Etiological Detection, Isolation, and Pathogenicity of Porcine Reproductive and Respiratory Syndrome Virus in China
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Sample Collection and Treatment
2.2. RNA Extraction and RT-PCR Detection
2.3. Virus Isolation and Growth Curve Detection
2.4. Indirect Immunofluorescence Assay (IFA)
2.5. Multiple Alignment, Phylogenetic, and Recombination Analyses
2.6. Animal Experiments
2.7. Statistical Analyses
3. Results
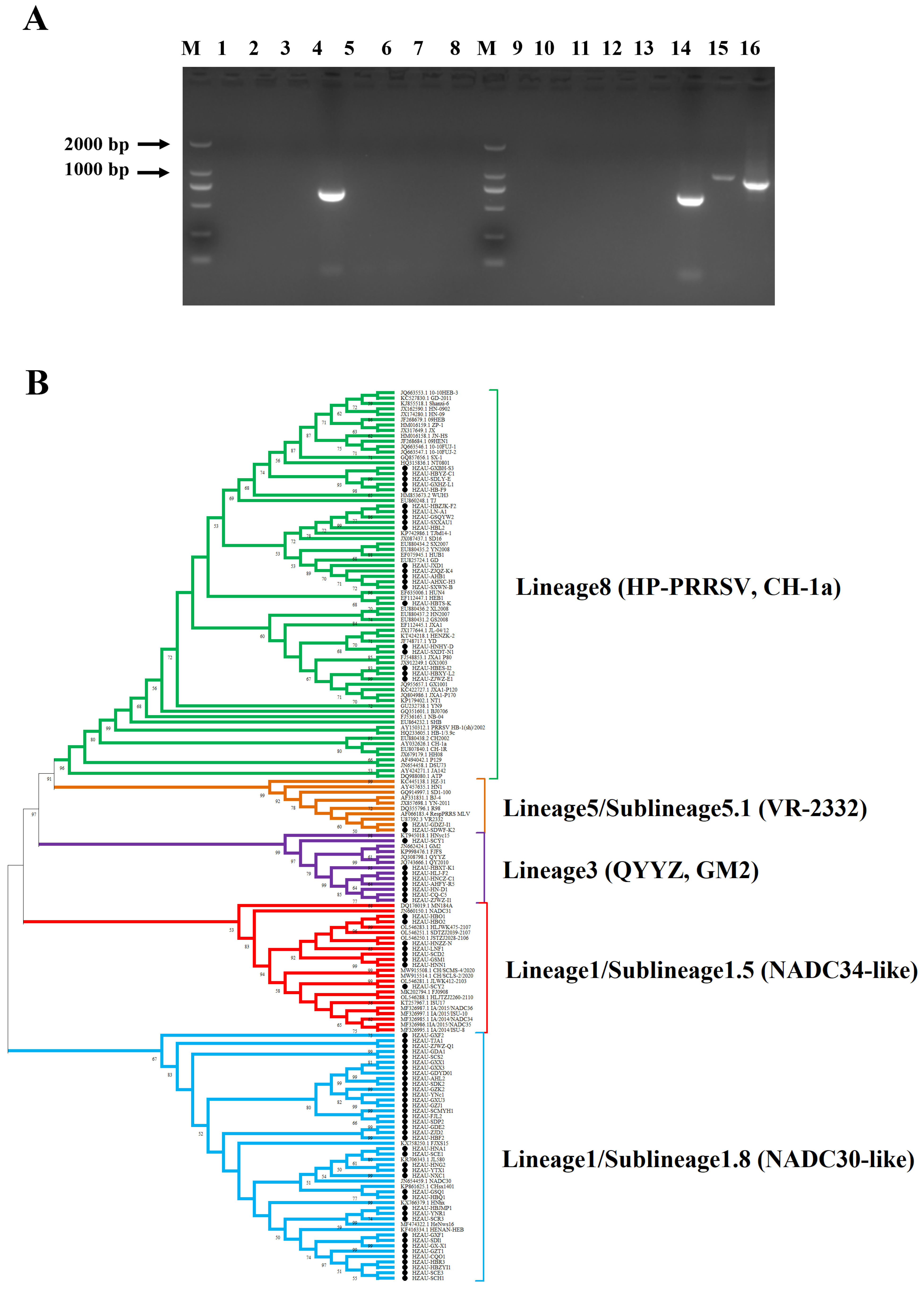
3.1. RT-PCR Detection and Phylogenetic Analysis of PRRSV in China
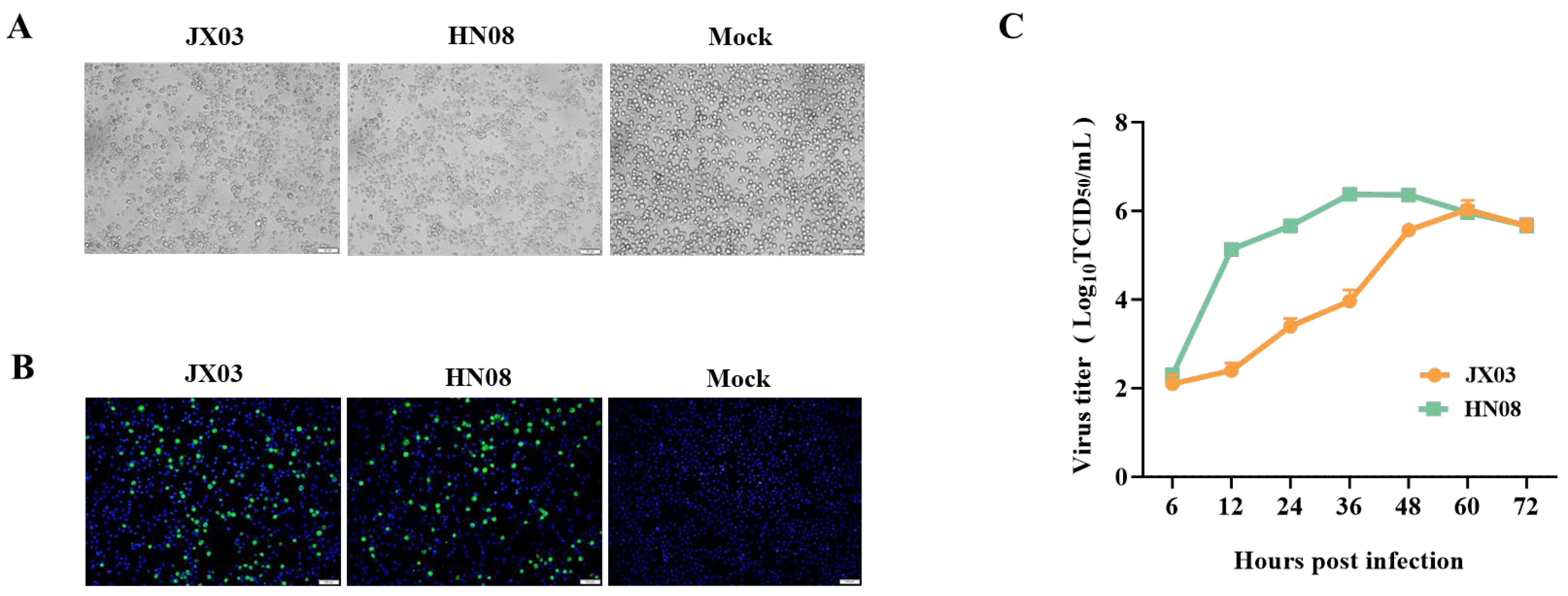
3.2. Isolation and Identification of PRRSV Epidemic Strains
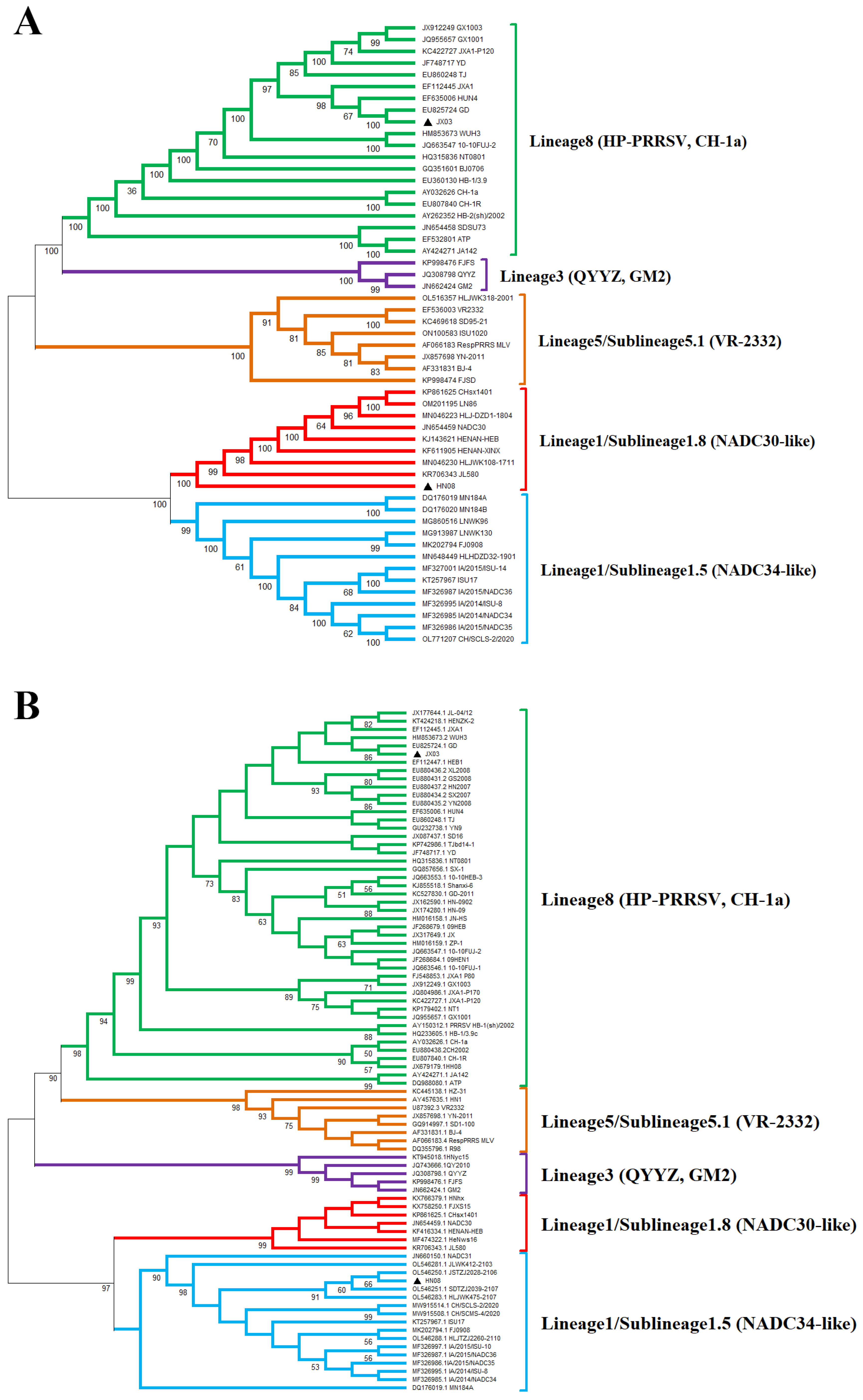
3.3. Whole-Genome Characterization and Phylogenetic Analysis of PRRSV Isolates
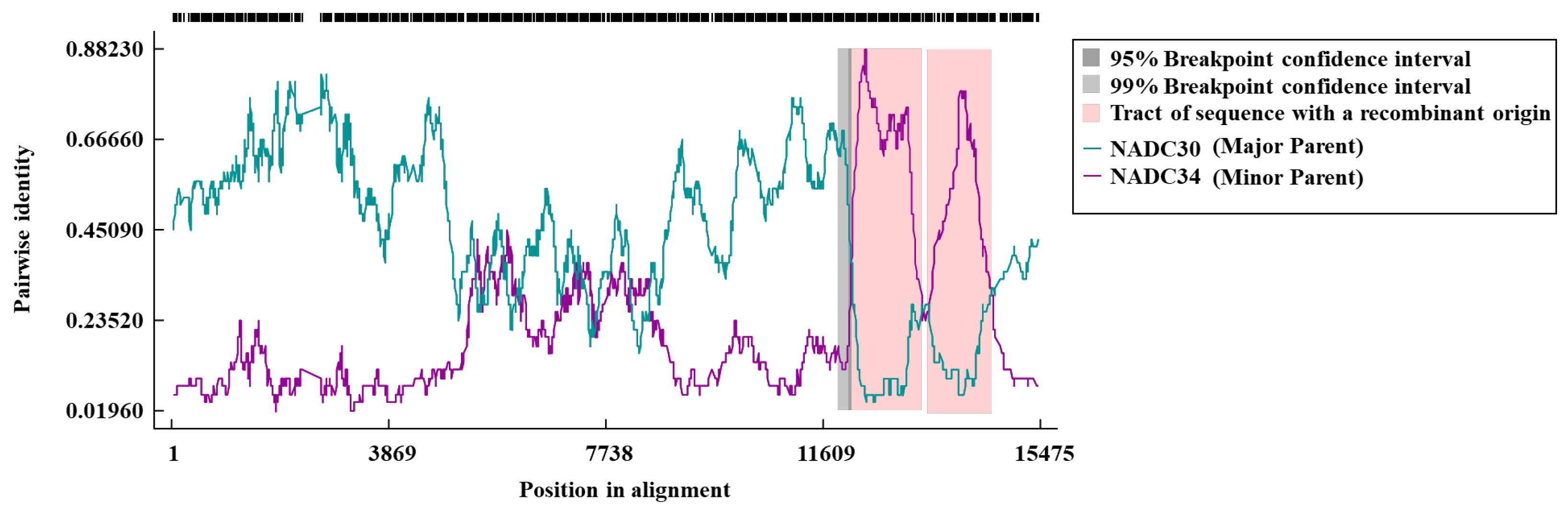
3.4. Gene Recombination Analysis of HN08
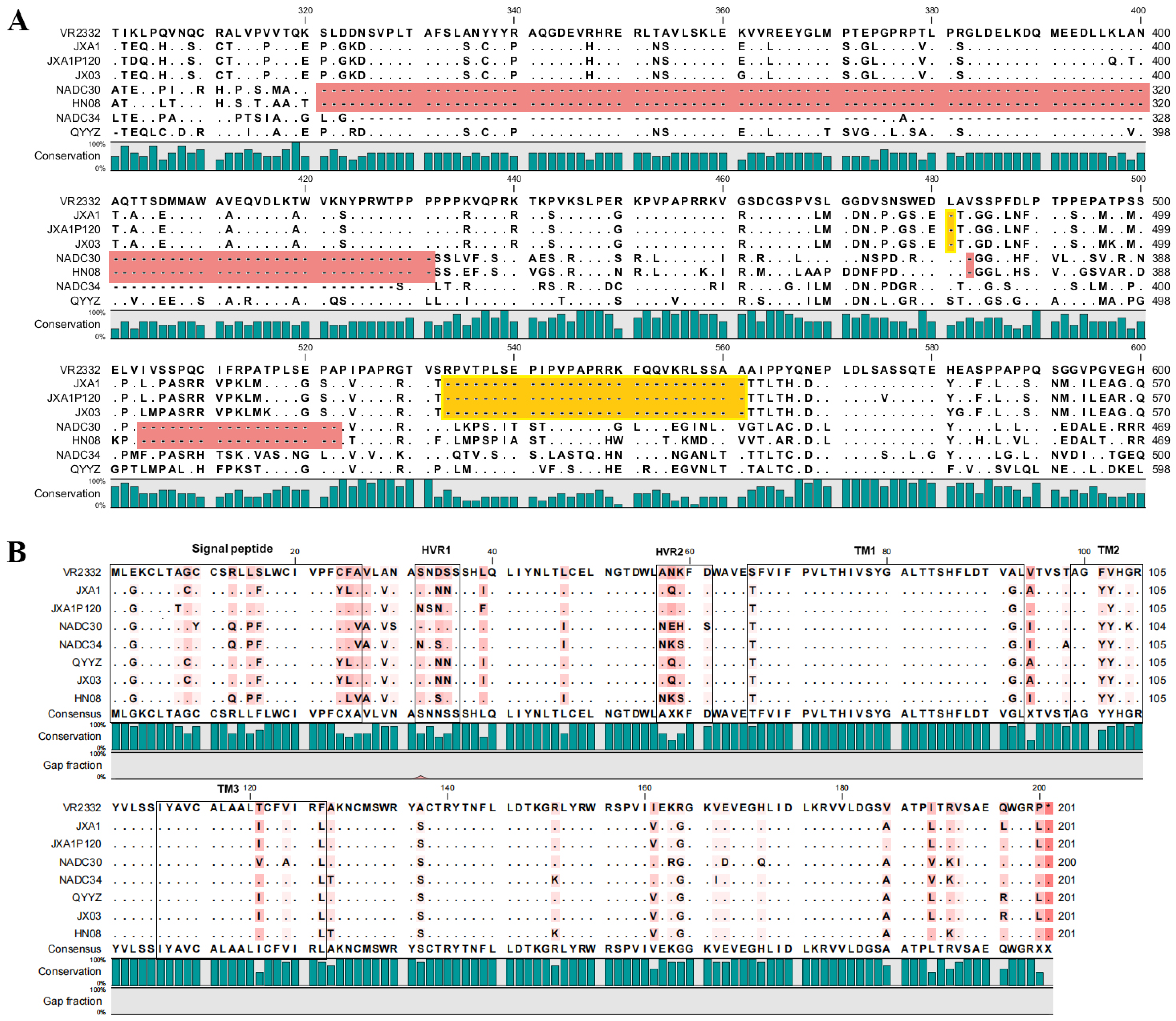
3.5. Amino Acid Variations in nsp2 and GP5 of PRRSV Isolates
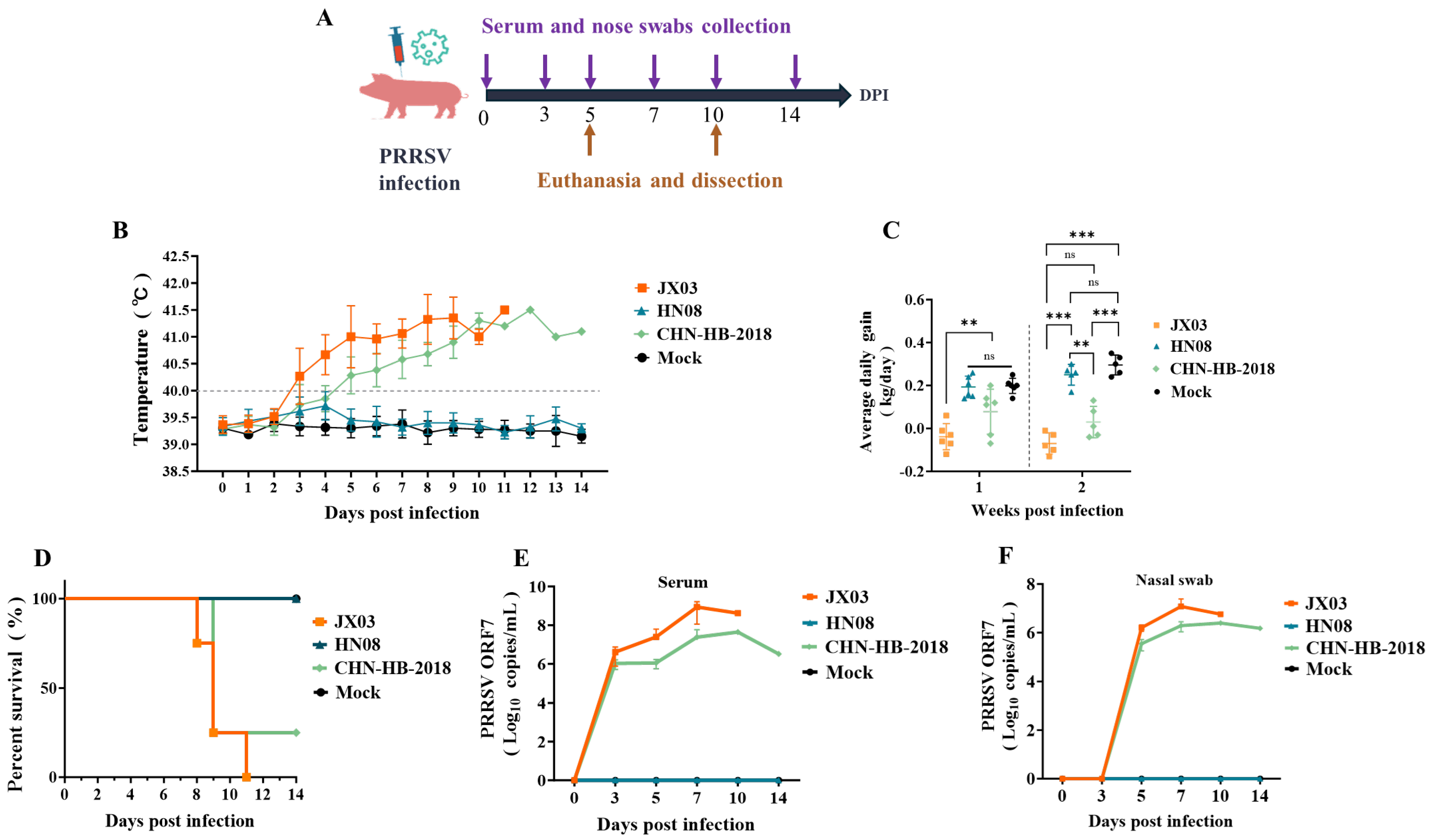
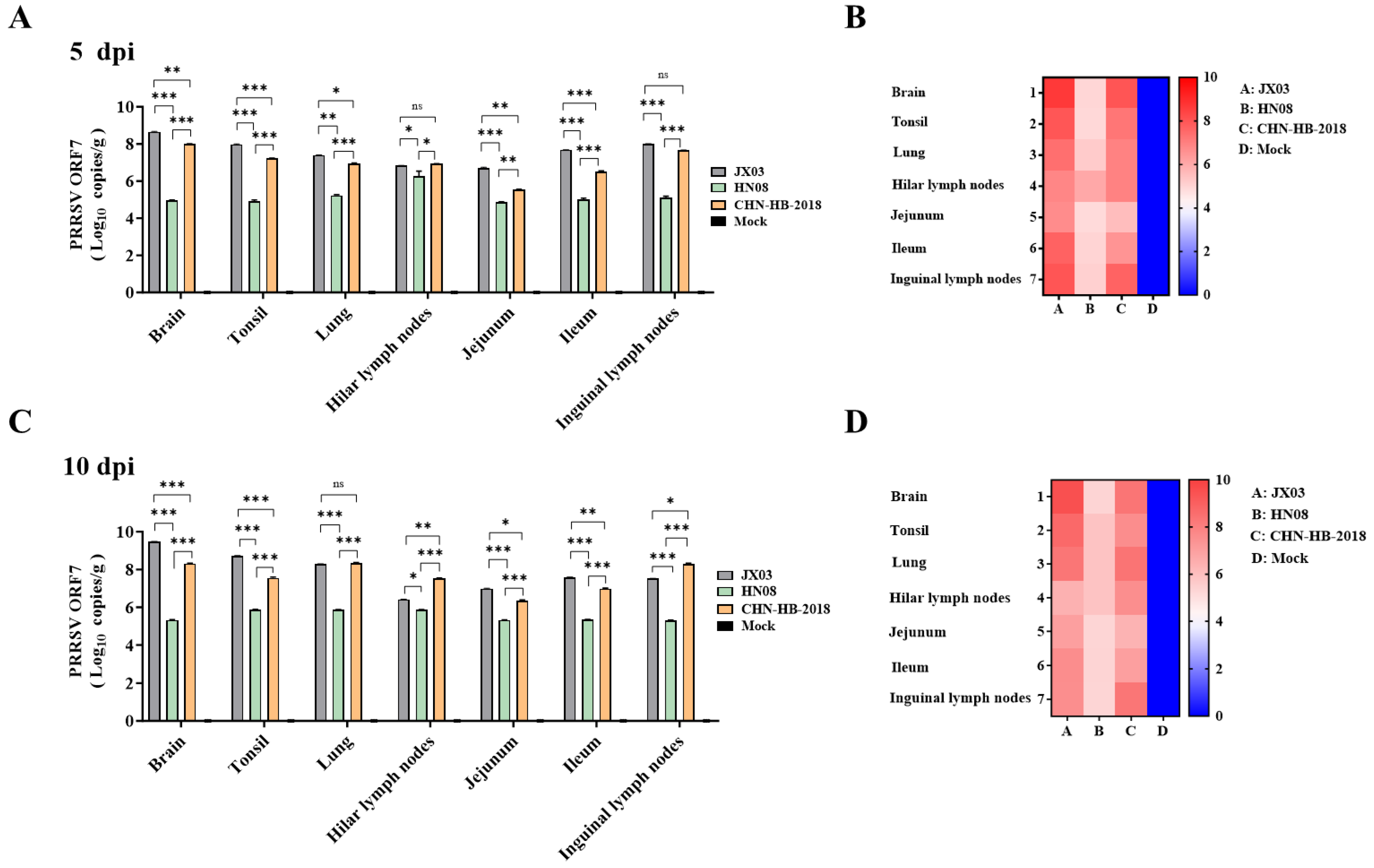
3.6. Analysis of Clinical Symptoms and Viral Loads in Piglets Infected with PRRSV
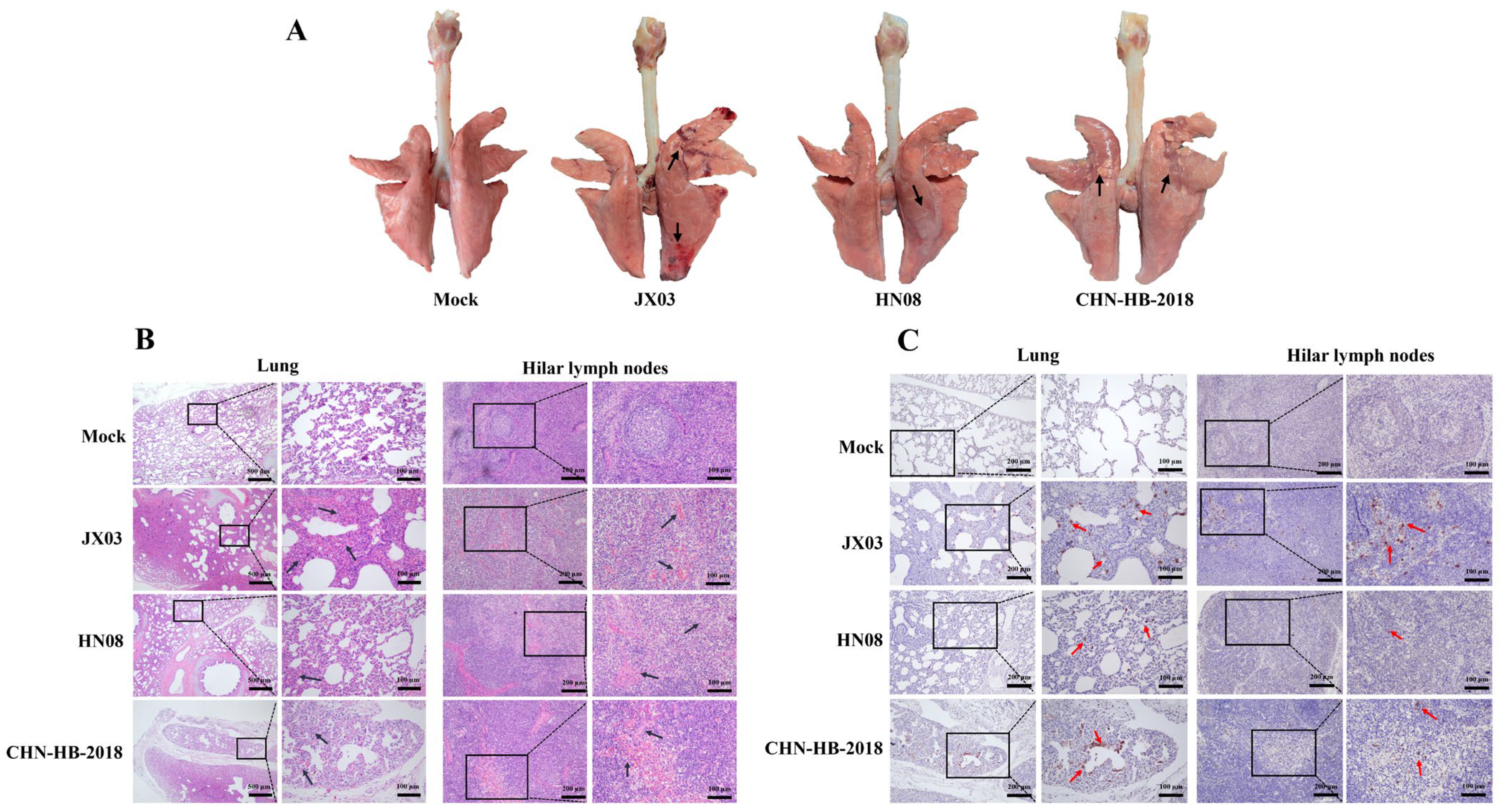
3.7. Pathological Analysis of Lungs and Hilar Lymph Nodes in Piglets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nathues, H.; Alarcon, P.; Rushton, J.; Jolie, R.; Fiebig, K.; Jimenez, M.; Geurts, V.; Nathues, C. Cost of porcine reproductive and respiratory syndrome virus at individual farm level—An economic disease model. Prev. Vet. Med. 2017, 142, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Díaz, J.A.C.; Fitzgerald, R.M.; Shalloo, L.; da Costa, M.R.; Niemi, J.; Leonard, F.C.; Kyriazakis, I.; Manzanilla, E.G. Financial analysis of herd status and vaccination practices for porcine reproductive and respiratory syndrome virus, swine influenza virus, and in farrow-to-finish pig farms using a bio-economic simulation model. Front. Vet. Sci. 2020, 7, 556674. [Google Scholar]
- Snijder, E.J.; Kikkert, M.; Fang, Y. Arterivirus molecular biology and pathogenesis. J. Gen. Virol. 2013, 94, 2141–2163. [Google Scholar] [CrossRef]
- Snijder, E.J.; Meulenberg, J.J. The molecular biology of arteriviruses. J. Gen. Virol. 1998, 79, 961–979. [Google Scholar] [CrossRef]
- Wang, L.H.; Hou, J.; Zhang, H.X.; Feng, W.H. Complete genome sequence of a novel highly pathogenic porcine reproductive and respiratory syndrome virus variant. J. Virol. 2012, 86, 13121. [Google Scholar] [CrossRef]
- Steinberger, J.; Kontaxis, G.; Rancan, C.; Skern, T. Comparison of self-processing of foot-and-mouth disease virus leader proteinase and porcine reproductive and respiratory syndrome virus leader proteinase nsp1α. Virology 2013, 443, 271–277. [Google Scholar] [CrossRef]
