PRRSV RNA Detection and Predictive Values Between Different Sow and Neonatal Litter Sample Types
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design Overview
2.2. Farm Eligibility Criteria
2.3. Study Herds Overview
2.4. Sample Collection
2.4.1. TOSc Collection from Sows
2.4.2. Blood Collection from Live Piglets
2.4.3. Tongue Fluids (TF) and Serum Collection from Dead Animals
2.5. Diagnostic Testing and PRRSV Status Definition for Different Subpopulations Within the Litter
2.6. Statistical Analysis
3. Results
3.1. Number and Rate of Positive Sows and Positive “Whole Litter” and “Live Litter”
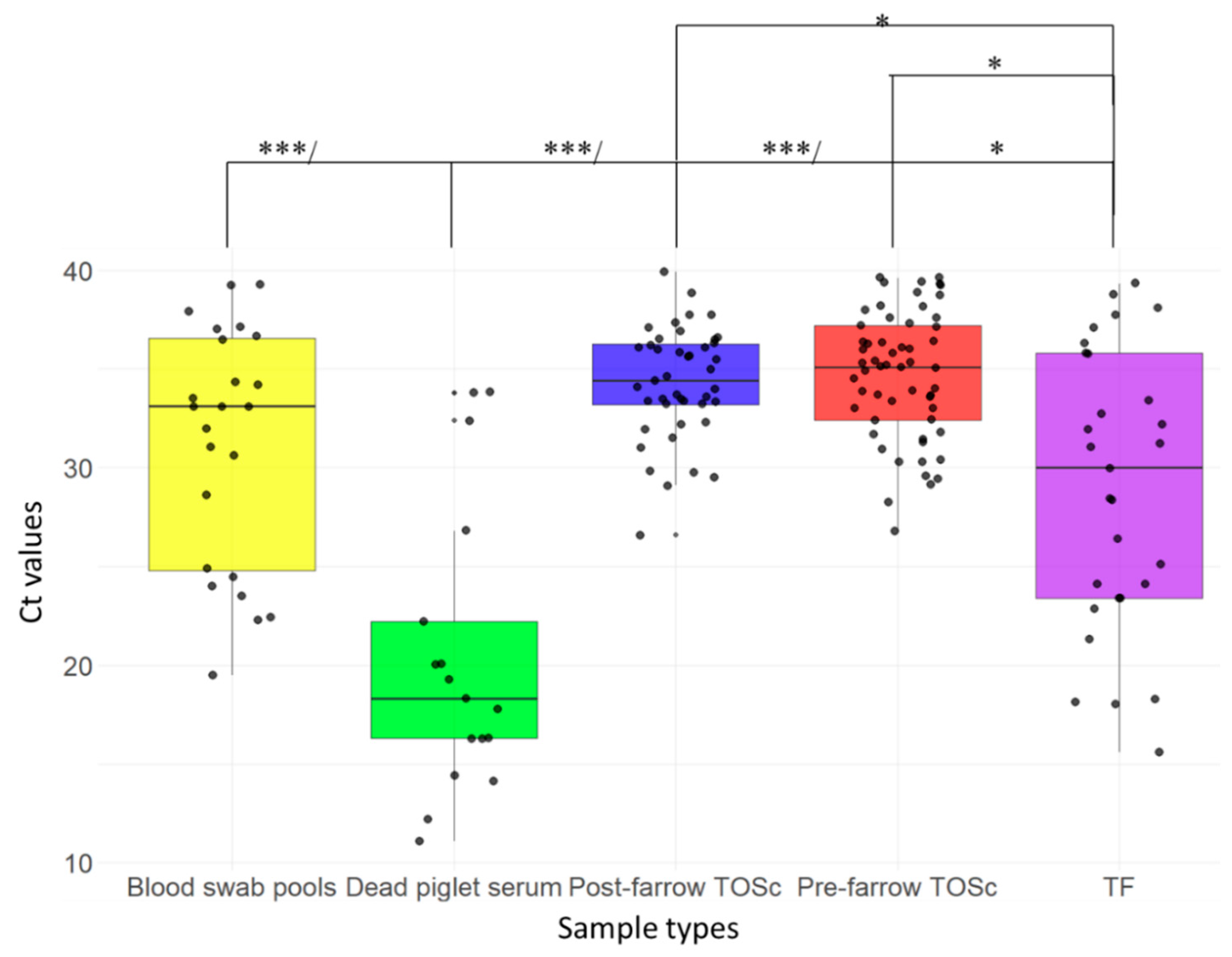
3.2. PRRSV RT-rtPCR Positivity and Ct Values Comparison Between Samples
3.3. Comparison of Number of Total Born and Live Piglets Between Positive “Whole Litter” and Negative “Whole Litter”
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PRRSV | Porcine reproductive and respiratory syndrome virus |
| AASV | American Association of Swine Veterinarians |
| TTS | time to stability |
| PF | processing fluid |
| FOF | family oral fluids |
| TOSc | tonsil oral scrubbing |
| TF | tongue fluid |
| PPV | positive predictive values |
| NPV | negative predictive values |
References
- Holtkamp, D.J. Proposed modifications to porcine reproductive and respiratory syndrome virus herd classification. J. Swine Health Prod. 2021, 29, 261–270. [Google Scholar] [CrossRef]
- Daniel, C.L.L.; Giovani, T.; Rodger, M.; Derald, H.; Leticia, L.; Cesar, C. Swine Disease Management Information Program # 20-109; The National Pork Board: Des Moines, IA, USA, 2022. [Google Scholar]
- Linhares, D.C.L.; Cano, J.P.; Torremorell, M.; Morrison, R.B. Comparison of time to PRRSv-stability and production losses between two exposure programs to control PRRSv in sow herds. Prev. Vet. Med. 2014, 116, 111–119. [Google Scholar] [CrossRef]
- Pileri, E.; Mateu, E. Review on the transmission porcine reproductive and respiratory syndrome virus between pigs and farms and impact on vaccination. Vet. Res. 2016, 47, 108. [Google Scholar] [CrossRef]
- Almeida, M.N.; Rotto, H.; Schneider, P.; Robb, C.; Zimmerman, J.J.; Holtkamp, D.J.; Rademacher, C.J.; Linhares, D.C.L. Collecting oral fluid samples from due-to-wean litters. Prev. Vet. Med. 2020, 174, 104810. [Google Scholar] [CrossRef] [PubMed]
- López, W.; Zimmerman, J.; Gauger, P.; Harmon, K.; Magtoto, R.; Bradner, L.; Holtkamp, D.; Zhang, M.; Zhang, J.; Ramirez, A.; et al. Considerations in the use of processing fluids for the detection of PRRSV RNA and antibody. J. Vet. Diagn. Investig. 2022, 34, 859–863. [Google Scholar] [CrossRef] [PubMed]
