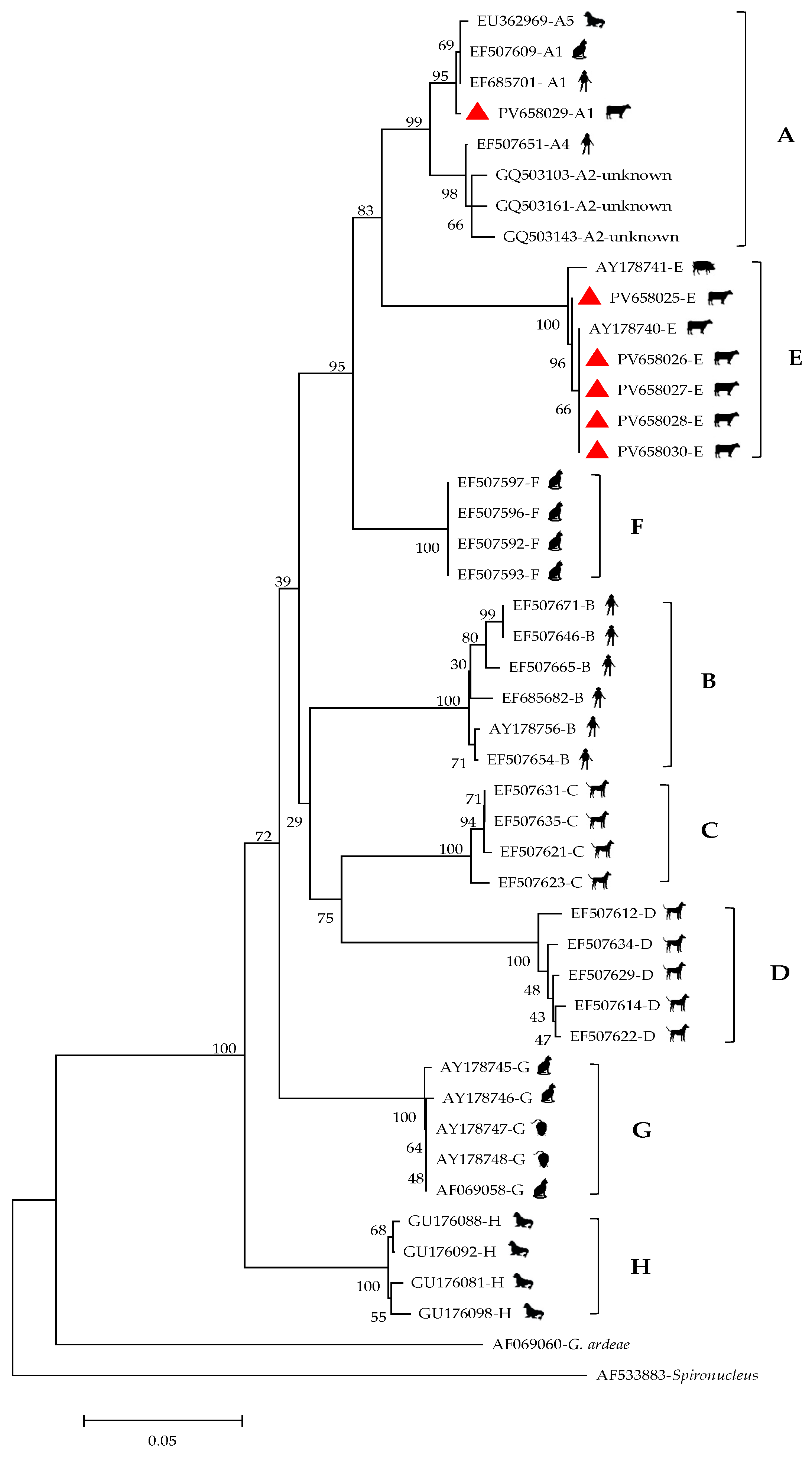
3.2. Prevalence and Genotypes of G. duodenalis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Matos, O.; Lobo, M.L.; Xiao, L. Epidemiology of Enterocytozoon bieneusi infection in humans. J. Parasitol. Res. 2012, 1, 981424. [Google Scholar]
- Stentiford, G.D.; Becnel, J.; Weiss, L.M.; Keeling, P.J.; Didier, E.S.; Williams, B.P.; Bjornson, S.; Kent, M.L.; Freeman, M.A.; Brown, M.J.F.; et al. Microsporidia–Emergent pathogens in the global food chain. Trends Parasitol. 2016, 32, 336–348. [Google Scholar] [CrossRef]
- Li, W.; Feng, Y.; Santin, M. Host Specificity of Enterocytozoon bieneusi and Public Health Implications. Trends Parasitol. 2019, 35, 436–451. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Feng, Y.; Xiao, L. Diagnosis and molecular typing of Enterocytozoon bieneusi: The significant role of domestic animals in transmission of human microsporidiosis. Res. Vet Sci. 2020, 133, 251–261. [Google Scholar] [CrossRef]
- Didier, E.S.; Weiss, L.M. Microsporidiosis: Not just in AIDS patients. Curr. Opin. Infect. Dis. 2011, 24, 490–495. [Google Scholar] [CrossRef]
- Lobo, M.L.; Xiao, L.H.; Antunes, F.; Matos, O. Microsporidia as emerging pathogens and the implication for public health: A 10-year study on HIV positive and negative patients. Int. J. Parasitol. 2012, 42, 197–205. [Google Scholar] [CrossRef]
- Galvan, A.L.; Sanchez, A.M.; Valentin, M.A.P.; Henriques-Gil, N.; Izquierdo, F.; Fenoy, S.; Del Aguila, C. First cases of microsporidiosis in transplant recipients in Spain and review of the literature. J. Clin. Microbiol. 2011, 49, 1301–1306. [Google Scholar] [CrossRef]
- Sak, B.; Brady, D.; Pelikánová, M.; Květoňová, D.; Rost, M.; Kostka, M.; Tolarová, V.; Hůzová, Z.; Kváč, M. Unapparent microsporidial infection among immunocompetent humans in the Czech Republic. J. Clin. Microbiol. 2011, 49, 1064–1070. [Google Scholar] [CrossRef] [PubMed]
- Nkinin, S.W.; Asonganyi, T.; Didier, E.S.; Kaneshiro, E.S. Microsporidian infection is prevalent in healthy people in Cameroon. J. Clin. Microbiol. 2007, 45, 2841–2846. [Google Scholar] [CrossRef]
- Cheng, C.; Cai, Y.; Xing, H.; Tao, J.; Cheng, D. Investigation of the Infection of Enterocytozoon bieneusi in Sheep and Goats in Jiangsu, China. Vet. Sci. 2024, 11, 327. [Google Scholar] [CrossRef]
- Meng, X.; Ou, Y.; Jiang, W.; Guo, Y.; Xiao, L.; Feng, Y.; Li, N. Identification of two new genetic loci for high-resolution genotyping of Enterocytozoon bieneusi. Parasite 2025, 32, 6. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Yu, S.; Feng, Y.; Zhang, L.; Santin, M.; Xiao, L.; Li, W. Widespread distribution of human-infective Enterocytozoon bieneusi genotypes in small rodents in northeast China and phylogeny and zoonotic implications revisited. Acta Trop. 2024, 253, 107160. [Google Scholar] [CrossRef] [PubMed]
- Santin, M.; Fayer, R. Enterocytozoon bieneusi genotype nomenclature based on the internal transcribed spacer sequence: A consensus. J. Eukaryot. Microbiol. 2009, 56, 34–38. [Google Scholar] [CrossRef]
- Zhang, Z.; Ma, J.; Huang, X.; Wen, X.; Jiang, W.; Chen, L.; Li, N.; Guo, Y.; Zhang, L.; Xiao, L.; et al. Population genetic analysis suggests genetic recombination is responsible for increased zoonotic potential of Enterocytozoon bieneusi from ruminants in China. One Health 2020, 11, 100184. [Google Scholar] [CrossRef]
