Rabbit Enteric Syndrome: The Implications of Rabbit Coronavirus (RbCoV) and Lapine Bocaparvovirus (LBoV) in Rabbitries of Spain
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study A: Frequency of Detection of RbCoV, LBoV and RKoB
2.1.1. Animals and Samples
2.1.2. qPCR Detection
2.2. Study B: Lesional Study
2.2.1. Animals and Samples
2.2.2. Histopathological Study
2.2.3. qPCR Detection
2.3. Study C: Epidemiological Situation of RbCoV and LBoV in Spain
3. Results
3.1. Study A: Frequency of Detection of RbCoV, LBoV and RKoB
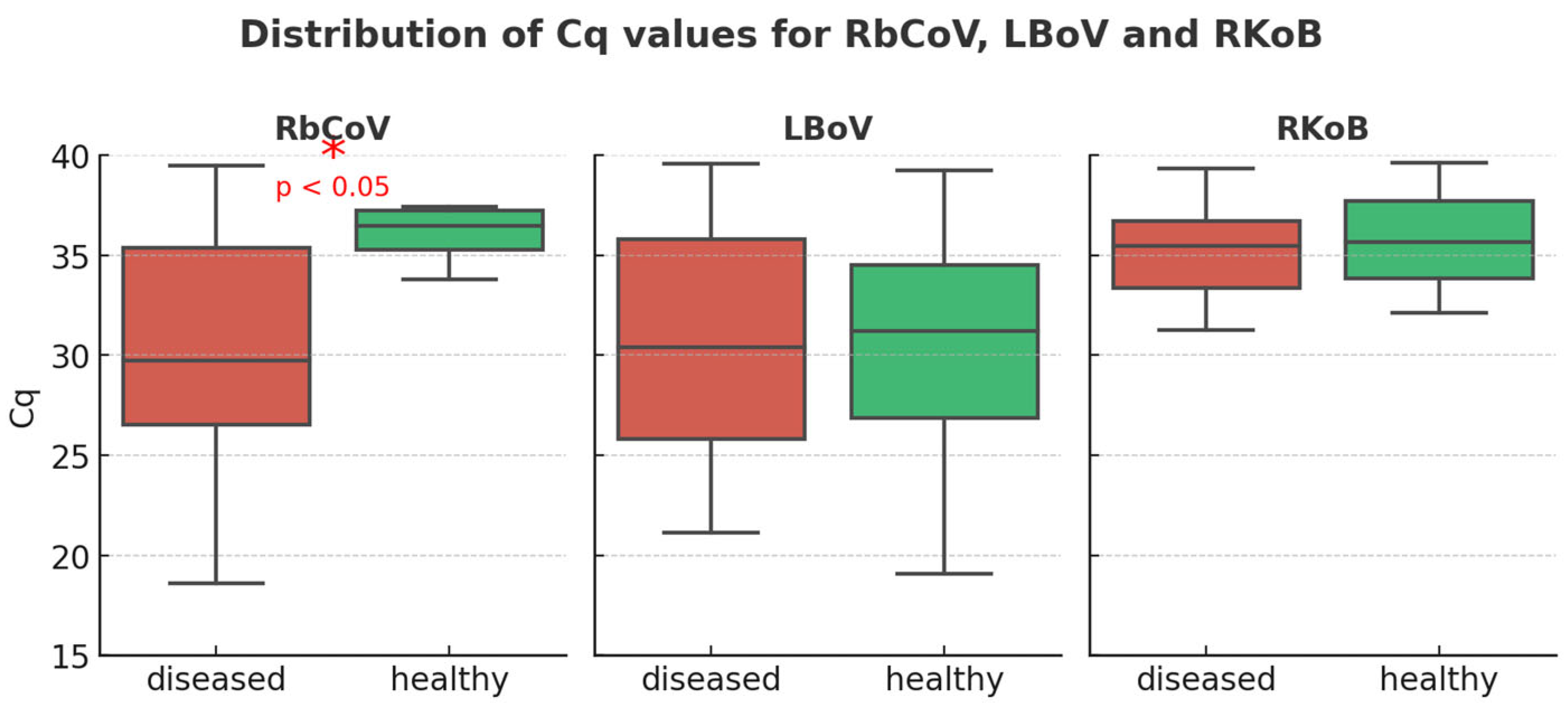
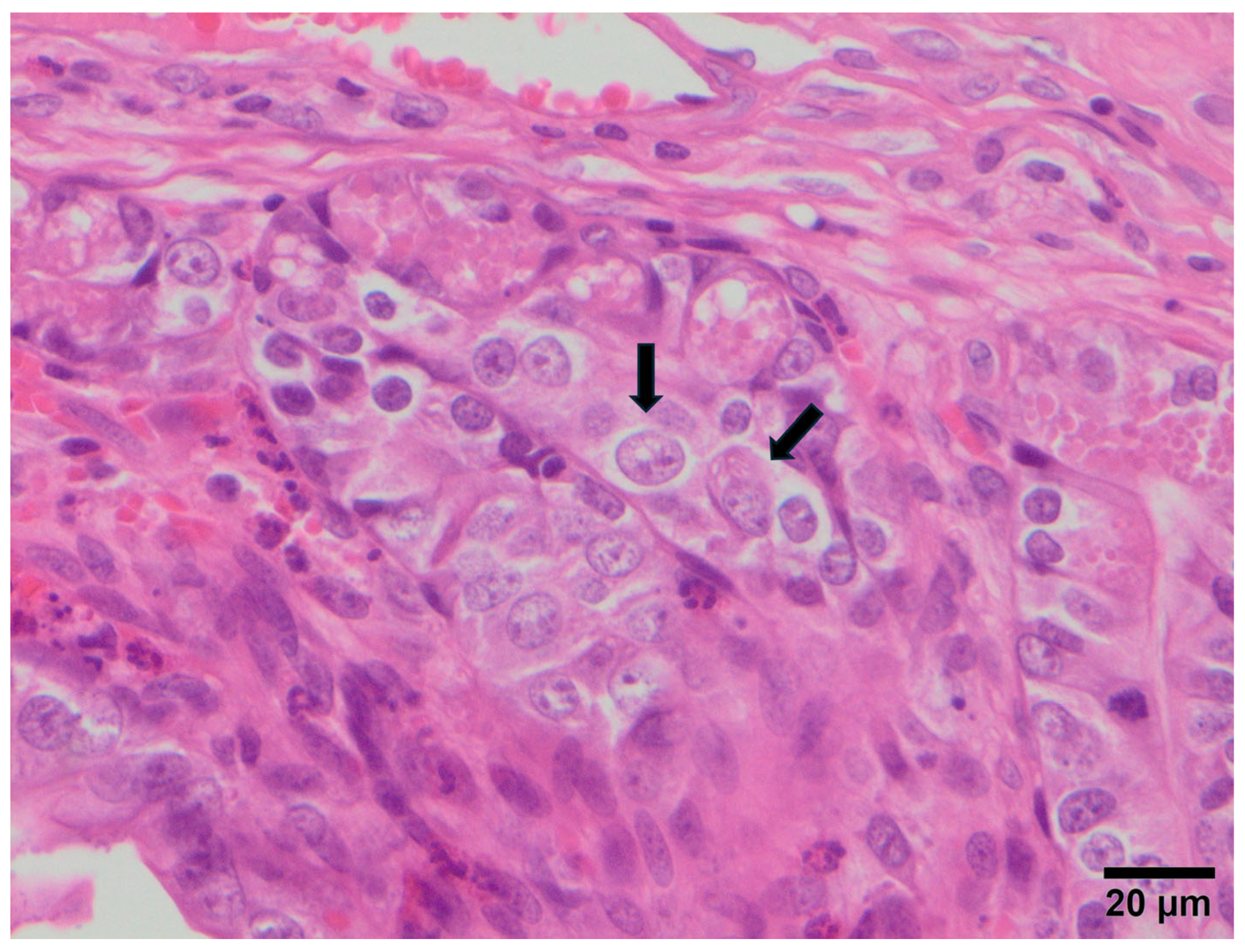
