Prevalence, Phylogenetic Distribution, Antimicrobial Resistance, and Genetic Relatedness of Extraintestinal Pathogenic E. coli (ExPEC) Strains Isolated from Beef Cattle and Slaughterhouse Environment
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Sample Collection
2.2. Sampling Procedure
2.3. Isolation of ExPEC in the Samples
2.4. Detection of Virulence Genes Associated with ExPEC
2.5. Antimicrobial Susceptibility Testing
2.6. Phylogenetic Group Determination
2.7. DNA Fingerprinting and Phylogenetic Analysis
2.8. Statistical Analyses
3. Results
3.1. Prevalence of ExPEC
3.2. Virulence Genes Associated with ExPEC
3.3. Antimicrobial Resistance
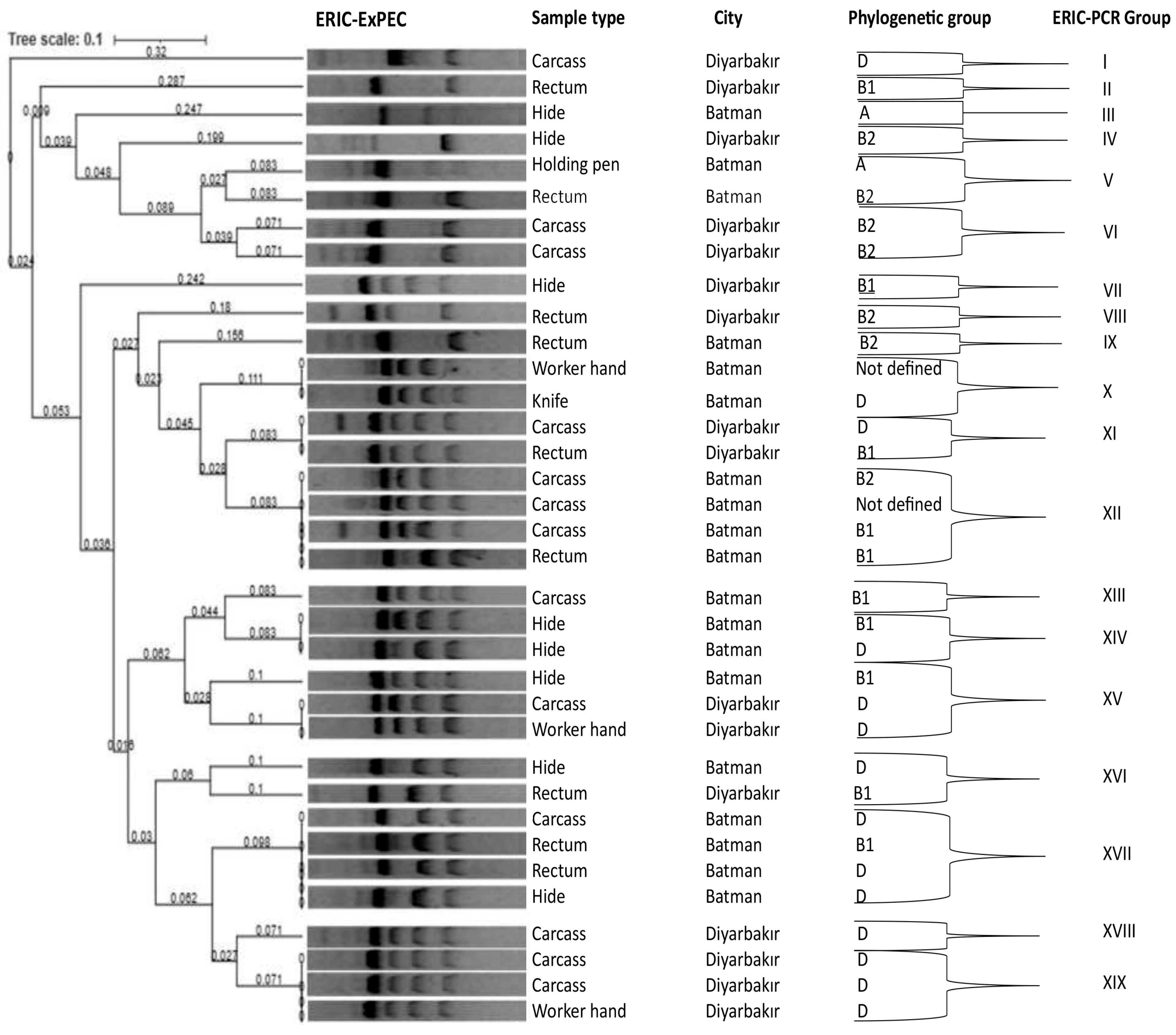
3.4. Phylogenetic Grouping and DNA Fingerprinting
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Banerjee, R.; Johnson, J.R. A New clone sweeps clean: The enigmatic emergence of Escherichia coli sequence type 131. Antimicrob. Agents Chemother. 2014, 58, 4997–5004. [Google Scholar] [CrossRef]
- Gomes, T.A.; Elias, W.P.; Scaletsky, I.C.; Guth, B.E.C.; Rodrigues, J.F.; Piazza, R.M.; Ferreira, L.; Martinez, M.B. Diarrheagenic Escherichia coli. Braz. J. Microbiol. 2016, 47, 3–30. [Google Scholar] [CrossRef] [PubMed]
- Pakbin, B.; Brück, W.M.; Rossen, J.W.A. Virulence factors of enteric pathogenic Escherichia coli: A review. Int. J. Mol. Sci. 2021, 22, 9922. [Google Scholar] [CrossRef] [PubMed]
- Enciso-Martínez, Y.; González-Aguilar, G.A.; Martínez-Téllez, M.A.; González-Pérez, C.J.; Valencia-Rivera, D.E.; Barrios-Villa, E.; Ayala-Zavala, J.F. Relevance of tracking the diversity of Escherichia coli pathotypes to reinforce food safety. Int. J. Food Microbiol. 2022, 374, 109736. [Google Scholar] [CrossRef] [PubMed]
- Singer, R.S. Urinary tract infections attributed to diverse ExPEC strains in food animals: Evidence and data gaps. Front. Microbiol. 2015, 6, 28. [Google Scholar] [CrossRef]
- Clermont, O.; Christenson, J.K.; Denamur, E.; Gordon, D.M. The Clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 2013, 5, 58–65. [Google Scholar] [CrossRef]