- Kuhn, J.H.; Lauck, M.; Bailey, A.L.; Shchetinin, A.M.; Vishnevskaya, T.V.; Bào, Y.M.; Ng, T.F.F.; LeBreton, M.; Schneider, B.S.; Gillis, A.; et al. Reorganization and expansion of the nidoviral family. Arch. Virol. 2016, 161, 755–768. [Google Scholar] [CrossRef]
- Yim-Im, W.; Anderson, T.K.; Paploski, I.A.D.; VanderWaal, K.; Gauger, P.; Krueger, K.; Shi, M.; Main, R.; Zhang, J. Refining PRRSV-2 genetic classification based on global ORF5 sequences and investigation of their geographic distributions and temporal changes. Microbiol. Spectr. 2023, 11, e0291623. [Google Scholar] [CrossRef]
- Yim-Im, W.; Anderson, T.K.; Böhmer, J.; Baliellas, J.; Stadejek, T.; Gauger, P.C.; Krueger, K.M.; Vermeulen, C.J.; Buter, R.; Kazlouski, A.; et al. Refining genetic classification of global porcine reproductive and respiratory syndrome virus type 1 (PRRSV-1) and investigating their geographic and temporal distributions. Vet. Microbiol. 2025, 302, 110413. [Google Scholar] [CrossRef]
- Guo, B.Q.; Chen, Z.S.; Liu, W.Q.; Cui, Y.Z. Isolation and identification of porcine reproductive and respiratory syndrome virus (PRRSV) from aborted fetuses suspected of PRRS. China. J. Prev. Vet. Med. 1996, 2, 1–5. [Google Scholar]
- Wu, J.; Li, J.; Tian, F.; Ren, S.; Yu, M.; Chen, J.; Lan, Z.; Zhang, X.; Yoo, D.; Wang, J. Genetic variation and pathogenicity of highly virulent porcine reproductive and respiratory syndrome virus emerging in China. Arch. Virol. 2009, 154, 1589–1597. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, J.; Yang, J.; Zeng, H.; Guo, L.; Ren, S.; Sun, W.; Chen, Z.; Cong, X.; Shi, J.; et al. Emergence of different recombinant porcine reproductive and respiratory syndrome viruses, China. Sci. Rep. 2018, 8, 4118. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wei, L.; Liu, X.; Liu, S.; Chen, H.; Chen, P.; Li, X.; Qian, P. Pathogenicity studies of NADC34-like porcine reproductive and respiratory syndrome virus LNSY-GY and NADC30-like porcine reproductive and respiratory syndrome virus GXGG-8011 in piglets. Viruses 2023, 15, 2247. [Google Scholar] [CrossRef]
- Tong, G.Z.; Zhou, Y.J.; Hao, X.F.; Tian, Z.J.; An, T.Q.; Qiu, H.J. Highly pathogenic porcine reproductive and respiratory syndrome, China. Emerg Infect Dis. 2007, 13, 1434–1436. [Google Scholar] [CrossRef]
- Zhou, L.; Wang, Z.; Ding, Y.; Ge, X.; Guo, X.; Yang, H. NADC30-like strain of porcine reproductive and respiratory syndrome virus, China. Emerg. Infect. Dis. 2015, 21, 2256–2257. [Google Scholar] [CrossRef]
- Zhang, H.L.; Zhang, W.L.; Xiang, L.R.; Leng, C.L.; Tian, Z.J.; Tang, Y.D.; Cai, X.-H. Emergence of novel porcine reproductive and respiratory syndrome viruses (ORF5 RFLP 1-7-4 viruses) in China. Vet. Microbiol. 2018, 222, 105–108. [Google Scholar] [CrossRef]
- Liu, J.; Wei, C.; Lin, Z.; Xia, W.; Ma, Y.; Dai, A.; Yang, X. Full genome sequence analysis of a 1-7-4-like PRRSV strain in Fujian province, China. PeerJ 2019, 7, e7859. [Google Scholar] [CrossRef]