- Jiang, Y.X.; Tao, W.; Wan, Q.; Li, Q.; Yang, Y.Q.; Lin, Y.; Zhang, S.; Li, W. Zoonotic and potentially host-adapted Enterocytozoon bieneusi genotypes in sheep and cattle in northeast China and an increasing concern about the zoonotic importance of previously considered ruminant-adapted genotypes. Appl. Environ. Microbiol. 2015, 81, 3326–3335. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, W.; Yang, F.; Zhang, L.; Wang, R.; Cao, J.; Shen, Y.; Liu, A. Enterocytozoon bieneusi in dairy cattle in the Northeast of China: Genetic diversity of ITS gene and evaluation of zoonotic transmission potential. J. Eukaryot. Microbiol. 2015, 62, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Del Coco, V.F.; Cordoba, M.A.; Bilbao, G.; Castro, P.D.; Basualdo, J.A.; Santín, M. First report of Enterocytozoon bieneusi from dairy cattle in Argentina. Vet. Parasitol. 2014, 199, 112–115. [Google Scholar] [CrossRef]
- Bulumulla, S.; Xiao, L.; Feng, Y.; Ash, A.; Ryan, U.; Barbosa, A.D. Update on transmission of zoonotic Giardia in cattle. Trends Parasitol. 2025, 41, 210–221. [Google Scholar] [CrossRef]
- Gao, Z.; Lu, Y.; Chong, Y.; Li, M.; Hong, J.; Wu, J.; Wu, D.; Xi, D.; Deng, W. Beef cattle genome project: Advances in genome sequencing, assembly, and functional genes discovery. Int. J. Mol. Sci. 2024, 25, 7147. [Google Scholar] [CrossRef]
- Song, H.Y.; Wang, K.S.; Yang, J.F.; Mao, H.M.; Pu, L.H.; Zou, Y.; Ma, J.; Zhu, X.Q.; Zou, F.C.; He, J.J. Prevalence and Novel Genotypes Identification of Enterocytozoon bieneusi in Dairy Cattle in Yunnan Province, China. Animals 2021, 11, 3014. [Google Scholar] [CrossRef]
- Heng, Z.J.; Yang, J.F.; Xie, X.Y.; Xu, C.R.; Chen, J.R.; Ma, J.; He, J.J.; Mao, H.M. Prevalence and multilocus genotyping of Giardia duodenalis in Holstein cattle in Yunnan, China. Front. Vet. Sci. 2022, 21, 949462. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.X.; Zou, Y.; Li, T.S.; Chen, H.; Wang, S.S.; Cao, F.Q.; Yang, J.F.; Sun, X.L.; Zhu, X.Q.; Zou, F.C. First report of the prevalence and genetic characterization of Giardia duodenalis and Cryptosporidium spp. in Yunling cattle in Yunnan Province, southwestern China. Microb. Pathog. 2021, 158, 105025. [Google Scholar] [CrossRef]
- Buckholt, M.A.; Lee, J.H.; Tzipori, S. Prevalence of Enterocytozoon bieneusi in swine: An 18-month survey at a slaughterhouse in Massachusetts. Appl. Environ. Microbiol. 2002, 68, 2595–2599. [Google Scholar] [CrossRef] [PubMed]
- Wegayehu, T.; Karim, M.R.; Ekro, B.; Zhang, L.; Tilahun, G. Multilocus genotyping of Giardia duodenalis isolates from calves in Oromia Special Zone, Central Ethiopia. Infect. Genet. Evol. 2016, 4, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Lasek-Nesselquist, E.; Welch, D.M.; Sogin, M.L. The identification of a new Giardia duodenalis assemblage in marine vertebrates and a preliminary analysis of G. duodenalis population biology in marine systems. Int. J. Parasitol. 2010, 40, 1063–1074. [Google Scholar] [CrossRef]
- Taghipour, A.; Bahadory, S.; Abdoli, A. A systematic review and meta-analysis on the global prevalence of cattle microsporidiosis with focus on Enterocytozoon bieneusi: An emerging zoonotic pathogen. Prev. Vet. Med. 2022, 200, 105581. [Google Scholar] [CrossRef]
- Qin, Y.; Chen, C.; Qin, Y.F.; Yang, X.B.; Li, M.H.; Meng, X.Z.; Zhao, Z.Y.; Ma, N.; Cai, Y.; Zhang, Y.; et al. Prevalence and related factors of Enterocytozoon bieneusi in cattle: A global systematic review and meta-analysis. Prev. Vet. Med. 2022, 208, 105775. [Google Scholar] [CrossRef]
- Ruan, Y.; Xu, X.; He, Q.; Li, L.; Guo, J.; Bao, J.; Pan, G.; Li, T.; Zhou, Z. The largest meta-analysis on the global prevalence of microsporidia in mammals, avian and water provides insights into the epidemic features of these ubiquitous pathogens. Parasit. Vectors 2021, 14, 186. [Google Scholar] [CrossRef]
- Qiu, L.; Xia, W.; Li, W.; Ping, J.; Ding, S.; Liu, H. The prevalence of microsporidia in China: A systematic review and meta-analysis. Sci. Rep. 2019, 9, 3174. [Google Scholar] [CrossRef]