- Manges, A.R.; Johnson, J.R. Food-borne origins of Escherichia coli causing extraintestinal infections. Clin. Infect. Dis. 2012, 55, 712–719. [Google Scholar] [CrossRef]
- Manges, A. Escherichia coli and urinary tract infections: The role of poultry-meat. Clin. Microbiol. Infect. 2016, 22, 122–129. [Google Scholar] [CrossRef]
- Wasiński, B. Extra-intestinal pathogenic Escherichia coli—Threat connected with food-borne infections. Ann. Agric. Environ. Med. 2019, 26, 532–537. [Google Scholar] [CrossRef]
- Meena, P.R.; Yadav, P.; Hemlata, H.; Tejavath, K.K.; Singh, A.P. Priyanka Poultry-origin extraintestinal Escherichia coli strains carrying the traits associated with urinary tract infection, sepsis, meningitis and avian colibacillosis in India. J. Appl. Microbiol. 2021, 130, 2087–2101. [Google Scholar] [CrossRef]
- Meena, P.R.; Priyanka, P.; Singh, A.P. Extraintestinal pathogenic Escherichia coli (ExPEC) reservoirs, and antibiotics resistance trends: A one-health surveillance for risk analysis from “farm-to-fork”. Lett. Appl. Microbiol. 2023, 76, ovac016. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Dong, W.; Ma, J.; Yuan, L.; Hejair, H.M.; Pan, Z.; Liu, G.; Yao, H. Characterization and virulence clustering analysis of extraintestinal pathogenic Escherichia coli isolated from swine in China. BMC Vet. Res. 2017, 13, 94. [Google Scholar] [CrossRef] [PubMed]
- De Campos, A.C.; Puño-Sarmiento, J.J.; Medeiros, L.P.; Gazal, L.E.; Maluta, R.P.; Navarro, A.; Kobayashi, R.K.; Fagan, E.P.; Nakazato, G. Virulence genes and antimicrobial resistance in Escherichia coli from cheese made from unpasteurized milk in Brazil. Foodborne Pathog. Dis. 2018, 15, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Boudjerda, D.; Lahouel, M. Virulence and antimicrobial resistance of Escherichia coli isolated from chicken meat, beef, and raw milk. Austral J. Vet. Sci. 2022, 54, 115–125. [Google Scholar] [CrossRef]
- Xia, X.; Meng, J.; Zhao, S.; Bodeis-Jones, S.; Gaines, S.A.; Ayers, S.L.; Mcdermott, P.F. Identification and antimicrobial resistance of extraintestinal pathogenic Escherichia coli from retail meats. J. Food Prot. 2011, 74, 38–44. [Google Scholar] [CrossRef]