- Li, C.; Zhuang, J.; Wang, J.; Han, L.; Sun, Z.; Xiao, Y.; Ji, G.; Li, Y.; Tan, F.; Li, X.; et al. Outbreak investigation of NADC30-like PRRSV in South-East China. Transbound. Emerg. Dis. 2016, 63, 474–479. [Google Scholar] [CrossRef]
- Zhao, J.; Xu, L.; Xu, Z.; Deng, H.; Li, F.; Sun, X.; Zhou, Y.; Zhu, L. Emergence and spread of NADC34-like PRRSV in Southwest China. Transbound. Emerg. Dis. 2022, 69, e3416–e3424. [Google Scholar] [CrossRef]
- Zhai, W.; Yu, S.; Zhang, P.; Lin, Y.; Ge, S.; Zhang, T.; Zhang, K.; He, S.; Hu, Q.; Tang, X.; et al. Epidemiology and genetic characteristics of porcine reproductive and respiratory syndrome virus in the Hunan and Hebei provinces of China. Vet. Sci. 2023, 10, 63. [Google Scholar] [CrossRef]
- Zhao, H.; Zhou, J.; Zhang, M.; Yang, X.; Xi, Z.; Gao, Q.; Fang, R.; Zhao, P.; Zhao, J. Epidemiological surveillance and economic impact analysis of different porcine reproductive and respiratory syndrome virus infection statuses in 23 breeding pig farms in Hubei, China. Anim. Dis. 2024, 4, 47. [Google Scholar] [CrossRef]
- Guo, J.; Li, C.; Lu, H.; Wang, B.; Zhang, L.; Ding, J.; Jiao, X.; Li, Q.; Zhu, S.; Wang, A.; et al. Reverse genetics construction and pathogenicity of a novel recombinant NADC30-like PRRSV isolated in China. Front. Vet. Sci. 2024, 11, 1434539. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.; Gao, X.; Wu, Y.; Wang, F.; Lai, M.; Zheng, J.; Qiu, Y.; He, Y.; Liang, X.; Yuan, K.; et al. Genomic and pathogenicity analysis of two novel highly pathogenic recombinant NADC30-like PRRSV strains in China, in 2023. Microbiol. Spectr. 2024, 12, e0036824. [Google Scholar] [CrossRef]
- Cui, M.; Qiu, M.; Yang, S.; Qiu, Y.; Qi, W.; Lin, H.; Sun, Z.; Zheng, W.; Zhu, J.; Chen, N. The replication efficacy of NADC34-like porcine reproductive and respiratory syndrome virus 2 is not directly associated with the pathogenicity. Vet. Microbiol. 2025, 301, 110367. [Google Scholar] [CrossRef]
- Tian, K.; Yu, X.; Zhao, T.; Feng, Y.; Cao, Z.; Wang, C.; Hu, Y.; Chen, X.; Hu, D.; Tian, X.; et al. Emergence of fatal PRRSV variants: Unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS ONE 2007, 2, e526. [Google Scholar] [CrossRef]
- Jiang, P.; Chen, P.Y.; Dong, Y.Y.; Cai, J.L.; Cai, B.X.; Jiang, Z.H. Isolation and genome characterization of porcine reproductive and respiratory syndrome virus in P.R. China. J. Vet. Diagn. Investig. 2000, 12, 156–158. [Google Scholar] [CrossRef]
- Li, L.; Zhao, Q.; Ge, X.; Teng, K.; Kuang, Y.; Chen, Y.; Guo, X.; Yang, H. Chinese highly pathogenic porcine reproductive and respiratory syndrome virus exhibits more extensive tissue tropism for pigs. Virol. J. 2012, 9, 203. [Google Scholar] [CrossRef]
- Zhao, K.; Ye, C.; Chang, X.B.; Jiang, C.G.; Wang, S.J.; Cai, X.H.; Tong, G.-Z.; Tian, Z.-J.; Shi, M.; An, T.-Q. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in China. J. Virol. 2015, 89, 10712–10716. [Google Scholar] [CrossRef]