- Wang, S.S.; Wang, R.J.; Fan, X.C.; Liu, T.L.; Zhang, L.X.; Zhao, G.H. Prevalence and genotypes of Enterocytozoon bieneusi in China. Acta Trop. 2018, 183, 142–152. [Google Scholar] [CrossRef]
- Li, J.; Luo, N.; Wang, C.; Qi, M.; Cao, J.; Cui, Z.; Huang, J.; Wang, R.; Zhang, L. Occurrence, molecular characterization and predominant genotypes of Enterocytozoon bieneusi in dairy cattle in Henan and Ningxia, China. Parasit. Vectors 2016, 9, 142. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, Z.; Su, Y.; Liang, X.; Sun, X.; Peng, S.; Lu, H.; Jiang, N.; Yin, J.; Xiang, M.; et al. Identification and genotyping of Enterocytozoon bieneusi in China. J. Clin. Microbiol. 2011, 49, 2006–2008. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, P.; Zhao, X.; Xu, H.; Wu, W.; Wang, Y.; Guo, Y.; Wang, L.; Feng, Y.; Xiao, L. Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Vet. Parasitol. 2015, 207, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Tao, W.F.; Ni, H.B.; Du, H.F.; Jiang, J.; Li, J.; Qiu, H.Y.; Li, Y.; Zhang, X.X. Molecular detection of Cryptosporidium and Enterocytozoon bieneusi in dairy calves and Sika deer in four provinces in Northern China. Parasitol. Res. 2020, 119, 105–114. [Google Scholar] [CrossRef]
- Tang, C.; Cai, M.; Wang, L.; Guo, Y.; Li, N.; Feng, Y.; Xiao, L. Genetic diversity within dominant Enterocytozoon bieneusi genotypes in pre-weaned calves. Parasit. Vectors 2018, 11, 170. [Google Scholar] [CrossRef]
- Wang, H.Y.; Qi, M.; Sun, M.F.; Li, D.F.; Wang, R.J.; Zhang, S.M.; Zhao, J.F.; Li, J.Q.; Cui, Z.H.; Chen, Y.C.; et al. Prevalence and Population Genetics Analysis of Enterocytozoon bieneusi in Dairy Cattle in China. Front. Microbiol. 2019, 10, 1399. [Google Scholar] [CrossRef]
- Liu, Y.Y.; Qin, R.L.; Mei, J.J.; Zou, Y.; Zhang, Z.H.; Zheng, W.B.; Liu, Q.; Zhu, X.Q.; Gao, W.W.; Xie, S.C. Molecular Detection and Genotyping of Enterocytozoon bieneusi in Beef Cattle in Shanxi Province, North China. Animals 2022, 12, 2961. [Google Scholar] [CrossRef]
- Wang, X.T.; Wang, R.J.; Ren, G.J.; Yu, Z.Q.; Zhang, L.X.; Zhang, S.Y.; Lu, H.; Peng, X.Q.; Zhao, G.H. Multilocus genotyping of Giardia duodenalis and Enterocytozoon bieneusi in dairy and native beef (Qinchuan) calves in Shaanxi Province, Northwestern China. Parasitol. Res. 2016, 115, 1355–1361. [Google Scholar] [CrossRef]
- Hu, S.; Liu, Z.; Yan, F.; Zhang, Z.; Zhang, G.; Zhang, L.; Jian, F.; Zhang, S.; Ning, C.; Wang, R. Zoonotic and host-adapted genotypes of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi in dairy cattle in Hebei and Tianjin, China. Vet. Parasitol. 2017, 248, 68–73. [Google Scholar] [CrossRef]
- Qi, M.; Jing, B.; Jian, F.; Wang, R.; Zhang, S.; Wang, H.; Ning, C.; Zhang, L. Dominance of Enterocytozoon bieneusi genotype J in dairy calves in Xinjiang, Northwest China. Parasitol. Int. 2017, 66, 960–963. [Google Scholar] [CrossRef]
- Xin, X.; Sun, L.; Liu, W.; Zhang, J.; Ma, S.; Fu, X.; Zhao, W.; Yan, B. Molecular prevalence and genotype identification of Enterocytozoon bieneusi in cattle and goats from Zhejiang Province, China. Front. Vet. Sci. 2024, 11, 1415813. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, N.; Jiang, W.; Guo, Y.; Wang, X.; Jin, Y.; Feng, Y.; Xiao, L. Infection patterns, clinical significance, and genetic characteristics of Enterocytozoon bieneusi and Giardia duodenalis in dairy cattle in Jiangsu, China. Parasitol. Res. 2019, 118, 3053–3060. [Google Scholar] [CrossRef]
- Zheng, X.L.; Zhou, H.H.; Ren, G.; Ma, T.M.; Cao, Z.X.; Wei, L.M.; Liu, Q.W.; Wang, F.; Zhang, Y.; Liu, H.L.; et al. Genotyping and zoonotic potential of Enterocytozoon bieneusi in cattle farmed in Hainan Province, the southernmost region of China. Parasite 2020, 27, 65. [Google Scholar] [CrossRef]
