Genetic Diversity and Population Structure of Fat-Tailed Coarse-Wooled Sheep Breeds Ovis aries from Kazakhstan
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Information
2.2. DNA Extraction, Amplification, and Sequencing
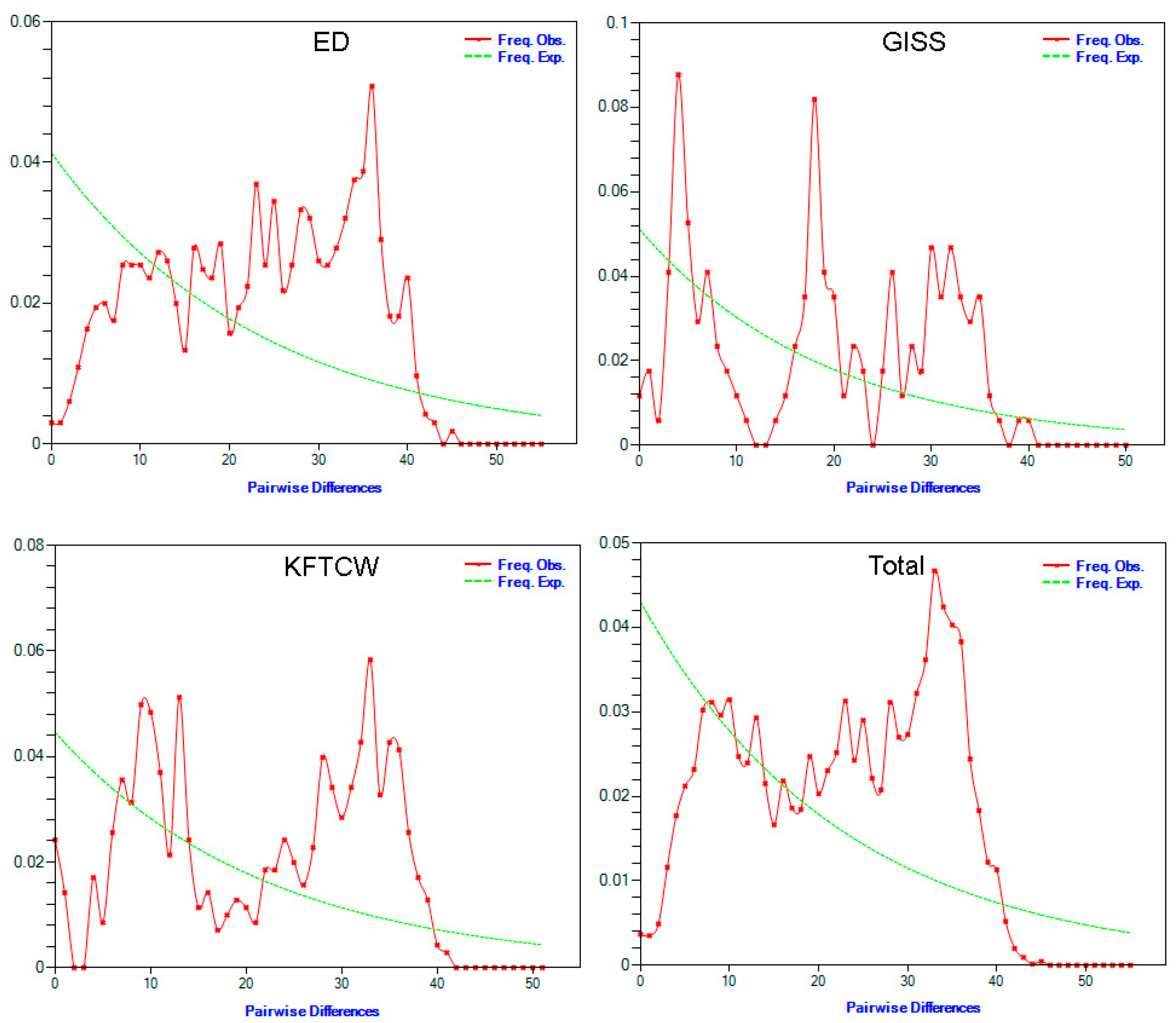
2.3. Molecular Diversity and Population Dynamics Analyses
2.4. Phylogenetic Analysis
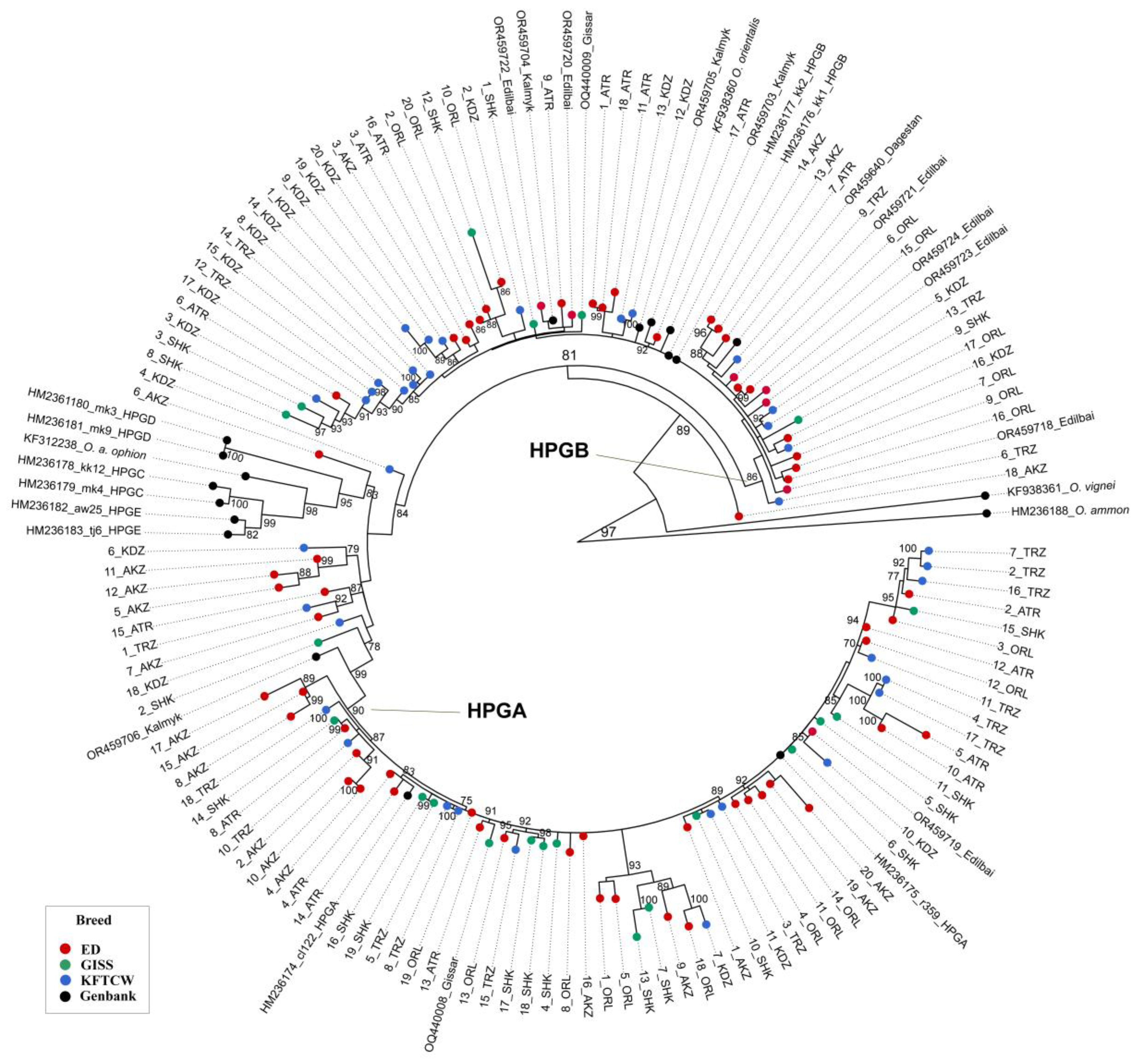
3. Results
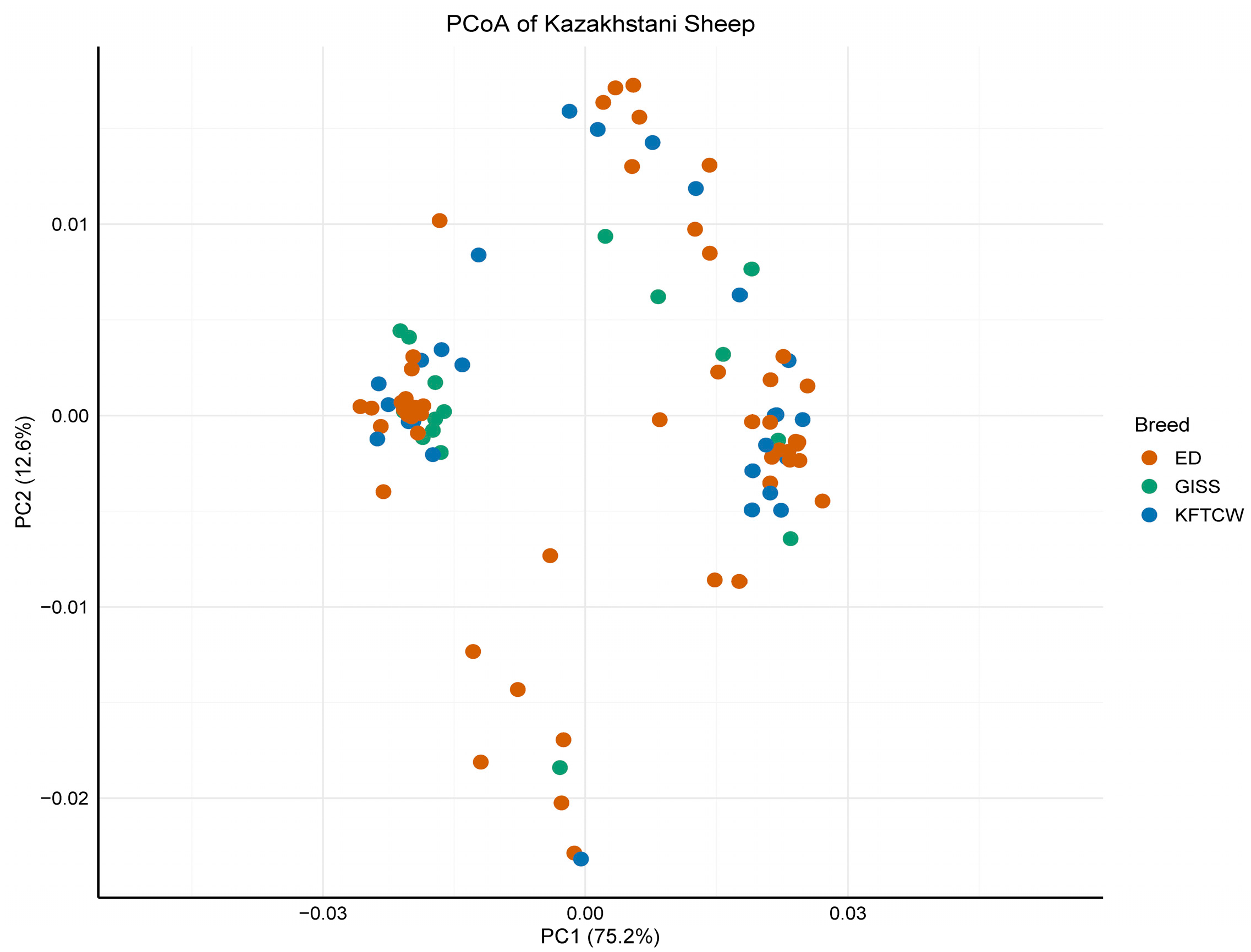
3.1. MtDNA Diversity and Population Structure of Kazakhstani Sheep Breeds
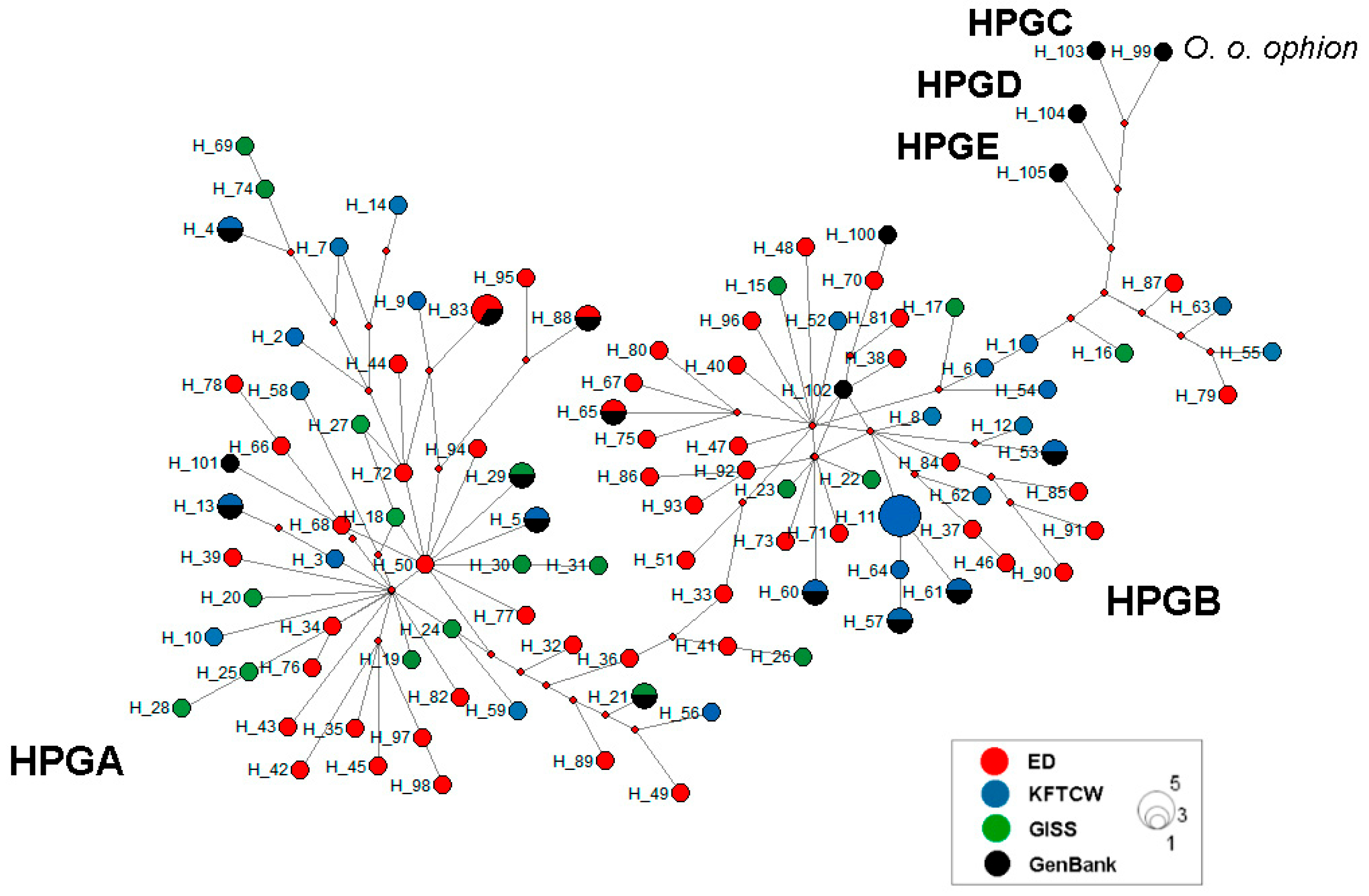
3.2. Phylogenetic Relationships
4. Discussion
4.1. Maternal Genetic Diversity and Regional Comparisons
4.2. Population Dynamics and Structure
4.3. Phylogenetic Relationships Among Kazakhstani Sheep Breeds