- Brockmeier, S.L.; Loving, C.L.; Vorwald, A.C.; Kehrli, M.E., Jr.; Baker, R.B.; Nicholson, T.L.; Lager, K.M.; Miller, L.C.; Faaberg, K.S. Genomic sequence and virulence comparison of four Type 2 porcine reproductive and respiratory syndrome virus strains. Virus Res. 2012, 169, 212–221. [Google Scholar] [CrossRef]
- Van Geelen, A.G.M.; Anderson, T.K.; Lager, K.M.; Das, P.B.; Otis, N.J.; Montiel, N.A.; Miller, L.C.; Kulshreshtha, V.; Buckley, A.C.; Brockmeier, S.L.; et al. Porcine reproductive and respiratory disease virus: Evolution and recombination yields distinct ORF5 RFLP 1-7-4 viruses with individual pathogenicity. Virology 2018, 513, 168–179. [Google Scholar] [CrossRef]
- Song, S.; Xu, H.; Zhao, J.; Leng, C.; Xiang, L.; Li, C.; Fu, J.; Tang, Y.-D.; Peng, J.; Wang, Q.; et al. Pathogenicity of NADC34-like PRRSV HLJDZD32-1901 isolated in China. Vet. Microbiol. 2020, 246, 108727. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Luo, Q.; Zheng, Y.; Sha, H.; Li, G.; Kong, W.; Huang, L.; Zhao, M. Genetic variability and recombination of the NSP2 gene of PRRSV-2 strains in China from 1996 to 2021. Vet. Sci. 2023, 10, 325. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; He, Y.; Xu, X.; Leng, X.; Li, S.; Wen, Y.; Wang, F.; Xia, M.; Cheng, S.; Wu, H. Pathological and immunological characteristics of piglets infected experimentally with a HP-PRRSV TJ strain. BMC Vet. Res. 2016, 12, 230. [Google Scholar] [CrossRef]
- Xu, H.; Li, C.; Li, W.; Zhao, J.; Gong, B.; Sun, Q.; Tang, Y.D.; Xiang, L.; Leng, C.; Peng, J.; et al. Novel characteristics of Chinese NADC34-like PRRSV during 2020–2021. Transbound. Emerg. Dis. 2022, 69, e3215–e3224. [Google Scholar] [CrossRef]
- Fang, Y.; Snijder, E.J. The PRRSV replicase: Exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res. 2010, 154, 61–76. [Google Scholar] [CrossRef]
- Sun, Y.F.; Yu, H.; Jiang, X.; Ma, J.F.; Xu, C.Q.; Yu, X.X.; Li, L.-A. Novel ORF5 deletion of NADC30-like porcine reproductive and respiratory syndrome viruses circulating in northern China from 2016 to 2018. J. Vet. Diagn. Investig. 2020, 32, 928–932. [Google Scholar] [CrossRef]
- Li, P.X.; Shen, Y.S.; Wang, T.L.; Li, J.; Li, Y.; Zhao, Y.R.; Liu, S.; Li, B.; Liu, M.; Meng, F. Epidemiological survey of PRRS and genetic variation analysis of the ORF5 gene in Shandong Province, 2020–2021. Front. Vet. Sci. 2022, 9, 987667. [Google Scholar] [CrossRef]
- Huang, B.; Xu, T.; Luo, Z.; Deng, L.; Jian, Z.; Lai, S.; Ai, Y.; Zhou, Y.; Ge, L.; Xu, Z.; et al. Prevalence and genetic diversity of PRRSV in Sichuan province of China from 2021 to 2023: Evidence of an ongoing epidemic transition. Virology 2024, 600, 110213. [Google Scholar] [CrossRef]