- Guzman-Hernandez, R.; Contreras-Rodriguez, A.; Hernandez-Velez, R.; Perez-Martinez, I.; Lopez-Merino, A.; Zaidi, M.B.; Estrada-Garcia, T. Mexican unpasteurised fresh cheeses are contaminated with Salmonella spp., non-O157 Shiga toxin producing Escherichia coli and potential uropathogenic E. coli strains: A public health risk. Int. J. Food Microbiol. 2016, 237, 10–16. [Google Scholar] [CrossRef]
- Sukkua, K.; Pomwised, R.; Rattanachuay, P.; Khianngam, S.; Sukhumungoon, P. Characterization of extraintestinal pathogenic Escherichia Coli from meat in Southern Thailand. Southeast Asian J. Trop. Med. Public Health 2017, 48, 98–108. [Google Scholar]
- Savin, M.; Bierbaum, G.; Kreyenschmidt, J.; Schmithausen, R.M.; Sib, E.; Schmoger, S.; Käsbohrer, A.; Hammerl, J.A. Clinically relevant Escherichia coli isolates from process waters and wastewater of poultry and pig slaughterhouses in germany. Microorganisms 2021, 9, 698. [Google Scholar] [CrossRef]
- Aslam, M.; Toufeer, M.; Narvaez Bravo, C.; Lai, V.; Rempel, H.; Manges, A.; Diarra, M.S. Characterization of extraintestinal pathogenic Escherichia coli isolated from retail poultry meats from Alberta, Canada. Int. J. Food Microbiol. 2014, 177, 49–56. [Google Scholar] [CrossRef]
- Schmidt, J.W.; Agga, G.E.; Bosilevac, J.M.; Brichta-Harhay, D.M.; Shackelford, S.D.; Wang, R.; Wheeler, T.L.; Arthur, T.M. Occurrence of antimicrobial-resistant Escherichia coli and Salmonella enterica in the beef cattle production and processing continuum. Appl. Environ. Microbiol. 2015, 81, 713–725. [Google Scholar] [CrossRef]
- Ghodousi, A.; Bonura, C.; Di Carlo, P.; van Leeuwen, W.B.; Mammina, C. Extraintestinal pathogenic Escherichia coli sequence type 131 H30-R and H30-Rx subclones in retail chicken meat, Italy. Int. J. Food Microbiol. 2016, 228, 10–13. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Antimicrobial Resistance Global Report on Surveillance. 2014. Available online: https://www.who.int/publications/i/item/9789241564748 (accessed on 9 November 2018).
- World Health Organization (WHO). WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 17 December 2022).