4.4. Implications for Breed Conservation and Genetic Management
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DNA | Deoxyribonucleic acid |
| KZ | Kazakhstan |
| D-loop | Displacement loop (mitochondrial control region) |
References
- Hiendleder, S.; Mainz, K.; Plante, Y.; Lewalski, H. Analysis of mitochondrial DNA sequences in domestic sheep: Phylogenetic and technical considerations. Genetics 2002, 160, 1651–1660. [Google Scholar] [CrossRef]
- Meadows, J.R.S.; Cemal, İ.; Karaca, O.; Gootwine, E.; Kijas, J.W. Five ovine mitochondrial lineages identified from sheep breeds of the Near East. Genetics 2007, 175, 1371–1379. [Google Scholar] [CrossRef]
- Zeder, M.A. The domestication of animals. J. Anthropol. Res. 2012, 68, 161–190. [Google Scholar] [CrossRef]
- Kamalakkannan, R.; Kumar, S.; Bhavana, K.; Prabhu, V.R.; Machado, C.B.; Singha, H.S.; Sureshgopi, D.; Vijay, V.; Nagarajan, M. Evidence for independent domestication of sheep mtDNA lineage A in India and introduction of lineage B through Arabian sea route. Sci. Rep. 2021, 11, 19733. [Google Scholar] [CrossRef] [PubMed]
- Mukhametzharova, I.; Islamov, Y.; Ganieva, G.; Bespalova, A. Genetic characterization of Kazakh native sheep breeds using mitochondrial DNA. Online J. Biol. Sci. 2018, 18, 341–348. [Google Scholar] [CrossRef]
- Meadows, J.R.S.; Hiendleder, S.; Kijas, J.W. Haplogroup relationships between domestic and wild sheep resolved using a mitogenome panel. Heredity 2011, 106, 700–706. [Google Scholar] [CrossRef]
- Demirci, S.; Koban Baştanlar, E.; Dağtaş, N.D.; Pişkin, E.; Engin, A.; Özer, F.; Yüncü, E.; Doğan, S.A.; Togan, I. Mitochondrial DNA diversity of modern, ancient and wild sheep (Ovis gmelinii anatolica) from Turkey: New insights on the evolutionary history of sheep. PLoS ONE 2013, 8, e81952. [Google Scholar] [CrossRef]
- Kerven, C.; Robinson, S.; Behnke, R. Pastoralism at scale on the Kazakh rangelands: From Clans to Workers to Ranchers. Front. Sustain. Food Syst. 2021, 4, 590401. [Google Scholar] [CrossRef]
- FAO. Domestic Animal Diversity Information System (DAD-IS). Food and Agriculture Organization of the United Nations. 2024. Available online: https://www.fao.org/dad-is/en/ (accessed on 19 October 2024).