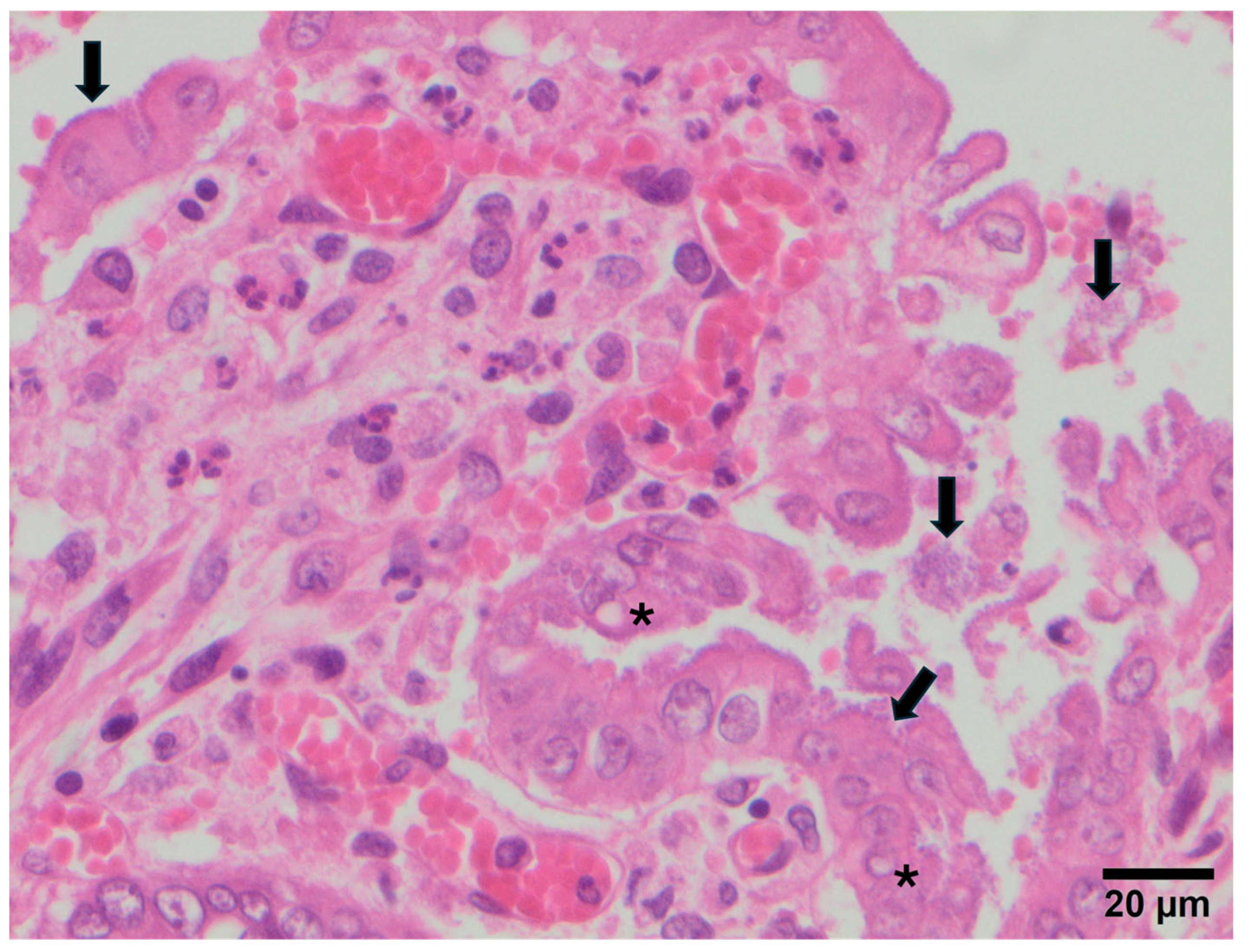
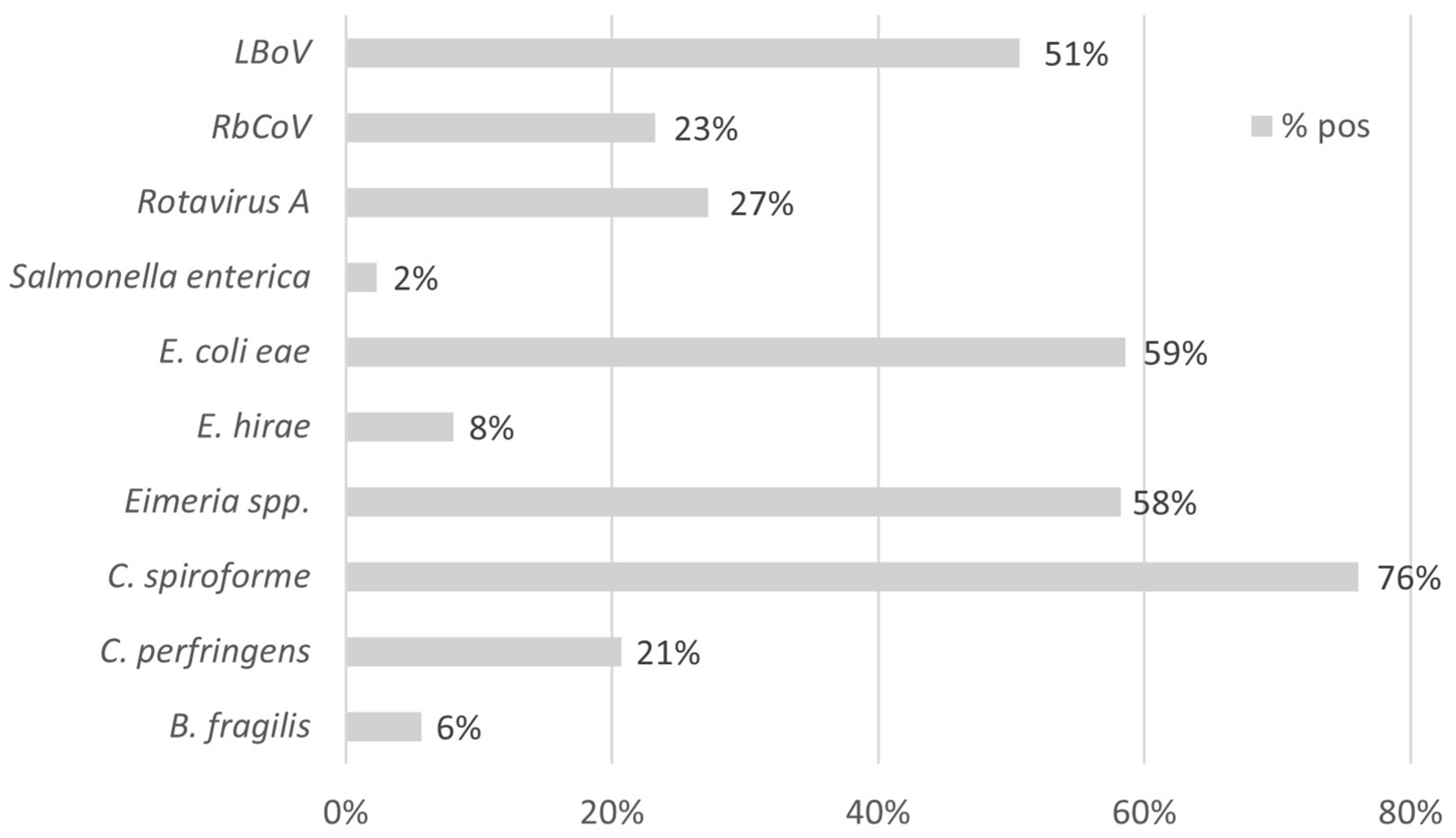
3.2. Study B: Lesional Study
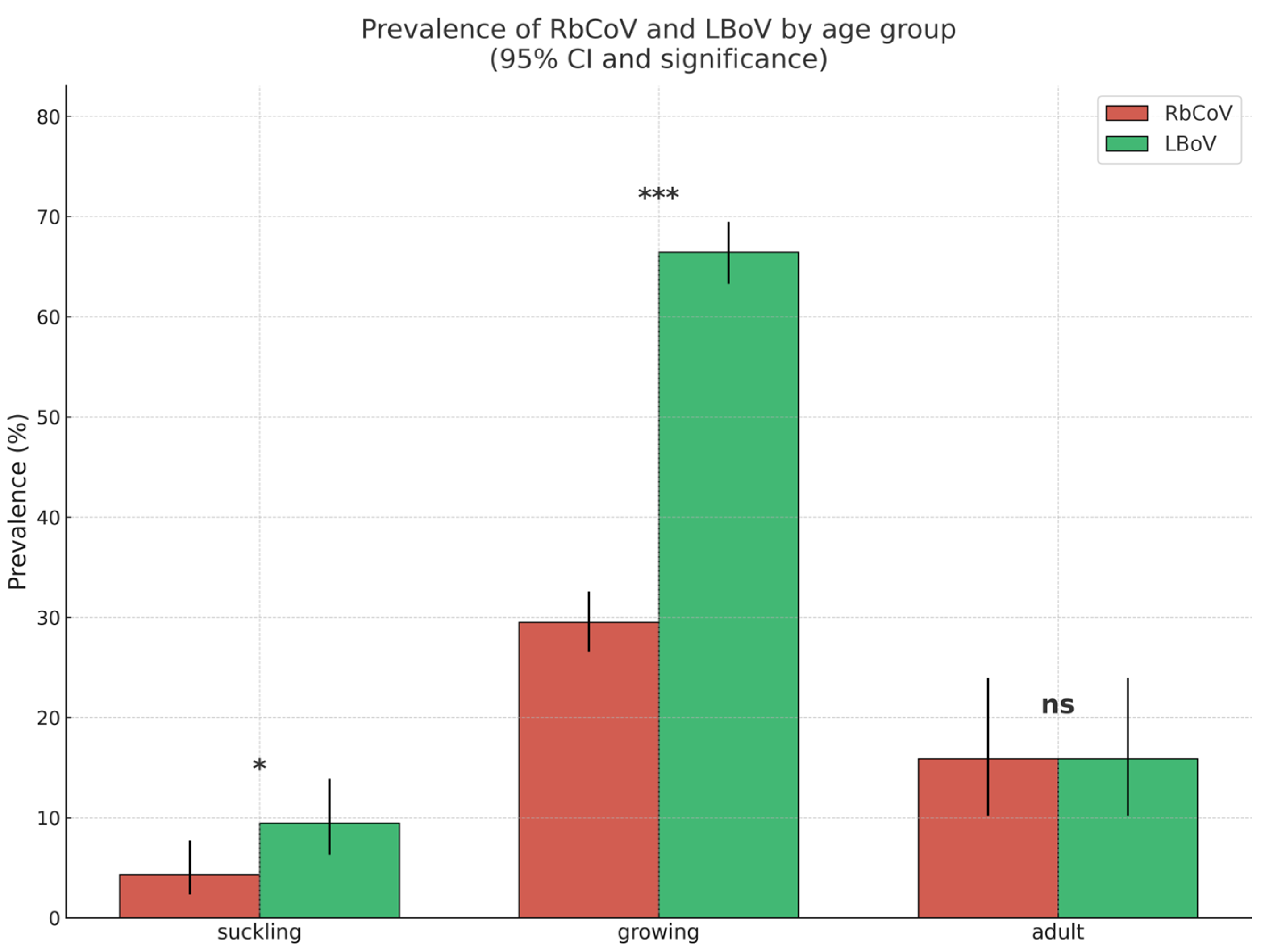
3.3. Study C: Epidemiological Situation de RbCoV and LBoV in Spain
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| RbCoV | Rabbit Coronavirus |
| LBoV | Lapine Bocaparvovirus |
| RKoB | Rabbit Kobuvirus |
| qPCR | Real-time PCR |
| RdRp | RNA-dependent RNA polymerase |
| NRACE | Non-Regenerative Atrophic Catarrhal Enteritis |
| RACE | Regenerative Atrophic Catarrhal Enteritis |
| WGS | Whole Genome Sequencing |
References