- Song, T. Analysis of PRRSV Genetic Variation and Quantitative Interactomics of Its Nsp2 Protein. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2016. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Methods Mol. Biol. 2000, 132, 71–91. [Google Scholar]
- Pang, Y.; Li, C.; Wang, Y.; Liu, J.; Su, G.; Duan, C.; Fang, L.; Zhou, Y.; Xiao, S. Porcine reproductive and respiratory syndrome virus infection manipulates central carbon metabolism. Vet. Microbiol. 2023, 279, 109674. [Google Scholar] [CrossRef] [PubMed]
- Yin, B.; Qi, S.; Sha, W.; Qin, H.; Liu, L.; Yun, J.; Zhu, J.; Li, G.; Sun, D. Molecular Characterization of the Nsp2 and ORF5 (ORF5a) Genes of PRRSV Strains in Nine Provinces of China During 2016–2018. Front. Vet. Sci. 2021, 8, 605832. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Y.; Du, Y.; Zhang, H.; Guo, J.; Sun, Z.; Luo, X.; Mei, X.; Xiao, S.; Fang, L.; Zhou, Y. Genetic characterization and pathogenicity of a recombinant porcine reproductive and respiratory syndrome virus strain in China. Viruses 2024, 16, 993. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Treffers, E.E.; Li, Y.H.; Tas, A.; Sun, Z.; van der Meer, Y.; de Ru, A.H.; van Veelen, P.A.; Atkins, J.F.; Snijder, E.J.; et al. Efficient-2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc. Natl. Acad. Sci. USA 2012, 109, E2920–E2928. [Google Scholar] [CrossRef]
- Gao, J.C.; Xiong, J.Y.; Ye, C.; Chang, X.B.; Guo, J.C.; Jiang, C.G.; Zhang, G.-H.; Tian, Z.-J.; Cai, X.-H.; Tong, G.-Z.; et al. Genotypic and geographical distribution of porcine reproductive and respiratory syndrome viruses in mainland China in 1996–2016. Vet. Microbiol. 2017, 208, 164–172. [Google Scholar] [CrossRef]
- Delputte, P.L.; Nauwynck, H.J. Porcine Arterivirus infection of alveolar macrophages is mediated by sialic acid on the virus. J. Virol. 2004, 78, 8094–8101. [Google Scholar] [CrossRef]
- Xie, J.X.; Vereecke, N.; Theuns, S.; Oh, D.; Vanderheijden, N.; Trus, I.; Sauer, J.; Vyt, P.; Bonckaert, C.; Lalonde, C.; et al. Comparison of primary virus isolation in pulmonary alveolar macrophages and four different continuous cell lines for type 1 and type 2 porcine reproductive and respiratory syndrome virus. Vaccines 2021, 9, 594. [Google Scholar] [CrossRef]
- Zhang, H.L.; Tang, Y.D.; Liu, C.X.; Xiang, L.R.; Zhang, W.L.; Leng, C.L.; Wang, Q.; An, T.-Q.; Peng, J.-M.; Tian, Z.-J.; et al. Adaptions of field PRRSVs in Marc-145 cells were determined by variations in the minor envelope proteins GP2a-GP3. Vet. Microbiol. 2018, 222, 46–54. [Google Scholar] [CrossRef]
- Osemeke, O.H.; Machado, I.; De Conti, E.; Musskopf, M.; Mil-Homens, M.P.; Stutzman, S.; Guo, B.; Petznick, T.; Silva, G.D.-S.; Gauger, P.; et al. Optimizing tongue fluid sampling and testing protocols for enhanced PRRSV isolation from perinatal swine mortalities. Viruses 2025, 17, 102. [Google Scholar] [CrossRef]