- Kanapin, K.K. Edilbay Sheep; Bastau: Almaty, Kazakhstan, 2009; 180p. (In Russian) [Google Scholar]
- Dossybayev, K.; Amandykova, M.; Abdrakhmanov, S.; Orakbayeva, A.; Yergali, K.; Torekhanov, A. Genetic diversity and relationships among sheep breeds in Kazakhstan based on 12 microsatellite loci. J. Anim. Plant Sci. 2019, 29, 1601–1609. [Google Scholar]
- Zhumadilla, K.; Zhumadillaev, N.K. State and prospects of development of Kurdy meat-sal sheep breeding in Kazakhstan. In Proceedings of the International Scientific and Practical Conference, Almaty, Kazakhstan, 29 October 2014; Zootechnical Science of Kazakhstan: Past, Present and Future. pp. 163–172. [Google Scholar]
- Ikromov, F. Methods for Increasing the Prolificacy of Gissar Ewes at Small- and Middle-Scale Households; Taj Livestock Research Institute: Tashkent, Uzbekistan, 2010; 7p. [Google Scholar]
- Wood, N.J.; Phua, S.H. Variation in the control region sequence of the sheep mitochondrial genome. Anim. Genet. 1996, 27, 25–33. [Google Scholar] [CrossRef]
- Pozharskiy, A.; Khamzina, A.; Gritsenko, D.; Khamzina, Z.; Kassymbekova, S.; Karimov, N.; Karymsakov, T.; Tlevlesov, N. SNP genotyping and population analysis of five indigenous Kazakh sheep breeds. Livest. Sci. 2020, 241, 104252. [Google Scholar] [CrossRef]
- Dossybayev, K.; Amandykova, M.; Orakbayeva, A.; Adylkanova, S.; Kozhakhmet, A.; Yergali, K.; Kulboldin, T.; Kulataev, B.; Torekhanov, A. Genome-wide association studies revealed several candidate genes of meat productivity in Saryarka fat-tailed coarse-wool sheep breed. Genes 2024, 15, 1549. [Google Scholar] [CrossRef]
- Tarlykov, P.; Atavliyeva, S.; Auganova, D.; Akhmetollayev, I.; Loshakova, T.; Varfolomeev, V.; Ramankulov, Y. Mitochondrial DNA analysis of ancient sheep from Kazakhstan: Evidence for early sheep introduction. Heliyon 2021, 7, e08011. [Google Scholar] [CrossRef]
- Tapio, M.; Marzanov, N.; Ozerov, M.; Ćinkulov, M.; Gonzarenko, G.; Kiselyova, T.; Kantanen, J. Sheep mitochondrial DNA variation in European, Caucasian, and Central Asian areas. Mol. Biol. Evol. 2006, 23, 1776–1783. [Google Scholar] [CrossRef]
- Hiendleder, S.; Mainz, K.; Plante, Y.; Lewalski, H. Analysis of mitochondrial DNA indicates that domestic sheep are derived from two different ancestral maternal sources: No evidence for contributions from urial and argali sheep. J. Hered. 1998, 89, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Othman, E.O.; Balabel, E.A.; Abdel-Samad, M.F. Mitochondrial DNA Diversity in Five Egyptian Sheep Breeds. Glob. Vet. 2014, 12, 369–375. [Google Scholar] [CrossRef]
- Zhao, Y.X.; Yang, J.; Lv, F.H.; Hu, X.J.; Xie, X.L.; Zhang, M.; Li, W.R.; Liu, M.J.; Wang, Y.T.; Li, J.Q.; et al. Genomic Reconstruction of the History of Native Sheep Reveals the Peopling Patterns of Nomads and the Expansion of Early Pastoralism in East Asia. Mol. Biol. Evol. 2017, 34, 2380–2395. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-L.; Gong, Y.-F.; Liu, Z.-Z.; Zheng, G.-R.; Zhou, R.-Y.; Jin, X.-M.; Li, L.-H.; Wang, H.-L. Study on tandem repeat sequence variation in sheep mtDNA D-loop region. Acta Genet. Sin. 2006, 33, 1087–1095. [Google Scholar]
- Burland, T.G. DNASTAR’s Lasergene sequence analysis software. Methods Mol. Biol. 2000, 132, 71–91. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Li, W.; Jakovlic, I.; Zou, H.; Zhang, J.; Wang, G. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Fluxus-Engineering. Network 10.2.0.0; Fluxus Technology Ltd.: Clare, Suffolk, UK, 2023; Available online: https://www.fluxus-engineering.com (accessed on 15 December 2024).