- Solans, L.; Arnal, J.L.; Sanz, C.; Benito, A.; Chacón, G.; Alzuguren, O.; Fernández, A.B. Rabbit Enteropathies on Commercial Farms in the Iberian Peninsula: Etiological Agents Identified in 2018–2019. Animals 2019, 9, 1142. [Google Scholar] [CrossRef] [PubMed]
- Marlier, D.; Dewrée, R.; Delleur, V.; Licois, D.; Lassence, C.; Poulipoulis, A.; Vindevogel, H. Descriptionm des principals étiologies des maladies digestives chez le lapin européen (Oryctolagus cuniculus). Ann. Méd. Vét. 2003, 147, 385–392. [Google Scholar]
- Cerioli, M.; Lavazza, A. Viral Enteritis of Rabbits. In Recent Advances in Rabbit Sciences; Maertens, L., Coudert, P., Eds.; Ilvo: Meerelbeke, Belgium, 2006; pp. 181–186. [Google Scholar]
- Lapierre, J.; Marsolais, G.; Pilon, P.; Descôteaux, J.P. Preliminary Report on the Observation of a Coronavirus in the Intestine of the Laboratory Rabbit. Can. J. Microbiol. 1980, 26, 1204–1208. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Woo, P.C.Y.; Yip, C.C.Y.; Fan, R.Y.Y.; Huang, Y.; Wang, M.; Guo, R.; Lam, C.S.F.; Tsang, A.K.L.; Lai, K.K.Y.; et al. Isolation and Characterization of a Novel Betacoronavirus Subgroup A Coronavirus, Rabbit Coronavirus HKU14, from Domestic Rabbits. J. Virol. 2012, 86, 5481–5496. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, Y.; Matsuno, S.; Mukoyama, J. Isolation and Characterization of a Parvovirus of Rabbits. Infect. Immun. 