- Zhang, Q.; Cai, J.; Li, P.; Wang, L.; Guo, Y.; Li, C.; Lei, M.; Feng, Y.; Xiao, L. Enterocytozoon bieneusi genotypes in Tibetan sheep and Yaks. Parasitol. Res. 2018, 117, 721–727. [Google Scholar] [CrossRef]
- Li, S.; Wang, P.; Zhu, X.Q.; Zou, Y.; Chen, X.Q. Prevalence and genotypes/subtypes of Enterocytozoon bieneusi and Blastocystis sp. in different breeds of cattle in Jiangxi Province, southeastern China. Infect. Genet. Evol. 2022, 98, 105216. [Google Scholar] [CrossRef]
- Liu, X.; Tang, L.; Li, W.; Li, C.; Gu, Y. Prevalence And molecular characterization of Cryptosporidium spp. And Enterocytozoon bieneusi from large-scale cattle farms in Anhui Province, China. J. Vet. Med. Sci. 2022, 84, 40–47. [Google Scholar] [CrossRef]
- Ma, J.G.; Zhang, N.Z.; Hou, J.L.; Zou, Y.; Hu, G.X.; Zhu, X.Q.; Zhou, D.H. Detection of Enterocytozoon bieneusi in white Yaks in Gansu Province, China. BioMed Res. Int. 2017, 2017, 5790181. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Qin, H.; Sun, M.; Fu, Y.; Lang, J.; Zhang, A.; Qin, Z.; Guo, Z.; Xu, H.; Li, X.; et al. Occurrence and genotypic identification of Blastocystis sp., Enterocytozoon bieneusi, and Giardia duodenalis in dairy cattle in Heilongjiang Province, China. Parasitol. Int. 2024, 100, 102871. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Chen, Y.; Chang, Y.; Zhang, X.; Li, D.; Wang, L.; Zheng, S.; Wang, R.; Zhang, S.; Li, J.; et al. Genotyping and identification of Cryptosporidium spp., Giardia duodenalis and Enterocytozoon bieneusi from free-range Tibetan yellow cattle and cattle–yak in Tibet, China. Acta Trop. 2020, 212, 105671. [Google Scholar] [CrossRef]
- Trout, J.M.; Santin, M.; Greiner, E.; Fayer, R. Prevalence and genotypes of Giardia duodenalis in post-weaned dairy calves. Vet. Parasitol. 2005, 130, 177–183. [Google Scholar] [CrossRef]
- Bartley, P.M.; Roehe, B.K.; Thomson, S.; Shaw, H.J.; Peto, F.; Innes, E.A.; Katzer, F. Detection of potentially human infectious assemblages of Giardia duodenalis in fecal samples from beef and dairy cattle in Scotland. Parasitology 2019, 146, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Santin, M.; Trout, J.M.; Fayer, R. A longitudinal study of Giardia duodenalis genotypes in dairy cows from birth to 2 years of age. Vet. Parasitol. 2009, 162, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.M.; Debnath, C.; Pramanik, A.K.; Xiao, L.; Nozaki, T.; Ganguly, S. Molecular evidence for zoonotic transmission of Giardia duodenalis among dairy farm workers in West Bengal, India. Vet. Parasitol. 2011, 178, 342–345. [Google Scholar] [CrossRef]
- Muhid, A.; Robertson, I.; Ng, J.; Yang, R.; Ryan, U. Prevalence of Giardia spp. infection in pre-weaned and weaned calves in relation to management factors. Vet. J. 2012, 191, 135–137. [Google Scholar] [CrossRef]
- Abeywardena, H.; Jex, A.R.; Nolan, M.J.; Haydon, S.R.; Stevens, M.A.; McAnulty, R.W.; Gasser, R.B. Genetic characterisation of Cryptosporidium and Giardia from dairy calves: Discovery of species/genotypes consistent with those found in humans. Infect. Genet. Evol. 2012, 12, 1984–1993. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.F.; Zhou, L.; Zhang, A.H.; Hou, M.R.; Liu, X.W.; Zhang, X.H.; Wang, J.W.; Wang, X.; Bai, X.; Jiao, C.L.; et al. Prevalence and Molecular Characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in Cattle in Heilongjiang Province, Northeast China. Animals 2024, 14, 1635. [Google Scholar] [CrossRef]