- Puvača, N.; Frutos, R.d.L. Antimicrobial resistance in Escherichia coli strains isolated from humans and pet animals. Antibiotics 2021, 10, 69. [Google Scholar] [CrossRef] [PubMed]
- Foxman, B. The epidemiology of urinary tract infection. Nat. Rev. Urol. 2010, 7, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Pitout, J.D.D. Extraintestinal pathogenic Escherichia coli: A combination of virulence with antibiotic resistance. Front. Microbiol. 2012, 3, 9. [Google Scholar] [CrossRef]
- Laxminarayan, R.; Chaudhury, R.R. Antibiotic resistance in India: Drivers and opportunities for action. PLoS Med. 2016, 13, e1001974. [Google Scholar] [CrossRef]
- Arthur, T.M.; Bosilevac, J.M.; Brichta-Harhay, D.M.; Guerini, M.N.; Kalchayanand, N.; Shackelford, S.D.; Wheeler, T.L.; Koohmaraie, M. Transportation and lairage environment effects on prevalence, numbers, and diversity of Escherichia coli O157:H7 on hides and carcasses of beef cattle at processing. J. Food Prot. 2007, 70, 280–286. [Google Scholar] [CrossRef]
- Zweifel, C.; Baltzer, D.; Stephan, R. Microbiological contamination of cattle and pig carcasses at five abattoirs determined by swab sampling in accordance with EU Decision 2001/471/EC. Meat Sci. 2005, 69, 559–566. [Google Scholar] [CrossRef]
- Letellier, A.; Beauchamp, G.; Guévremont, E.; D’ALlaire, S.; Hurnik, D.; Quessy, S. Risk factors at slaughter associated with presence of salmonella on hog carcasses in Canada. J. Food Prot. 2009, 72, 2326–2331. [Google Scholar] [CrossRef]
- Elsharawy, N.T.; Al-Zahrani, H.A.A.; El-Waseif, A.A. Phenotypic and genotypic characterization of antimicrobial resistance in Escherichia coli isolates from chicken meat. J. Food Nutr. Res. 2022, 10, 98–104. [Google Scholar] [CrossRef]
- Queipo-Ortuño, M.I.; Colmenero, J.D.D.; Macias, M.; Bravo, M.J.; Morata, P. Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in real-time pcr for serum samples from patients with brucellosis. Clin. Vaccine Immunol. 2008, 15, 293–296. [Google Scholar] [CrossRef]
- Wang, G.C.Y.; Wang, Y. The frequency of chimeric molecules as a consequence of PCR co-amplification of 16S rRNA genes from different bacterial species. Microbiology 1996, 142, 1107–1114. [Google Scholar] [CrossRef]
- Johnson, J.R.; Murray, A.C.; Gajewski, A.; Sullivan, M.; Snippes, P.; Kuskowski, M.A.; Smith, K.E. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chemother. 2003, 47, 2161–2168. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed]
- Versalovic, J.; Koeuth, T.; Lupski, R. Distribution of repetitive DNA sequences in eubacteria and application to finerpriting of bacterial enomes. Nucleic Acids Res. 1991, 19, 6823–6831. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Garcia-Vallve, S.; Palau, J.; Romeu, A. Horizontal gene transfer in glycosyl hydrolases inferred from codon usage in Escherichia coli and Bacillus subtilis. Mol. Biol. Evol. 1999, 16, 1125–1134. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, 256–259. [Google Scholar] [CrossRef]
- Bergeron, C.R.; Prussing, C.; Boerlin, P.; Daignault, D.; Dutil, L.; Reid-Smith, R.J.; Zhanel, G.G.; Manges, A.R. Chicken as Reservoir for extraintestinal pathogenic Escherichia coli in Humans, Canada. Emerg. Infect. Dis. 2012, 18, 415–421. [Google Scholar] [CrossRef]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global extraintestinal pathogenic Escherichia coli (ExPEC) lineages. Clin. Microbiol. Rev. 2019, 32, e00135-18. [Google Scholar] [CrossRef]