- Shi, C.X.; Liu, Y.L.; Ding, Y.Z.; Zhang, Y.G.; Zhang, J. PRRSV receptors and their roles in virus infection. Arch. Microbiol. 2015, 197, 503–512. [Google Scholar] [CrossRef]
- Xie, J.; Trus, I.; Oh, D.; Kvisgaard, L.K.; Rappe, J.C.F. A Triple Amino Acid Substitution at Position 88/94/95 in Glycoprotein GP2a of Type 1 Porcine Reproductive and Respiratory Syndrome Virus (PRRSV1) Is Responsible for Adaptation to MARC-145 Cells. Viruses 2019, 11, 36. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Zhang, Z.; Zhu, Z.; Sun, Z.; Tian, K.; Li, X. A Live-Attenuated Chimeric Vaccine Candidate Against the Emerging NADC34-Like PRRSV. Vet. Sci. 2025, 12, 290. [Google Scholar] [CrossRef]
- Zhou, L.; Han, J.; Yang, H. The evolution and diversity of porcine reproductive and respiratory syndrome virus in China. Vet. Microbiol. 2024, 298, 110252. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Peng, O.; Xu, Q.; Li, Q.; Li, W.; Lin, L.; Zhou, Q.; Cai, X.; Hu, G.; He, Z.; et al. Characterization and Pathogenicity of Two Novel PRRSVs Recombined by NADC30-like and NADC34-like Strains in China. Viruses 2022, 14, 2174. [Google Scholar] [CrossRef]
- Liu, J.; Liu, C.; Xu, Y.; Yang, Y.; Li, J.; Dai, A.; Huang, C.; Luo, M.; Wei, C. Molecular characteristics and pathogenicity of a novel recombinant porcine reproductive and respiratory syndrome virus strain from NADC30-, NADC34-, and JXA1-like strains that emerged in China. Microbiol. Spectr. 2022, 10, e0266722. [Google Scholar] [CrossRef]
- Zhao, H.Z.; Liu, C.Y.; Meng, H.; Sun, C.L.; Yang, H.W.; Wang, H.; Zou, J.; Li, P.; Han, F.Y.; Qi, G.; et al. Evolution characterization and pathogenicity of an NADC34-like PRRSV isolated from Inner Mongolia, China. Viruses 2024, 16, 683. [Google Scholar] [CrossRef]
- Xie, C.Z.; Ha, Z.; Zhang, H.; Zhang, Y.; Xie, Y.B.; Zhang, H.; Nan, F.-L.; Wang, Z.; Zhang, P.; Xu, W.; et al. Pathogenicity of porcine reproductive and respiratory syndrome virus (ORF5 RFLP 1-7-4 viruses) in China. Transbound. Emerg. Dis. 2020, 67, 2065–2072. [Google Scholar] [CrossRef]
- Jelsma, T.; Wijnker, J.J.; van der Poel, W.H.M.; Wisselink, H.J. Intestinal Viral Loads and Inactivation Kinetics of Livestock Viruses Relevant for Natural Casing Production: A Systematic Review and Meta-Analysis. Pathogens 2021, 10, 173. [Google Scholar] [CrossRef]
- Zhao, J.; Wan, S.; Sun, N.; Sun, P.; Sun, Y.; Khan, A.; Guo, J.; Zheng, X.; Fan, K.; Yin, W.; et al. Damage to intestinal barrier integrity in piglets caused by porcine reproductive and respiratory syndrome virus infection. Vet. Res. 2021, 52, 93. [Google Scholar] [CrossRef]
| Clinical Samples | Positive Numbers | Positive Samples | Positive Rate | Percentage a | ||||
|---|---|---|---|---|---|---|---|---|
| HP- PRRSV | NADC30/ 34-like | Classical | HP- PRRSV | NADC30/ 34-like | Classical | |||
| 1044 | 118 | 187 | 14 | 311 | 29.8% | 37.94% | 60.13% | 4.50% |