- Zhao, L.; Chai, H.L.; Wang, M.Y.; Zhang, Z.S.; Han, W.X.; Yang, B.; Wang, Y.; Zhang, S.; Zhao, W.H.; Ma, Y.M.; et al. Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle in Central Inner Mongolia, Northern China. BMC Vet. Res. 2023, 19, 134. [Google Scholar] [CrossRef]
- Dan, J.; Zhang, X.; Ren, Z.; Wang, L.; Cao, S.; Shen, L.; Deng, J.; Zuo, Z.; Yu, S.; Wang, Y.; et al. Occurrence and multilocus genotyping of Giardia duodenalis from post-weaned dairy calves in Sichuan province, China. PLoS ONE 2019, 14, e0224627. [Google Scholar] [CrossRef]
- Fu, Y.; Dong, H.; Bian, X.; Qin, Z.; Han, H.; Lang, J.; Zhang, J.; Zhao, G.; Li, J.; Zhang, L. Molecular characterizations of Giardia duodenalis based on multilocus genotyping in sheep, goats, and beef cattle in Southwest Inner Mongolia, China. Parasite 2022, 29, 33. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, T.; Koehler, A.V.; Hu, M.; Gasser, R.B. Molecular investigation of Cryptosporidium and Giardia in pre- and post-weaned calves in Hubei Province, China. Parasites Vectors 2017, 10, 519. [Google Scholar] [CrossRef]
- Qi, M.; Wang, H.; Jing, B.; Wang, R.; Jian, F.; Ning, C.; Zhang, L. Prevalence and multilocus genotyping of Giardia duodenalis in dairy calves in Xinjiang, Northwestern China. Parasites Vectors 2016, 9, 546. [Google Scholar] [CrossRef] [PubMed]
- Jian, Y.; Zhang, X.; Li, X.; Karanis, G.; Ma, L.; Karanis, P. Prevalence and molecular characterization of Giardia duodenalis in cattle and sheep from the Qinghai-Tibetan Plateau Area (QTPA), northwestern China. Vet. Parasitol. 2018, 250, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhao, G.; Chen, G.; Jian, F.; Zhang, S.; Feng, C.; Wang, R.; Zhu, J.; Dong, H.; Hua, J.; et al. Multilocus genotyping of Giardia duodenalis in dairy cattle in Henan, China. PLoS ONE 2014, 9, e100453. [Google Scholar] [CrossRef]
- Liu, G.; Su, Y.; Zhou, M.; Zhao, J.; Zhang, T.; Ahmad, W.; Lu, H.; Jiang, N.; Chen, Q.; Xiang, M.; et al. Prevalence and molecular characterization of Giardia duodenalis isolates from dairy cattle in northeast China. Exp. Parasitol. 2015, 154, 20–24. [Google Scholar] [CrossRef]
- Cui, Z.; Wang, L.; Cao, L.; Sun, M.; Liang, N.; Wang, H.; Chang, Y.; Lin, X.; Yu, L.; Wang, R.; et al. Genetic characteristics and geographic segregation of Giardia duodenalis in dairy cattle from Guangdong Province, southern China. Infect. Genet. Evol. 2018, 66, 95–100. [Google Scholar] [CrossRef]
- Huang, J.; Yue, D.; Qi, M.; Wang, R.; Zhao, J.; Li, J.; Shi, K.; Wang, M.; Zhang, L. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Ningxia, northwestern China. BMC Vet. Res. 2014, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, J.; Chang, Y.; Yu, F.; Zhang, S.; Wang, R.; Zhang, L. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Gansu, northwest China. Parasite 2020, 27, 62. [Google Scholar] [CrossRef]
- Thellier, M.; Breton, J. Enterocytozoon bieneusi in human and animals, focus on laboratory identification and molecular epidemiology, Paris. Parasite 2008, 15, 349–358. [Google Scholar] [CrossRef]
- Karim, M.R.; Wang, R.; Dong, H.; Zhang, L.; Li, J.; Zhang, S.; Rume, F.I.; Qi, M.; Jian, F.; Sun, M.; et al. Genetic polymorphism and zoonotic potential of Enterocytozoon bieneusi from nonhuman primates in China. Appl. Environ. Microbiol. 2014, 80, 1893–1898. [Google Scholar] [CrossRef]
- Karim, M.R.; Wang, R.; He, X.; Zhang, L.; Li, J.; Rume, F.I.; Dong, H.; Qi, M.; Jian, F.; Zhang, S.; et al. Multilocus sequence typing of Enterocytozoon bieneusi in nonhuman primates in China. Vet. Parasitol. 2014, 200, 13–23. [Google Scholar] [CrossRef]
- Yu, F.; Li, D.; Chang, Y.; Wu, Y.; Guo, Z.; Jia, L.; Xu, J.; Li, J.; Qi, M.; Wang, R.; et al. Molecular characterization of three intestinal protozoans in hospitalized children with different disease backgrounds in Zhengzhou, central China. Parasites Vectors 2019, 12, 543. [Google Scholar] [CrossRef]