1977, 18, 495–500. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, Y.; Chino, F. Experimental Infection of Young Rabbits with Rabbit Parvovirus. Arch. Virol. 1981, 68, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Lanave, G.; Martella, V.; Farkas, S.L.; Marton, S.; Fehér, E.; Bodnar, L.; Lavazza, A.; Decaro, N.; Buonavoglia, C.; Bányai, K. Novel Bocaparvoviruses in Rabbits. Vet. J. 2015, 206, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Pankovics, P.; Boros, Á.; Bíró, H.; Horváth, K.B.; Phan, T.G.; Delwart, E.; Reuter, G. Novel Picornavirus in Domestic Rabbits (Oryctolagus cuniculus var. domestica). Infect. Genet. Evol. 2016, 37, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Martín-Alonso, J.M.; Skilling, D.E.; González-Molleda, L.; del Barrio, G.; Machín, A.; Keefer, N.K.; Matson, D.O.; Iversen, P.L.; Smith, A.W.; Parra, F. Isolation and Characterization of a New Vesivirus from Rabbits. Virology 2005, 337, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Ministerio de Agriculturaa, Pesca y Alimentación. Available online: https://www.mapa.gob.es/es/estadistica/temas/estadisticas-agrarias/ganaderia/encuesta-nacional-cunicultura (accessed on 31 July 2025).