- López, W.A.; Zimmerman, J.J.; Gauger, P.C.; Harmon, K.M.; Bradner, L.; Zhang, M.; Giménez-Lirola, L.; Ramirez, A.; Cano, J.P.; Linhares, D.C.L. Practical aspects of PRRSV RNA detection in processing fluids collected in commercial swine farms. Prev. Vet. Med. 2020, 180, 105021. [Google Scholar] [CrossRef] [PubMed]
- Rossow, K.D.; Collins, J.E.; Goyal, S.M.; Nelson, E.A.; Christopher-Hennings, J.; Benfield, D.A. Pathogenesis of porcine reproductive and respiratory syndrome virus infection in gnotobiotic pigs. Vet. Pathol. 1995, 32, 361–373. [Google Scholar] [CrossRef]
- Dee, S.A.; Bierk, M.D.; Deen, J.; Molitor, T.W. An evaluation of test and removal for the elimination of porcine reproductive and respiratory syndrome virus from 5 swine farms. Can. J. Vet. Res. 2001, 65, 22–27. [Google Scholar]
- Nielsen, E.O.; Lauritsen, K.T.; Friis, N.F.; Enøe, C.; Hagedorn-Olsen, T.; Jungersen, G. Use of a novel serum ELISA method and the tonsil-carrier state for evaluation of Mycoplasma hyosynoviae distributions in pig herds with or without clinical arthritis. Vet. Microbiol. 2005, 111, 41–50. [Google Scholar] [CrossRef]
- Tousignant, S.J.P.; Bruner, L.; Schwartz, J.; Vannucci, F.; Rossow, S.; Marthaler, D.G. Longitudinal study of Senecavirus a shedding in sows and piglets on a single United States farm during an outbreak of vesicular disease. BMC Vet. Res. 2017, 13, 277. [Google Scholar] [CrossRef]
- Wills, R.W.; Doster, A.R.; Galeota, J.A.; Sur, J.H.; Osorio, F.A. Duration of infection and proportion of pigs persistently infected with porcine reproductive and respiratory syndrome virus. J. Clin. Microbiol. 2003, 41, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Silva, A.P.S.P.; de Almeida Moraes, D.C.; Yeske, P.; Osemeke, O.H.; Magalhães, E.S.; Gustavo De Sousa, E.S.; Linhares, D.C.L. Comparison of a novel rapid sampling method to serum and tonsil scraping to detect PRRSV in acutely infected sows. Prev. Vet. Med. 2024, 223, 106082. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Silva, A.P.P.; Tong, H.; Yeske, P.; Dalquist, L.; Kelly, J.; Finch, M.; Reever, A.V.A.; Reicks, D.L.; Connor, J.F.; et al. Characterizing best practices for tonsil-oral-scrubbing (TOSc) collection for PRRSV RNA detection in sows. Porc. Health Manag. 2024, 10, 37. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Petznick, T.; Pratt, E.; Cezar, G.; Will, K.; Mil-Homens, M.; Tong, H.; Machado, I.; Moraes, D.C.A.; Paiva, R.C.; et al. Comparison of tonsil-oral-scrubbing with serum, oral fluid, and tonsil scraping to detect PRRSV RNA in sows over time following live virus inoculation. Front. Vet. Sci. 2024, 11, 1506995. [Google Scholar] [CrossRef] [PubMed]
- Baliellas, J.; Novell, E.; Enric-Tarancón, V.; Vilalta, C.; Fraile, L. Porcine Reproductive and Respiratory Syndrome Surveillance in breeding Herds and Nurseries Using Tongue Tips from Dead Animals. Vet. Sci. 2021, 8, 259. [Google Scholar] [CrossRef]