- Yu, F.; Wu, Y.; Li, T.; Cao, J.; Wang, J.; Hu, S.; Zhu, H.; Zhang, S.; Wang, R.; Ning, C.; et al. High prevalence of Enterocytozoon bieneusi zoonotic genotype D in captive golden snub-nosed monkey (Rhinopithecus roxellanae) in zoos in China. BMC Vet. Res. 2017, 13, 158. [Google Scholar] [CrossRef]
- Zhang, X.X.; Cong, W.; Liu, G.H.; Ni, X.T.; Ma, J.G.; Zheng, W.B.; Zhao, Q.; Zhu, X.Q. Prevalence and genotypes of Enterocytozoon bieneusi in sika deer in Jilin Province, Northeastern China. Acta Parasitol. 2016, 61, 382–388. [Google Scholar] [CrossRef]
- Santín, M.; Fayer, R. Enterocytozoon bieneusi, Giardia, and Cryptosporidium Infecting White-tailed Deer. J. Eukaryot. Microbiol. 2015, 62, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Reete, J.; Rinder, H.; Thomschke, A.; Manke, H.; Schwebs, M.; Bruderek, A. First detection of the microsporidium Enterocytozoon bieneusi in non-mammalian hosts (chickens). Int. J. Parasitol. 2002, 32, 785–787. [Google Scholar]
- Pirestani, M.; Sadraei, J.; Forouzandeh, M. Molecular characterization and genotyping of human related microsporidia in free-ranging and captive pigeons of Tehran, Iran. Infect. Genet. Evol. 2013, 20, 495–499. [Google Scholar] [CrossRef]
- Santin, M.; Dargatz, D.; Fayer, R. Prevalence and genotypes of Enterocytozoon bieneusi in weaned beef calves on cow-calf operations in the USA. Parasitol. Res. 2012, 110, 2033–2041. [Google Scholar] [CrossRef] [PubMed]
- Nourrisson, C.; Lavergne, R.A.; Moniot, M.; Morio, F.; Poirier, P. Enterocytozoon bieneusi, a human pathogen. Emerg. Microbes Infect. 2024, 13, 2406276. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.J.; Zhang, S.; Wang, L.F.; Ding, Y.L.; Liu, H.X.; Wang, M.Y.; Wang, Y.; Chai, H.L.; Zhang, Z.S.; Yi, C.; et al. Prevalence and genotyping of Enterocytozoon bieneusi in cattle from Shanxi and Inner Mongolia, China. Sci. Rep. 2025, 15, 6818. [Google Scholar] [CrossRef]
- Ma, J.; Cai, J.; Ma, J.; Feng, Y.; Xiao, L. Enterocytozoon bieneusi genotypes in Yaks (Bos grunniens) and their public health potential. J. Eukaryot. Microbiol. 2015, 62, 21–25. [Google Scholar] [CrossRef]
- Li, S.; Zou, Y.; Zhang, X.L.; Wang, P.; Chen, X.Q.; Zhu, X.Q. Prevalence and Multilocus Genotyping of Giardia lamblia in Cattle in Jiangxi Province, China: Novel Assemblage E Subtypes Identified. Korean J. Parasitol. 2020, 58, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Ning, X.; Yue, Z.; Jian, F.; Li, D.; Lang, J.; Lu, S.; Ning, C. Unveiling the presence and genotypic diversity of Giardia duodenalis on large-scale sheep farms: Insights from the Henan and Ningxia Regions, China. Parasit. Vectors 2024, 17, 312. [Google Scholar] [CrossRef]
- Abdel-Moein, K.A.; Saeed, H. The zoonotic potential of Giardia intestinalis assemblage E in rural settings. Parasitol. Res. 2016, 115, 3197–3202. [Google Scholar] [CrossRef]
- Fantinatti, M.; Bello, A.R.; Fernandes, O.; Da-Cruz, A.M. Identification of Giardia lamblia assemblage E in humans points to a new anthropozoonotic cycle. J. Infect. Dis. 2016, 214, 1256–1259. [Google Scholar] [CrossRef] [PubMed]
- Garcia, R.J.; Ogbuigwe, P.; Pita, A.B.; Velathanthiri, N.; Knox, M.A.; Biggs, P.J.; French, N.P.; Hayman, D.T.S. First report of novel assemblages and mixed infections of Giardia duodenalis in human isolates from New Zealand. Acta Trop. 2021, 220, 105969. [Google Scholar] [CrossRef]
- Li, J.; Wang, H.; Wang, R.; Zhang, L. Giardia duodenalis Infections in Humans and Other Animals in China. Front. Microbiol. 2017, 8, 2004. [Google Scholar] [CrossRef]