- Fu, Y.-X.; Li, W.-H. Statistical tests of neutrality of mutations. Genetics 1993, 133, 693–709. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.1): An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2005, 1, 47–50. [Google Scholar] [CrossRef]
- Paradis, E.; Schliep, K. ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Koshkina, O.; Deniskova, T.; Dotsev, A.; Kunz, E.; Selionova, M.; Medugorac, I.; Zinovieva, N. Phylogenetic analysis of Russian native sheep breeds based on mtDNA sequences. Genes 2023, 14, 1701. [Google Scholar] [CrossRef]
- Lv, F.-H.; Wang, X.-X.; Liu, D.; Peng, W.-F.; Yang, J.; Zhao, Y.-X.; Li, W.-R.; Liu, M.-J.; Ma, Y.-H.; Zhao, Q.-J.; et al. Mitogenomic meta-analysis identifies two phases of migration in the history of eastern Eurasian sheep. Mol. Biol. Evol. 2015, 32, 2515–2533. [Google Scholar] [CrossRef]
- Sanna, D.; Barbato, M.; Hadjisterkotis, E.; Cossu, P.; Decandia, L.; Trova, S.; Pirastru, M.; Leoni, G.G.; Naitana, S.; Francalacci, P.; et al. The first mitogenome of the Cyprus mouflon (Ovis gmelina ophion): New insights into the phylogeny of the genus Ovis. PLoS ONE 2015, 10, e0144257. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; Haeseler, A.V. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates and new developments. Nucleic Acids Res. 2024, 52, W38–W47. [Google Scholar] [CrossRef]
- Mereu, P.; Pirastru, M.; Sana, D.; Bassu, G.; Naitana, S.; Leoni, G.G. Phenotype transition from wild mouflon to domestic sheep. Genet. Sel. Evol. 2024, 56, 1. [Google Scholar] [CrossRef]
- Nigussie, H.; Mwacharo, J.M.; Osama, S.; Agaba, M.; Mekasha, Y.; Kebede, K.; Abegaz, S.; Pal, S.K. Genetic diversity and matrilineal genetic origin of fat-rumped sheep in Ethiopia. Trop. Anim. Health Prod. 2019, 51, 1393–1404. [Google Scholar] [CrossRef]
- Meadows, J.R.S.; Li, K.; Kantanen, J.; Tapio, M.; Sipos, W.; Pardeshi, V.; Gupta, V.; Calvo, J.H.; Whan, V.; Norris, B.; et al. Mitochondrial sequence reveals high levels of gene flow between breeds of domestic sheep from Asia and Europe. J. Hered. 2005, 96, 494–501. [Google Scholar] [CrossRef]
- Kalaydzhiev, G.; Palova, N.; Dundarova, H.; Lozanova, L.; Mehandjyiski, I.; Radoslavov, G.; Hristov, P. Mitochondrial diversity and phylogenetic relationship of eight native Bulgarian sheep Breeds. Animals 2023, 13, 3655. [Google Scholar] [CrossRef]
- Kim, Y.S.; Tseveen, K.; Batsukh, B.; Seong, J.; Kong, H.S. Origin-related study of genetic diversity and heteroplasmy of Mongolian sheep (Ovis aries) using mitochondrial DNA. J. Anim. Reprod. Biotechnol. 2020, 35, 198–206. [Google Scholar] [CrossRef]