- Machado, I.F.; Magalhães, E.S.; Poeta Silva, A.P.S.; Moraes, D.C.A.; Cezar, G.; Mil-Homens, M.P.; Osemeke, O.H.; Paiva, R.; Moura, C.A.A.; Gauger, P.; et al. Porcine reproductive and respiratory syndrome virus RNA detection in tongue tips from dead animals. Front. Vet. Sci. 2022, 9, 993442. [Google Scholar] [CrossRef]
- Kang, I.; Ha, Y.; Kim, D.; Oh, Y.; Cho, K.D.; Lee, B.H.; Lim, J.; Kim, S.H.; Kwon, B.; Chae, C. Localization of porcine reproductive and respiratory syndrome virus in mammary glands of experimentally infected sows. Res. Vet. Sci. 2010, 88, 304–306. [Google Scholar] [CrossRef]
- Cano, J.P.; Dee, S.A.; Murtaugh, M.P.; Rovira, A.; Morrison, R.B. Infection dynamics and clinical manifestations following experimental inoculation of gilts at 90 days of gestation with a low dose of porcine reproductive and respiratory syndrome virus. Can. J. Vet. Res. 2009, 73, 303–307. [Google Scholar]
- Osemeke, O.H. A cross-sectional assessment of PRRSV nucleic acid detection by RT-qPCR in serum, ear-vein blood swabs, nasal swabs, and oral swabs from weaning-age pigs under field conditions. Front. Vet. Sci. 2023, 10, 1200376. [Google Scholar] [CrossRef]
- Prieto, C.; Suárez, P.; Simarro, I.; García, C.; Fernández, A.; Castro, J.M. Transplacental infection following exposure of gilts to porcine reproductive and respiratory syndrome virus at the onset of gestation. Vet. Microbiol. 1997, 57, 301–311. [Google Scholar] [CrossRef]
- Christianson, W.T.; Choi, C.S.; Collins, J.E.; Molitor, T.W.; Morrison, R.B.; Joo, H.S. Pathogenesis of porcine reproductive and respiratory syndrome virus infection in mid-gestation sows and fetuses. Can. J. Vet. Res. 1993, 57, 262–268. [Google Scholar] [PubMed]
- Harding, J.C.S.; Ladinig, A.; Novakovic, P.; Detmer, S.E.; Wilkinson, J.M.; Yang, T.; Lunney, J.K.; Plastow, G.S. Novel insights into host responses and reproductive pathophysiology of porcine reproductive and respiratory syndrome caused by PRRSV-2. Vet. Microbiol. 2017, 209, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Osemeke, O.H.; Donovan, T.; Dion, K.; Holtkamp, D.J.; Linhares, D.C. Characterization of changes in productivity parameters as breeding herds transitioned through the 2021 PRRSV Breeding Herd Classification System. J. Swine Health Prod. 2022, 30, 145–148. [Google Scholar] [CrossRef] [PubMed]

| Farm | PRRSV RFLP and Lineage | Herd Type | Sow Inventory | Production Type | AASV PRRSV Status Before Outbreak | Screened Population | Screening Timepoint (2 Weeks Before Farrow) | Reproductive Data Collected |
|---|---|---|---|---|---|---|---|---|
| A | 1-8-4, 1H | Breed-to-wean | 2500 | 4-week batch | II-Vx | 280 pregnant sows with parities 1–9 | 106 days after LVI | No |