- Feng, Y.; Xiao, L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin. Microbiol. Rev. 2011, 24, 110–140. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Ryan, U.; Xiao, L.; Feng, Y. Zoonotic giardiasis: An update. Parasitol. Res. 2021, 120, 4199–4218. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, X.; Zhu, H.; Zhang, L.; Feng, Y.; Jian, F.; Ning, C.; Qi, M.; Zhou, Y.; Fu, K.; et al. Genetic characterizations of Cryptosporidium spp. and Giardia duodenalis in humans in Henan, China. Exp. Parasitol. 2011, 127, 42–45. [Google Scholar] [CrossRef]
- Sousa, M.C.; Morais, J.B.; Machado, J.E.; Poiares-da-Silva, J. Genotyping of Giardia lamblia human isolates from Portugal by PCR-RFLP and sequencing. J. Eukaryot. Microbiol. 2006, 53, S174–S176. [Google Scholar] [CrossRef]
- Lalle, M.; Jimenez-Cardosa, E.; Caccio, S.M.; Pozio, E. Genotyping of Giardia duodenalis from humans and dogs from Mexico using a beta-giardin nested polymerase chain reaction assay. J. Parasitol. 2005, 91, 203–205. [Google Scholar] [CrossRef] [PubMed]
- Volotao, A.C.; Costa-Macedo, L.M.; Haddad, F.S.; Brandao, A.; Peralta, J.M.; Fernandes, O. Genotyping of Giardia duodenalis from human and animal samples from Brazil using beta-giardin gene: A phylogenetic analysis. Acta Trop. 2007, 102, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Ryan, U.; Zahedi, A.; Paparini, A. Cryptosporidium in humans and animals-a one health approach to prophylaxis. Parasite Immunol. 2016, 38, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.W.; Shu, F.F.; Pu, L.H.; Zou, Y.; Yang, J.F.; Zou, F.C.; Zhu, X.Q.; Li, Z.; He, J.J. Occurrence and Molecular Characterization of Cryptosporidium spp. in Dairy Cattle and Dairy Buffalo in Yunnan Province, Southwest China. Animals 2022, 12, 1031. [Google Scholar] [CrossRef]
- Deng, M.L.; Heng, Z.J.; Li, L.J.; Yang, J.F.; He, J.J.; Zou, F.C.; Shu, F.F. Cryptosporidium spp. Infection and Genotype Identification in Pre-Weaned and Post-Weaned Calves in Yunnan Province, China. Animals 2024, 14, 1907. [Google Scholar] [CrossRef]
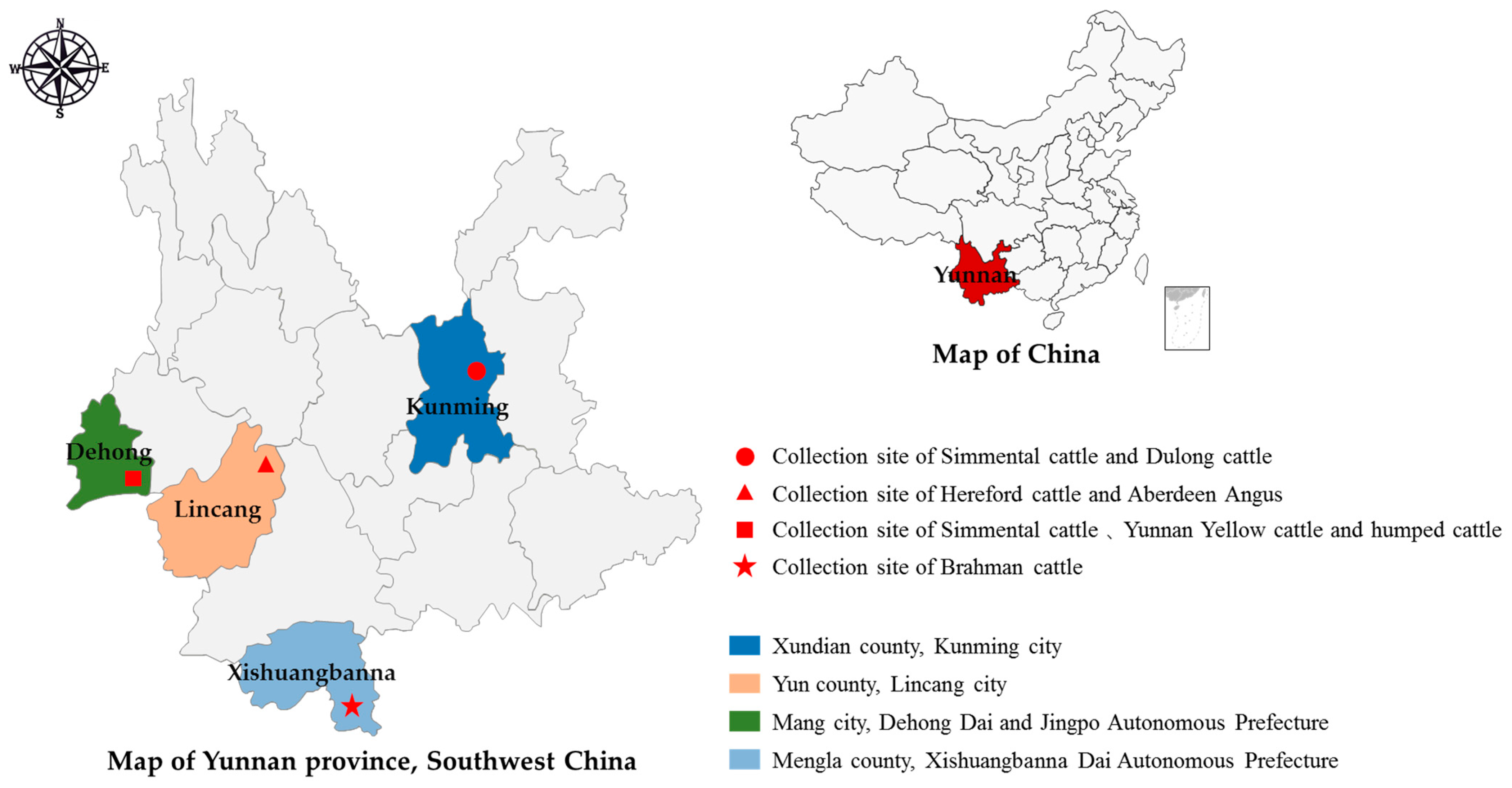
| Location | Geographical Coordinates | Altitude (m) | No. of Samples | Total | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Simmental Cattle | Brahman Cattle | Aberdeen Angus Cattle | Yunnan Yellow Cattle | Humped Cattle | Dulong Cattle | Hereford Cattle | ||||
| Dehong | 98°48′ E, 24°33′ N | 835–2800 | 87 | - | - | 39 | 36 | - | - | 162 |
| Kunming | 102°91′ E, 25°63′ N | 2100–2900 | 129 | - | - | - | - | 31 | - | 160 |
| Xishuangbanna | 101°55′ E, 21°52′ N | 640–1200 | - | 106 | - | - | - | - | - | 106 |
| Lincang | 99°95′ E, 24°14′ N | 740–1400 | - | - | 80 | - | - | - | 21 | 101 |
| Total | - | - | 216 | 106 | 80 | 39 | 36 | 31 | 21 | 529 |
| Species | Loci | Primer ID | Primer Sequences (5′-3′) | Fragment Length (bp) | Temperature Annealing (°C) | Reference |
|---|---|---|---|---|---|---|
| Enterocytozoon bieneusi | ITS gene | EB-F1 | GGTCATAGGGATGAAGAG | 392 | 55 °C | [23] |
| EB-R1 | TTCGAGTTCTTTCGCGCTC | |||||
| EB-F2 | GCTCTGAATATCTATGGCT | |||||
| EB-R2 | ATCGCCGACGGATCCAAGTG | |||||
| Giardia duodenalis | gdh gene | gdh-F1 | TTCCGTRTYCAGTACAACTC | 530 | 59 °C | [24] |
| gdh-R1 | ACCTCGTTCTGRGTGGCGCA | |||||
| gdh-F2 | ATGACYGAGCTYCAGAGGCACGT | |||||
| gdh-R2 | GTGGCGCARGGCATGATGCA |
| Variable | Category | No. Tested | No. Positive | Prevalence (%) (95% CI) | OR (95%, CI) | p-Value | Genotype |
| Region | Kunming | 160 | 7 | 4.4 (1.17–7.58) | 1.49 (0.38–5.92) | 0.207 | I (4), BEB8 (3) |
| Dehong | 162 | 6 | 3.7 (0.76–6.64) | 1.26 (0.31–5.14) | J (3), I (2), BEB4 (1) | ||
| Xishuangbanna | 106 | - | - | - | - | ||
| Lincang | 101 | 3 | 3.0 (−0.40–6.34) | Reference | I (2), J (1) | ||