| JX03 | CH-1a | CH-1R | JXA1 | NADC30 | NADC34 | GM2 | QYYZ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | |
| Complete Genome | 94.3 | 94.1 | 99.0 | 81.8 | 81.7 | 86.3 | 86.4 | |||||||
| 5′UTR | 96.8 | 95.3 | 99.5 | 92.1 | 90.0 | 94.7 | 94.7 | |||||||
| nsp1 | 94.3 | 92.4 | 94.2 | 92.4 | 99 | 98.7 | 84.8 | 85.4 | 84.4 | 87.2 | 88 | 89.1 | 88.5 | 89.1 |
| nsp2 | 86.5 | 78.4 | 86.7 | 78.6 | 98.9 | 97.3 | 63.7 | 46.3 | 66.3 | 50.3 | 77.3 | 62.8 | 77.5 | 62.9 |
| nsp3 | 94.8 | 98.0 | 94.8 | 97.8 | 99.0 | 98.9 | 83.3 | 90.6 | 82.8 | 90.8 | 81.8 | 88.1 | 82.0 | 88.1 |
| nsp4 | 94.9 | 96.1 | 95.1 | 96.6 | 98.4 | 97.5 | 84.2 | 92.6 | 85.5 | 93.1 | 84.8 | 92.2 | 84.6 | 91.7 |
| nsp5 | 94.5 | 94.1 | 93.9 | 93.5 | 98.9 | 98.8 | 90.0 | 92.9 | 84.3 | 88.8 | 82.7 | 90.0 | 82.4 | 90.0 |
| nsp6 | 94.5 | 94.1 | 93.9 | 93.5 | 98.8 | 98.8 | 90.0 | 92.9 | 84.3 | 88.8 | 82.7 | 90.0 | 82.4 | 90.0 |
| nsp7 | 94.5 | 94.1 | 93.9 | 93.5 | 98.8 | 98.8 | 90.0 | 92.9 | 84.3 | 88.8 | 82.7 | 90.0 | 82.4 | 90.0 |
| nsp8 | 97.1 | 100.0 | 97.1 | 100.0 | 97.8 | 100.0 | 89.1 | 95.7 | 88.4 | 93.5 | 92.8 | 100.0 | 92.0 | 100.0 |
| nsp9 | 96.4 | 98.6 | 96.3 | 98.3 | 98.9 | 99.1 | 87.2 | 97.2 | 87.4 | 96.9 | 98.9 | 99.1 | 91.3 | 96.9 |
| nsp10 | 95.5 | 97.7 | 95.4 | 97.7 | 99.5 | 99.1 | 86.0 | 95.5 | 85.8 | 95.2 | 90.3 | 96.4 | 90.6 | 94.3 |
| nsp11 | 94.9 | 96.9 | 95.2 | 97.8 | 99.0 | 99.1 | 91.2 | 96.4 | 87.2 | 95.5 | 89.0 | 94.6 | 88.8 | 93.8 |
| nsp12 | 95.9 | 97.4 | 95.9 | 97.4 | 98.3 | 99.5 | 89.4 | 96.1 | 82.7 | 91.6 | 87.3 | 96.8 | 86.6 | 96.1 |
| ORF2 | 96.4 | 96.1 | 95.7 | 95.7 | 99.2 | 98.8 | 86.5 | 86.4 | 87.4 | 86.0 | 89.9 | 88.7 | 89.9 | 89.1 |
| ORF3 | 95.3 | 92.5 | 95.0 | 92.9 | 99.0 | 98.0 | 83.7 | 82.0 | 84.2 | 83.5 | 90.6 | 89.8 | 90.8 | 89.4 |
| ORF4 | 96.5 | 96.5 | 96.3 | 96.3 | 97.6 | 97.6 | 87.5 | 87.5 | 86.8 | 86.8 | 95.0 | 95.0 | 94.8 | 94.8 |
| ORF5 | 94.9 | 93.0 | 94.0 | 91.5 | 99.3 | 99.5 | 85.7 | 86.1 | 87.1 | 87.1 | 83.3 | 82.1 | 83.6 | 82.6 |
| ORF6 | 97.3 | 97.1 | 97.0 | 96.6 | 99.8 | 99.4 | 89.1 | 93.7 | 89.3 | 94.3 | 91.6 | 96.6 | 91.0 | 97.1 |
| ORF7 | 95.7 | 94.4 | 95.7 | 95.2 | 99.5 | 98.4 | 90.6 | 90.3 | 89.2 | 90.3 | 87.9 | 88.7 | 89.2 | 91.1 |
| 3′UTR | 96.1 | 97.2 | 99.4 | 90.4 | 89.9 | 88.8 | 89.9 | |||||||
| HN08 | CH-1a | CH-1R | JXA1 | NADC30 | NADC34 | GM2 | QYYZ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | nt | aa | |
| Complete Genome | 82.7 | 82.8 | 82.5 | 90.0 | 85.3 | 79.6 | 79.8 | |||||||
| 5′UTR | 90.8 | 89.2 | 90.3 | 96.4 | 89.7 | 88.2 | 88.2 | |||||||
| nsp1 | 81.2 | 83.6 | 81.3 | 83.6 | 80.5 | 83.9 | 91.0 | 91.1 | 83.2 | 84.9 | 79.2 | 82.8 | 79.7 | 82.6 |
| nsp2 | 60.6 | 42.2 | 60.8 | 42.4 | 61.0 | 42.7 | 90.7 | 80.6 | 74.5 | 55.5 | 59.6 | 40.0 | 59.6 | 40.3 |
| nsp3 | 84.4 | 92.6 | 84.5 | 92.4 | 84.3 | 91.7 | 89.8 | 94.8 | 84.2 | 90.8 | 79.7 | 87.4 | 79.6 | 87.4 |