| B | 1-4-4, L1C.5 | Breed-to-wean | 4500 | Continuous | IV | 275 pregnant gilts | 93 days after LVI | Yes |
| Farm | Number of Positive Sows Identified/Total Number of Sows Screened | Number and Rate of Positive Sows Selected |
|---|---|---|
| A | 17/280 | 14 (43.8%) |
| B | 45/275 | 43 (38.7%) |
| Pre-Farrow TOSc | Post-Farrow TOSc | Blood Swab Pools | TF | DEAD Piglet Serum | |
|---|---|---|---|---|---|
| PRRSV RNA positivity (95% CI) | 34.3% (20.6–51.1%) a | 21.0% (11.2–35.9%) ab | 8.0% (3.5–17.5%) c | 10.9% (5.0–22.0%) bc | 4.7% (1.8–11.8%) c |
| Ct value mean (range) | 34.7 (26–39.6) | 34.2 (26.6–39.9) | 31.2 (19.5–39.3) | 28.9 (15.6–39.3) | 20.3 (11.1–33.8) |
| Pre-Farrow TOSc PRRSV Status | Post-Farrow TOSc PRRSV Status | Parallel TOSc PRRSV Status | TF PRRSV Status | |
|---|---|---|---|---|
| NPV with 95% CI | 87.2% (80.2–94.3%) | 89.0% (82.9–95.1%) | 91.7% (85.5–98.1%) | 95.4% (88.8–100%) |
| PPV with 95% CI | 22.8% (11.9–33.7%) | 30.2% (16.5–44.0%) | 25.7% (15.5–36.0%) | 58.3% (38.6–78.1%) |
| Average Number of Total Born with 95% CI | Average Number of Live Piglets with 95% CI | |
|---|---|---|
| Positive “whole litter” | 13.9 (11.3–16.5) a | 9.0 (7.3–11.0) a |
| Negative “whole litter” | 14.8 (14.1–15.6) a | 13.5(12.8–14.2) b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, P.; Machado, I.; Petznick, T.; Pratt, E.; Xiao, J.; Sievers, C.; Yeske, P.; Jayaraman, S.; Moraes, D.C.A.; Cezar, G.; et al. PRRSV RNA Detection and Predictive Values Between Different Sow and Neonatal Litter Sample Types. Vet. Sci. 2025, 12, 150. https://doi.org/10.3390/vetsci12020150
Li P, Machado I, Petznick T, Pratt E, Xiao J, Sievers C, Yeske P, Jayaraman S, Moraes DCA, Cezar G, et al. PRRSV RNA Detection and Predictive Values Between Different Sow and Neonatal Litter Sample Types. Veterinary Sciences. 2025; 12(2):150. https://doi.org/10.3390/vetsci12020150
Chicago/Turabian StyleLi, Peng, Isadora Machado, Thomas Petznick, Emily Pratt, Jinnan Xiao, Chris Sievers, Paul Yeske, Swami Jayaraman, Daniel C. A. Moraes, Guilherme Cezar, and et al. 2025. "PRRSV RNA Detection and Predictive Values Between Different Sow and Neonatal Litter Sample Types" Veterinary Sciences 12, no. 2: 150. https://doi.org/10.3390/vetsci12020150
APA StyleLi, P., Machado, I., Petznick, T., Pratt, E., Xiao, J., Sievers, C., Yeske, P., Jayaraman, S., Moraes, D. C. A., Cezar, G., Mil-Homens, M., Tong, H., Will, K., Reicks, D., Kelly, J., Osemeke, O. H., Silva, G. S., & Linhares, D. C. L. (2025). PRRSV RNA Detection and Predictive Values Between Different Sow and Neonatal Litter Sample Types. Veterinary Sciences, 12(2), 150. https://doi.org/10.3390/vetsci12020150