| Breed | Simmental cattle | 216 | 12 | 5.6 (2.48–8.63) | 2.06 (0.26–16.34) | 0.088 | I (6), J (3), BEB8 (3) |
| Brahman cattle | 106 | - | - | - | - | ||
| Aberdeen Angus | 80 | 3 | 3.8 (−0.50–8.00) | 1.36 (0.14–13.58) | I (2), J (1) | ||
| Yunnan Yellow cattle | 39 | - | - | - | - | ||
| Humped cattle | 36 | 1 | 2.8 (−2.86–8.42) | Reference | BEB4 (1) | ||
| Dulong cattle | 31 | - | - | - | - | ||
| Hereford cattle | 21 | - | - | - | - | ||
| Gender | Female | 248 | 10 | 4.0 (1.57–6.50) | 1.93 (0.69–5.38) | 0.204 | I (5), J (2), BEB8 (3) |
| Male | 281 | 6 | 2.1 (0.43–3.84) | Reference | I (3),J (2), BEB4 (1) | ||
| Age | Pre-weaned (0–60 days) | 29 | 6 | 20.7 (5.01–36.37) | 31.39 (7.37–133.64) | <0.001 | I (4), BEB8 (2) |
| Post-weaned (61–180 days) | 62 | 2 | 3.2 (−1.30–7.75) | 4.01 (0.66–24.51) | J (1), BEB8 (1) | ||
| Juvenile cattle (7–18 months) | 74 | 5 | 6.8 (0.90–12.61) | 8.72 (2.04–37.34) | J (3), I (2) | ||
| Adult cattle (>18 months) | 364 | 3 | 0.8 (−0.11–1.76) | Reference | I (2), BEB4 (1) | ||
| Total | 529 | 16 | 3.0 (1.56–4.49) | - | - | I (8), J (4), BEB8 (3), BEB4 (1) | |
| Variable | Category | No. Tested | No. Positive | Prevalence (%) (95% CI) | OR (95%, CI) | p-Value | Assemblages |
|---|---|---|---|---|---|---|---|
| Region | Kunming | 160 | 12 | 7.5 (3.37–11.63) | 8.51 (1.09–66.48) | <0.001 | E (12) |
| Dehong | 162 | - | - | - | - | ||
| Xishuangbanna | 106 | 1 | 0.9 (−0.93–2.81) | Reference | E (1) | ||
| Lincang | 101 | 6 | 5.9 (1.25–10.63) | 6.63 (0.78–56.09) | E (5), A (1) | ||
| Breed | Simmental cattle | 216 | 12 | 5.6 (2.48–8.63) | 6.18 (0.79–48.15) | 0.116 | E (12) |
| Brahman cattle | 106 | 1 | 0.9 (−0.93–2.81) | Reference | E (1) | ||
| Aberdeen Angus | 80 | 5 | 6.3 (0.83–11.67) | 7.00 (0.80–61.15) | E (4), A (1) | ||
| Yunnan Yellow cattle | 39 | - | - | - | - | ||
| humped cattle | 36 | - | - | - | - | ||
| Dulong cattle | 31 | - | - | - | - | ||
| Hereford cattle | 21 | 1 | 4.8 (−5.17–14.70) | 5.25 (0.32–87.44) | E (1) | ||
| Gender | Female | 248 | 11 | 4.4 (1.86–7.02) | 1.58 (0.63–4.00) | 0.327 | E (11) |
| Male | 281 | 8 | 2.8 (0.89–4.80) | Reference | E (7), A (1) | ||
| Age | Pre-weaned (0–60 days) | 29 | 7 | 24.1 (7.57–40.70) | 28.64 (7.79–105.25) | <0.001 | E (7) |
| Post-weaned (61–180 days) | 62 | 4 | 6.5 (0.16–12.74) | 6.21 (1.51–25.51) | E (3), A (1) | ||
| Juvenile cattle (7–18 months) | 74 | 4 | 5.4 (0.13–10.68) | 5.14 (1.26–21.05) | E (4) | ||
| Adult cattle (>18 months) | 364 | 4 | 1.1 (0.02–2.17) | Reference | E (4) | ||
| Total | 529 | 19 | 3.6 (2.00–5.18) | - | - | E (18), A (1) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, F.; Cheng, W.; Yang, J.; He, J.; Li, L.; Zou, F.; Shu, F. Investigation of Infection of Enterocytozoon bieneusi and Giardia duodenalis in Beef Cattle in Yunnan, China. Vet. Sci. 2025, 12, 552. https://doi.org/10.3390/vetsci12060552
Yang F, Cheng W, Yang J, He J, Li L, Zou F, Shu F. Investigation of Infection of Enterocytozoon bieneusi and Giardia duodenalis in Beef Cattle in Yunnan, China. Veterinary Sciences. 2025; 12(6):552. https://doi.org/10.3390/vetsci12060552
Chicago/Turabian StyleYang, Fan, Wenjie Cheng, Jianfa Yang, Junjun He, Liujia Li, Fengcai Zou, and Fanfan Shu. 2025. "Investigation of Infection of Enterocytozoon bieneusi and Giardia duodenalis in Beef Cattle in Yunnan, China" Veterinary Sciences 12, no. 6: 552. https://doi.org/10.3390/vetsci12060552
APA StyleYang, F., Cheng, W., Yang, J., He, J., Li, L., Zou, F., & Shu, F. (2025). Investigation of Infection of Enterocytozoon bieneusi and Giardia duodenalis in Beef Cattle in Yunnan, China. Veterinary Sciences, 12(6), 552. https://doi.org/10.3390/vetsci12060552