| nsp4 | 92.3 | 95.1 | 92.5 | 95.6 | 94.3 | 96.6 | 84.2 | 92.6 | 84.0 | 94.6 | 84.0 | 92.2 | 83.8 | 91.7 |
| nsp5 | 90.0 | 92.4 | 89.8 | 91.8 | 92.7 | 93.5 | 87.3 | 92.4 | 81.6 | 88.8 | 81.2 | 90.0 | 80.8 | 90.6 |
| nsp6 | 91.7 | 100.0 | 91.7 | 100.0 | 93.8 | 100.0 | 87.5 | 93.8 | 81.2 | 87.5 | 91.7 | 100.0 | 91.7 | 100.0 |
| nsp7 | 94.5 | 95.0 | 94.1 | 94.6 | 97.4 | 98.1 | 81.5 | 83.8 | 81.2 | 85.3 | 90.3 | 91.1 | 92.7 | 94.2 |
| nsp8 | 97.8 | 100.0 | 97.8 | 100.0 | 97.1 | 100.0 | 89.9 | 95.7 | 89.1 | 93.5 | 93.5 | 100.0 | 92.8 | 100.0 |
| nsp9 | 91.8 | 98.0 | 91.7 | 97.7 | 92.7 | 98.1 | 90.8 | 97.7 | 87.5 | 96.9 | 89.3 | 97.0 | 88.5 | 96.6 |
| nsp10 | 84.2 | 93.2 | 84.2 | 93.2 | 83.8 | 93.4 | 94.1 | 97.5 | 89.8 | 97.0 | 84.8 | 94.8 | 83.6 | 93.6 |
| nsp11 | 89.9 | 95.5 | 90.2 | 96.4 | 87.8 | 96.0 | 93.9 | 97.3 | 85.6 | 94.6 | 85.7 | 93.8 | 85.6 | 92.9 |
| nsp12 | 88.6 | 94.8 | 88.8 | 94.8 | 88.8 | 95.5 | 93.5 | 96.8 | 85.5 | 90.3 | 86.8 | 94.8 | 86.4 | 94.2 |
| ORF2 | 85.1 | 82.9 | 85.1 | 82.5 | 84.8 | 81.3 | 83.7 | 81.7 | 94.8 | 92.2 | 84.4 | 81.3 | 84.6 | 81.7 |
| ORF3 | 83.8 | 80.4 | 84.1 | 80.8 | 83.4 | 81.2 | 84.6 | 83.1 | 92.9 | 92.9 | 83.4 | 81.6 | 83.1 | 82.4 |
| ORF4 | 86.4 | 88.3 | 86.4 | 87.7 | 85.5 | 87.2 | 94.8 | 95.5 | 94.8 | 96.6 | 85.5 | 86.0 | 85.7 | 86.6 |
| ORF5 | 86.4 | 88.1 | 86.1 | 86.6 | 86.2 | 89.6 | 87.2 | 91.0 | 95.9 | 97.0 | 83.6 | 84.1 | 83.6 | 83.6 |
| ORF6 | 86.9 | 92.6 | 86.9 | 92.0 | 87.2 | 93.1 | 94.7 | 96.0 | 94.3 | 95.4 | 89.7 | 93.1 | 88.8 | 93.1 |
| ORF7 | 90.3 | 91.9 | 90.3 | 91.9 | 90.3 | 90.3 | 95.4 | 95.2 | 93.8 | 92.7 | 84.9 | 86.3 | 86.6 | 89.5 |
| 3′UTR | 89.3 | 88.8 | 90.4 | 97.8 | 95.5 | 88.8 | 89.9 | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, Y.; Chen, J.; Ren, T.; Xie, C.; Zhang, Y.; Fang, L.; Zhou, Y. Etiological Detection, Isolation, and Pathogenicity of Porcine Reproductive and Respiratory Syndrome Virus in China. Vet. Sci. 2025, 12, 530. https://doi.org/10.3390/vetsci12060530
Du Y, Chen J, Ren T, Xie C, Zhang Y, Fang L, Zhou Y. Etiological Detection, Isolation, and Pathogenicity of Porcine Reproductive and Respiratory Syndrome Virus in China. Veterinary Sciences. 2025; 12(6):530. https://doi.org/10.3390/vetsci12060530
Chicago/Turabian StyleDu, Yingbin, Jingyi Chen, Tianze Ren, Chunying Xie, Yiye Zhang, Liurong Fang, and Yanrong Zhou. 2025. "Etiological Detection, Isolation, and Pathogenicity of Porcine Reproductive and Respiratory Syndrome Virus in China" Veterinary Sciences 12, no. 6: 530. https://doi.org/10.3390/vetsci12060530
APA StyleDu, Y., Chen, J., Ren, T., Xie, C., Zhang, Y., Fang, L., & Zhou, Y. (2025). Etiological Detection, Isolation, and Pathogenicity of Porcine Reproductive and Respiratory Syndrome Virus in China. Veterinary Sciences, 12(6), 530. https://doi.org/10.3390/vetsci12060530