- Deniskova, T.; Dotsev, A.; Lushihina, E.; Shakhin, A.; Kunz, E.; Medugorac, I.; Rever, H.; Wimmers, K.; Khayatzadeh, N.; Solkner, J.; et al. Population Structure and Genetic Diversity of Sheep Breeds in the Kyrgyzstan. Front Genet. 2019, 10, 1311. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, X.; Hu, Y.; Li, H.; Peng, Z.; Xiang, H.; Zhou, X.; Zhao, X. Ancient mitochondrial genome depicts sheep maternal dispersal and migration in Eastern Asia. J. Genet. Genom. 2024, 51, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Naderi, S.; Rezaei, H.R.; Pompanon, F.; Blum, M.G.B.; Negrini, R.; Naghash, H.R.; Balkiz, Ö.; Mashkour, M.; Gaggiotti, O.E.; Ajmone-Marsan, P.; et al. The goat domestication process inferred from large-scale mitochondrial DNA analysis of wild and domestic individuals. Proc. Natl. Acad. Sci. USA 2008, 105, 17659–17664. [Google Scholar] [CrossRef] [PubMed]
- Pedrosa, S.; Uzun, M.; Arranz, J.J.; Gutiérrez-Gil, B.; San Primitivo, F.; Bayón, Y. Evidence of three maternal lineages in Near Eastern sheep supporting multiple domestication events. Proc. R. Soc. B Biol. Sci. 2005, 272, 2211–2217. [Google Scholar] [CrossRef]
- Ganbold, O.; Lee, S.-H.; Seo, D.; Paek, W.K.; Manjula, P.; Munkhbayar, M.; Lee, J.H. Genetic diversity and the origin of Mongolian native sheep. Livest. Sci. 2019, 220, 17–25. [Google Scholar] [CrossRef]
- Sharma, R.; Ahlawat, S.; Sharma, H.; Sharma, P.; Panchal, P.; Arora, R.; Tantia, M.S. Microsatellite and mitochondrial DNA analyses unveil the genetic structure of native sheep breeds from three major agro-ecological regions of India. Sci. Rep. 2020, 10, 20422. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Nadeem, M.S.; Johansson, A.M.; Jonas, E.; Muhammad, K.; Mehmood, S.A. Molecular diversity and phylogenetic analysis of ten sheep breeds from Khyber Pakhtunkhwa, Pakistan, based on mitochondrial D-loop sequences. Mamm. Genome 2025, 36, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Moradi, M.H.; Phua, S.H.; Hedayat, N.; Khodaei-Motlagh, M.; Razmkabir, M. Haplotype and Genetic Diversity of the mDNA in indigenous Iranian sheep and an insight into the history of sheep domestication. J. Agric. Sci. Technol. 2017, 19, 591–601. [Google Scholar]
- Machová, K.; Málková, A.; Vostrý, L. Sheep Post-Domestication Expansion in the Context of Mitochondrial and Y Chromosome Haplogroups and Haplotypes. Genes 2022, 13, 613. [Google Scholar] [CrossRef]
- Peng, W.; Zhang, Y.; Gao, L.; Feng, C.; Yang, Y.; Li, B.; Wu, L.; Wu, A.; Wang, S.; Ren, X.; et al. Analysis of World-Scale Mitochondrial DNA Reveals the Origin and Migration Route of East Asia Goats. Front. Genet. 2022, 13, 796979. [Google Scholar] [CrossRef]
- Radoslavov, G.; Kalaydzhiev, G.; Yordanov, G.; Palova, N.; Salkova, D.; Hristov, P. Genetic diversity of native Bulgarian sheep breeds based on the mitochondrial D-loop sequence analysis. Small Rumin. Res. 2025, 249, 107527. [Google Scholar] [CrossRef]
- Kaptan, D.; Atağ, G.; Vural, K.B.; Morell Miranda, P.; Akbaba, A.; Yüncü, E.; Buluktaev, A.; Abazari, M.F.; Yorulmaz, S.; Kazancı, D.D.; et al. The Population History of Domestic Sheep Revealed by Paleogenomes. Mol. Biol. Evol. 2024, 41, msae158. [Google Scholar] [CrossRef]
- Deniskova, T.E.; Dotsev, A.V.; Selionova, M.I.; Kunz, E.; Medugorac, I.; Reyer, H.; Barbato, M.; Traspov, A.A.; Brem, G.; Zinovieva, N.A. Population structure and genetic diversity of 25 Russian sheep breeds based on whole-genome genotyping. Genet. Sel. Evol. 2018, 50, 29. [Google Scholar] [CrossRef]