- Martínez, N.; Brandão, P.E.; de Souza, S.P.; Barrera, M.; Santana, N.; de Arce, H.D.; Pérez, L.J. Molecular and Phylogenetic Analysis of Bovine Coronavirus Based on the Spike Glycoprotein Gene. Infect. Genet. Evol. 2012, 12, 1870–1878. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, J.B.; Lederman, M.; Stout, E.R.; Bates, R.C. Natural Parvovirus Infection in Laboratory Rabbits. Am. J. Vet. Res. 1989, 50, 1048–1051. [Google Scholar] [CrossRef] [PubMed]
- Deeb, B.J.; DiGiacomo, R.F.; Evermann, J.F.; Thouless, M.E. Prevalence of Coronavirus Antibodies in Rabbits. Lab. Anim. Sci. 1993, 43, 431–433. [Google Scholar] [PubMed]
- Zhou, L.; Lu, X.; Zhao, C.; Zhang, Y.; Ning, S.; Zhang, E. Characterization of a Novel Picornavirus Prevalent in Experimental Rabbits (Oryctolagus cuniculus). Heliyon 2023, 9, e15702. [Google Scholar] [CrossRef] [PubMed]
- Swennes, A.G.; Buckley, E.M.; Madden, C.M.; Byrd, C.P.; Donocoff, R.S.; Rodriguez, L.; Parry, N.M.A.; Fox, J.G. Enteropathogenic Escherichia coli Prevalence in Laboratory Rabbits. Vet. Microbiol. 2013, 163, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.V. Digestive Disorders. In Texbook of Rabbit Medicine, 3rd ed.; Smith, M.V., Ed.; Elsevier: Warsaw, Poland, 2023; pp. 156–191. [Google Scholar]
- Balicka-Ramisz, A.; Laurans, L.; Pohorecki, K.; Batko, M.; Ramisz, A. Prevalence of Eimeria spp. Infection in Domestic Rabbits of Polish Farms. World Rabbit. Sci. 2020, 28, 181–185. [Google Scholar] [CrossRef]
- Fernández, G. Enfermedades Infecciosas que Cursan con Procesos Digestivos en Conejos. Bol. De Cunicult. 2006, 144, 23–40. [Google Scholar]
- Oglesbee, B.L.; Jenkins, J.R. Gastrointestinal Diseases. In Ferrets, Rabbits and Rodent. Clinical Medicine and Surgery, 3rd ed.; Katherine, E.Q., James, W.C., Eds.; Elsevier Saunders: St. Louis, MO, USA, 2012; pp. 193–204. [Google Scholar]
- Krogstad, A.P.; Simpson, J.E.; Korte, S.W. Viral Diseases of the Rabbit. Vet. Clin. Exot. Anim. 2005, 8, 123–138. [Google Scholar] [CrossRef]
| HP | qPCR | |||||||
|---|---|---|---|---|---|---|---|---|
| Rabbitry and Sample | Lesion | LBoV | RbCoV | Lapine Rotavirus | Eimeria spp. | C. spiroforme | C. perfringens | E. coli (eae) |
| R1-1 | a + c (NRACE) | 35.27 | 21.02 | N/A | N/A | 30.33 | N/A | 21.41 |
| R1-2 | a + c (NRACE) | 25.77 | 27.68 | 34.89 | 33.99 | 36.54 | N/A | 22.63 |
| R1-3 | d | 24.09 | 39.26 | 32.95 | 36.09 | N/A | N/A | 21.15 |
| R1-4 | d | 34.18 | 34.15 | N/A | N/A | N/A | 36.99 | 21.73 |
| R1-5 | a | 22.90 | 24.44 | N/A | N/A | 34.07 | N/A | 22.53 |
| R2-1 | a + c (NRACE) | 30.78 | 30.99 | N/A | 35.01 | N/A | N/A | 20.38 |
| R2-2 | W/A | 20.92 | N/A | N/A | 31.09 | 29.51 | 36.27 | 36.38 |