- Stromberg, Z.R.; Johnson, J.R.; Fairbrother, J.M.; Kilbourne, J.; Van Goor, A.; Curtiss, R.; Mellata, M. Evaluation of Escherichia coli isolates from healthy chickens to determine their potential risk to poultry and human health. PLoS ONE 2017, 12, e0180599. [Google Scholar] [CrossRef]
- Lyhs, U.; Ikonen, I.; Pohjanvirta, T.; Raninen, K.; Perko-Mäkelä, P.; Pelkonen, S. Extraintestinal pathogenic Escherichia coli in poultry meat products on the Finnish retail market. Acta Vet. Scand. 2012, 54, 64. [Google Scholar] [CrossRef]
- Santo, E.; Rodolpho, D.; Marin, J.M. Presence of extraintestinal pathogenic Escherichia coli in butcheries in Taquaritinga, SP, Brazil. Braz. J. Microbiol. 2007, 38, 591–593. [Google Scholar] [CrossRef]
- Ovuru, K.F.; Izah, S.C.; Ogidi, O.I.; Imarhiagbe, O.; Ogwu, M.C. Slaughterhouse facilities in developing nations: Sanitation and hygiene practices, microbial contaminants and sustainable management system. Food Sci. Biotechnol. 2024, 33, 519–537. [Google Scholar] [CrossRef] [PubMed]
- Sora, V.M.; Meroni, G.; Martino, P.A.; Soggiu, A.; Bonizzi, L.; Zecconi, A. Extraintestinal pathogenic Escherichia coli: Virulence factors and antibiotic resistance. Pathogens 2021, 10, 1355. [Google Scholar] [CrossRef] [PubMed]
- Micenková, L.; Bosák, J.; Vrba, M.; Ševčíková, A.; Šmajs, D. Human extraintestinal pathogenic Escherichia coli strains differ in prevalence of virulence factors, phylogroups, and bacteriocin determinants. BMC Microbiol. 2016, 16, 218. [Google Scholar] [CrossRef]
- Zou, M.; Ma, P.-P.; Liu, W.-S.; Liang, X.; Li, X.-Y.; Li, Y.-Z.; Liu, B.-T. Prevalence and antibiotic resistance characteristics of extraintestinal pathogenic Escherichia coli among healthy chickens from farms and live poultry markets in China. Animals 2021, 11, 1112. [Google Scholar] [CrossRef]
- Bhave, S.; Kolhe, R.; Mahadevaswamy, R.; Bhong, C.; Jadhav, S.; Nalband, S.; Gandhale, D. Phylogrouping and antimicrobial resistance analysis of extraintestinal pathogenic Escherichia coli isolated from poultry species. Turk. J. Vet. Anim. Sci. 2019, 43, 117–126. [Google Scholar] [CrossRef]
- Ojima, T.; Hirano, K.; Honda, K.; Kusumoto, M. Development of a multiplex PCR assay for rapid virulence factor profiling of extraintestinal pathogenic Escherichia coli isolated from cattle. J. Microbiol. Methods 2016, 128, 31–33. [Google Scholar] [CrossRef]
- Massella, E.; Giacometti, F.; Bonilauri, P.; Reid, C.J.; Djordjevic, S.P.; Merialdi, G.; Bacci, C.; Fiorentini, L.; Massi, P.; Bardasi, L.; et al. Antimicrobial resistance profile and ExPEC virulence potential in commensal Escherichia coli of multiple sources. Antibiotics 2021, 10, 351. [Google Scholar] [CrossRef]
- Sun, M.; Gao, X.; Zhao, K.; Ma, J.; Yao, H.; Pan, Z. Insight into the virulence related secretion systems, fimbriae, and toxins in O2:K1 Escherichia coli isolated from bovine mastitis. Front. Vet. Sci. 2021, 8, 622725. [Google Scholar] [CrossRef]
- Gu, X.; Wu, Q.; Chai, Y.; Huang, X.; Zhou, X.; Han, M.; Wu, T.; Zhang, X.; Zhong, F. Epidemiological and molecular characteristics of extraintestinal pathogenic Escherichia coli isolated from diseased cattle and sheep in Xinjiang, China from 2015 to 2019. BMC Vet. Res. 2025, 21, 42. [Google Scholar] [CrossRef]