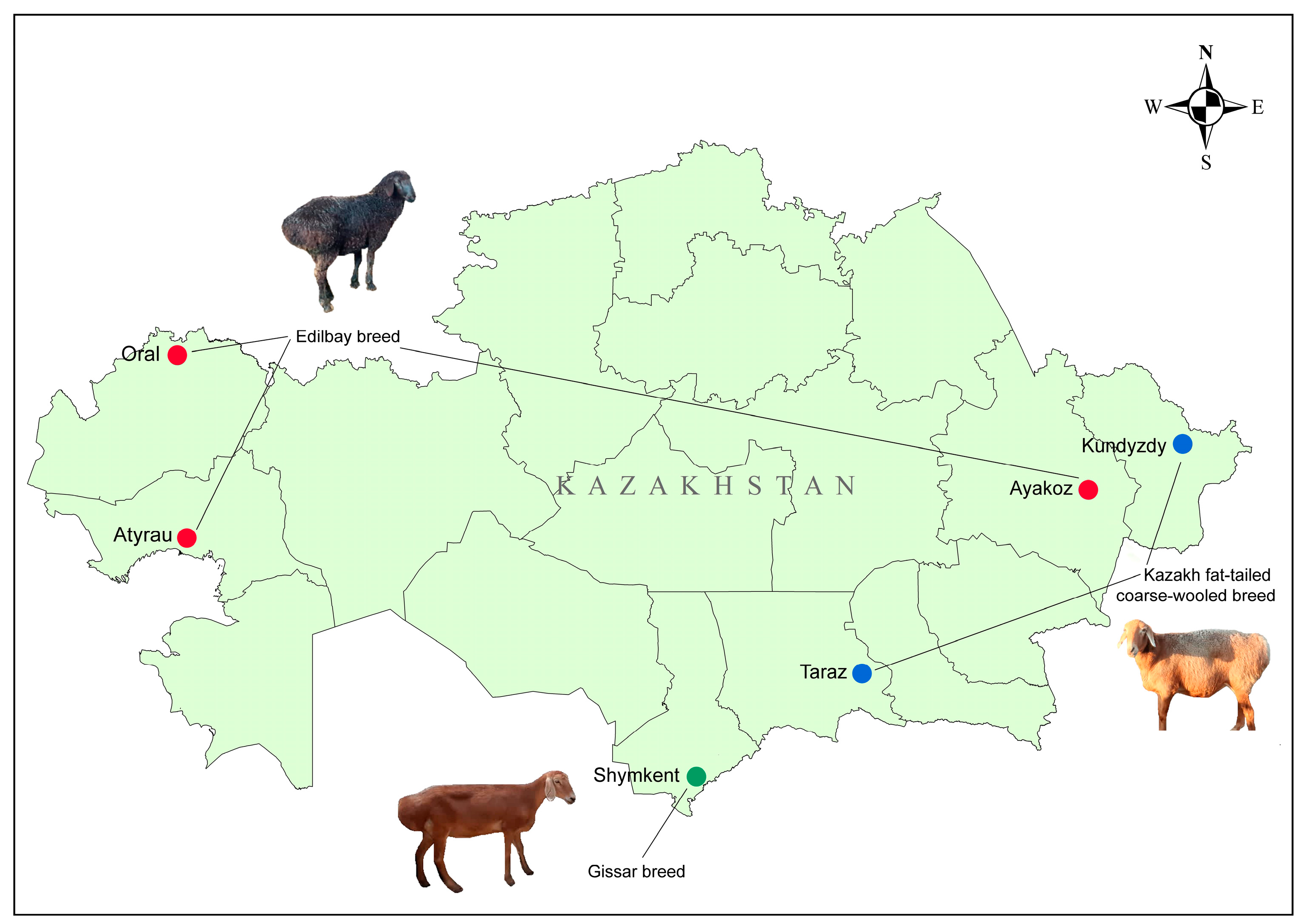
| Breed | Flock | Location | Ns | Farm |
|---|---|---|---|---|
| Edilbay (ED) | ATR | Atyrau region, WKZ | 18 | “Suyindik” |
| ORL | Birlik village, Zhangaly district, WKZ | 20 | “Birlik” | |
| AKZ | Ayakoz town, EKZ | 20 | “Aigul” | |
| Kazakh fat-tailed coarse-wooled (KFTCW) | KDZ | Kundyzdy village, Abay region, EKZ | 20 | “Kundyzdy” |
| TRZ | Shokpar village, Shu district, Zhambyl region, SKZ | 18 | “Aksunkar” | |
| Gissar (GISS) | SHK | Darbaza village, Turkestan region, SKZ | 19 | “Darbaza” |
| Breed | Ns | S | m | k | Nh | Hd ± st.dev. | π ± st.dev. |
|---|---|---|---|---|---|---|---|
| GISS | 58 | 98 | 99 | 23.200 | 54 | 0.997 ± 0.004 | 0.02187 ± 0.00275 |
| KFTCW | 38 | 79 | 79 | 21.438 | 27 | 0.976 ± 0.014 | 0.02528 ± 0.00115 |
| ED | 19 | 57 | 57 | 18.549 | 17 | 0.988 ± 0.021 | 0.02736 ± 0.00074 |
| Total | 115 | 118 | 121 | 22.255 | 98 | 0.996 ± 0.002 | 0.02624 ± 0.00048 |
| Breed | Fu and Li’s D | Fu and Li’s F | Rag | R2 |
|---|---|---|---|---|
| ED | 0.37616 | 0.41342 | 0.0024 | 0.1155 |
| KFTCW | 0.44014 | 0.55362 | 0.0058 | 0.1313 |
| GISS | 0.42196 | 0.53936 | 0.0151 | 0.1524 |
| Total | −0.11543 | −0.11554 | 0.0012 | 0.0927 |
| Source of Variation | d.f. | Sum of Squares | Variance Components | Percentage of Variation |
|---|---|---|---|---|
| Among groups | 2 | 43.814 | −0.34252 Va | −3.06 |
| Among populations within groups | 3 | 102.001 | 1.23505 Vb | 11.03 |
| Within populations | 109 | 1122.758 | 10.30054 Vc | 92.03 |
| Total | 114 | 1268.574 | 11.19307 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dossybayev, K.; Ualiyeva, D.; Kapassuly, T.; Amandykova, M.; Kozhahmet, A.; Bekmanov, B.; Amzeyev, R.; Naruya, S. Genetic Diversity and Population Structure of Fat-Tailed Coarse-Wooled Sheep Breeds Ovis aries from Kazakhstan. Vet. Sci. 2025, 12, 988. https://doi.org/10.3390/vetsci12100988
Dossybayev K, Ualiyeva D, Kapassuly T, Amandykova M, Kozhahmet A, Bekmanov B, Amzeyev R, Naruya S. Genetic Diversity and Population Structure of Fat-Tailed Coarse-Wooled Sheep Breeds Ovis aries from Kazakhstan. Veterinary Sciences. 2025; 12(10):988. https://doi.org/10.3390/vetsci12100988
Chicago/Turabian StyleDossybayev, Kairat, Daniya Ualiyeva, Tilek Kapassuly, Makpal Amandykova, Altynay Kozhahmet, Bakytzhan Bekmanov, Rauan Amzeyev, and Saitou Naruya. 2025. "Genetic Diversity and Population Structure of Fat-Tailed Coarse-Wooled Sheep Breeds Ovis aries from Kazakhstan" Veterinary Sciences 12, no. 10: 988. https://doi.org/10.3390/vetsci12100988
APA StyleDossybayev, K., Ualiyeva, D., Kapassuly, T., Amandykova, M., Kozhahmet, A., Bekmanov, B., Amzeyev, R., & Naruya, S. (2025). Genetic Diversity and Population Structure of Fat-Tailed Coarse-Wooled Sheep Breeds Ovis aries from Kazakhstan. Veterinary Sciences, 12(10), 988. https://doi.org/10.3390/vetsci12100988