| R2-3 | a | 20.28 | N/A | N/A | 33.15 | 29.83 | N/A | N/A |
| R2-4 | a | 35.11 | N/A | N/A | 28.58 | 31.86 | N/A | 22.89 |
| R2-5 | a | 29.66 | N/A | N/A | 31.92 | N/A | N/A | N/A |
| R3-1 | a + c (NRACE) | N/A | 38.11 | 38,63 | N/A | 33.49 | N/A | 20.92 |
| R3-2 | W/A | 36.99 | 32.37 | 35.37 | N/A | N/A | 39.93 | 21.90 |
| R3-3 | a | 31.25 | 35.09 | 37.71 | 31.04 | N/A | N/A | 22.75 |
| R3-4 | a + c (NRACE) | 36.76 | 38.50 | N/A | 35.91 | N/A | 38.67 | 20.17 |
| R3-5 | a + b (RACE) | 29.28 | 38.81 | 39,72 | N/A | 33.09 | N/A | 23.73 |
| R4-1 | a + c (NRACE) | 33.97 | 36.38 | N/A | 37.61 | 29.40 | N/A | 23.30 |
| R4-2 | W/A | 37.91 | 35.50 | N/A | N/A | 35.02 | N/A | 22.85 |
| R4-3 | a | 33.22 | 25.96 | N/A | N/A | N/A | N/A | 23.04 |
| R4-4 | W/A | 29.40 | 35.13 | N/A | N/A | 32.13 | N/A | 23.08 |
| R4-5 | a + b (RACE) | 28.24 | 32.07 | N/A | N/A | 37.70 | N/A | 26.90 |
| R5-1 | a | 25.30 | N/A | 36.44 | N/A | 29.74 | N/A | 21.58 |
| R5-2 | W/A | 29.64 | N/A | 38.19 | N/A | 34.20 | N/A | 20.25 |
| R5-3 | a + c (NRACE) | 27.85 | N/A | 39.68 | N/A | N/A | N/A | 21.29 |
| R5-4 | a | 36.74 | N/A | 36.66 | N/A | N/A | N/A | 20.12 |
| R5-5 | a + b (RACE) | 35.18 | N/A | N/A | N/A | 29.17 | N/A | 19.12 |
| R6-1 | a | 32.55 | N/A | 28.77 | N/A | 29.10 | 38.26 | N/A |
| R6-2 | a + b (RACE) | 23.89 | N/A | 29.25 | N/A | 27.45 | N/A | N/A |
| R6-3 | W/A | 34.02 | N/A | 35.86 | 31.60 | 23.71 | N/A | N/A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arnal, J.L.; Pallarés, F.J.; Sanz, C.; Álvarez-Delgado, C.; Chacón, G.; Carrasco, L. Rabbit Enteric Syndrome: The Implications of Rabbit Coronavirus (RbCoV) and Lapine Bocaparvovirus (LBoV) in Rabbitries of Spain. Vet. Sci. 2025, 12, 1037. https://doi.org/10.3390/vetsci12111037
Arnal JL, Pallarés FJ, Sanz C, Álvarez-Delgado C, Chacón G, Carrasco L. Rabbit Enteric Syndrome: The Implications of Rabbit Coronavirus (RbCoV) and Lapine Bocaparvovirus (LBoV) in Rabbitries of Spain. Veterinary Sciences. 2025; 12(11):1037. https://doi.org/10.3390/vetsci12111037
Chicago/Turabian StyleArnal, José Luis, Francisco José Pallarés, Celia Sanz, Carmen Álvarez-Delgado, Gema Chacón, and Librado Carrasco. 2025. "Rabbit Enteric Syndrome: The Implications of Rabbit Coronavirus (RbCoV) and Lapine Bocaparvovirus (LBoV) in Rabbitries of Spain" Veterinary Sciences 12, no. 11: 1037. https://doi.org/10.3390/vetsci12111037
APA StyleArnal, J. L., Pallarés, F. J., Sanz, C., Álvarez-Delgado, C., Chacón, G., & Carrasco, L. (2025). Rabbit Enteric Syndrome: The Implications of Rabbit Coronavirus (RbCoV) and Lapine Bocaparvovirus (LBoV) in Rabbitries of Spain. Veterinary Sciences, 12(11), 1037. https://doi.org/10.3390/vetsci12111037