- Maluta, R.P.; Logue, C.M.; Casas, M.R.T.; Meng, T.; Guastalli, E.A.L.; Rojas, T.C.G.; Montelli, A.C.; Sadatsune, T.; de Carvalho Ramos, M.; Nolan, L.K.; et al. overlapped sequence types (STS) and serogroups of avian pathogenic (APEC) and human extra-intestinal pathogenic (ExPEC) Escherichia coli Isolated in Brazil. PLoS ONE 2014, 9, e105016. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, A. Characterization of Escherichia coli phylogenetic groups associated with extraintestinal infections in South Indian population. Ann. Med. Health Sci. Res. 2015, 5, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Tayh, G.; Elmanama, A.; Selmi, R.; Ben Slama, K. Antibiotic resistance profile and molecular characterization of extraintestinal pathogenic Escherichia coli (ExPEC) from human clinical samples in gaza strip, palestine. Lett. Appl. Microbiol. 2023, 76, ovac033. [Google Scholar] [CrossRef] [PubMed]
- Kasimanickam, V.; Kasimanickam, M.; Kasimanickam, R. Antibiotics use in food animal production: Escalation of antimicrobial resistance: Where are we now in combating AMR? Med. Sci. 2021, 9, 14. [Google Scholar] [CrossRef]
- Kalyoncu, B.N.; Koçoğlu, E.; Özekïncï, T.; Bïçer, R.T.; Aydin, G.; Önder, N.; Özmen, M. Istanbul’da bir şehir hastanesinde izole edilen üriner sistem patojenleri ve antibiyotik direnç profillerinin değerlendirilmesi*. ANKEM Derg. 2023, 37, 18–27. [Google Scholar] [CrossRef]
- Rayasam, S.D.G.; Ray, I.; Smith, K.R.; Riley, L.W. Extraintestinal pathogenic Escherichia coli and antimicrobial drug resistance in a maharashtrian drinking water system. Am. J. Trop. Med. Hyg. 2019, 100, 1101–1104. [Google Scholar] [CrossRef]
- Caruso, G.; Giammanco, A.; Cardamone, C.; Oliveri, G.; Mascarella, C.; Capra, G.; Fasciana, T. Extra-Intestinal Fluo-roquinolone-Resistant Escherichia coli Strains Isolated from Meat. BioMed Res. Int. 2018, 2018, 8714975. [Google Scholar] [CrossRef]
- Gautam, H.; Maheshwari, B.; Mohapatra, S.; Sood, S.; Dhawan, B.; Kapil, A.; Tezpur, B. Clonal relationship among Acinetobacter baumannii isolates from different clinical specimens by ERIC-PCR. Int. J. Infect. Dis. 2022, 116, S18–S19. [Google Scholar] [CrossRef]
- Sarowska, J.; Olszak, T.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Futoma-Koloch, B.; Gawel, A.; Drulis-Kawa, Z.; Choroszy-Krol, I. Comparative Characteristics and pathogenic potential of Escherichia coli isolates originating from poultry farms, retail meat, and human urinary tract infection. Life 2022, 12, 845. [Google Scholar] [CrossRef]
- Beghain, J.; Bridier-Nahmias, A.; Le Nagard, H.; Denamur, E.; Clermont, O. ClermonTyping: An easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 2018, 4, e000192. [Google Scholar] [CrossRef]
| Samples | Diyarbakır | Batman | Number of Samples |
|---|---|---|---|
| Carcass | 62 | 71 | 133 |
| Hide | 62 | 71 | 133 |
| Rectum | 62 | 71 | 133 |
| Knife | 9 | 9 | 18 |
| Workers’ hand | 6 | 6 | 12 |
| Holding pen | 6 | 6 | 12 |
| Water | 3 | 3 | 6 |
| Total | 210 | 237 | 447 |
| Target Gene | Primer Sequence (5′–3′) | Amplicon Length (bp) | PCR Analysis (Method) | Reference |
|---|---|---|---|---|
| E. coli 16S rRNA | F: GACCTCGGTTTAGTTCACAGA R: CACACGCTGACGCTGACCA | 585 | E.coli confirmation (simplex PCR) | [33] |
| papA | F: ATGGCAGTGGTGTCTTTTGGTG R: CGTCCCACCATACGTGCTCTTC | 717 | ExPEC virulence gene analysis (Multiplex PCR) | [12,34] |
| papC | F: GTGGCAGTATGAGTAATGACCGTTA R: ATATCCTTTCTGCAGGGATGCAATA | 203 | ||
| iutA | F: ATCGGCTGGACATCATGGGAAC R: CGCATTTACCGTCGGGAACGG | 314 | ||
| kpsMTII | F: GCGCATTTGCTGATACTGTTG R: CATCCAGAC GATAAGCATGAGCA | 272 | ||
| fimH | F: TGCAGAACGGATAAGCCGTGG R: GCAGTCACCTGCCCTCCGGTA | 508 | ||
| chuA | F: GACGAACCA ACGGTCAGGAT R: TGCCGCCAGTACC AAAGACA | 279 | Phylogenetic group analysis (Triplex PCR) | [35] |
| yjaA | F: TGAAGTGTCAGGAGACGCT G R: ATGGAGAATGCGTTCCTCAAC | 211 | ||
| TspE4.C2 | F: GAGTAATGTCGGGGCATTCA R: CGCGCCAACAAAGTATTACG | 152 | ||
| ERIC | ERIC1: ATGTAAGCTCCTGGGGATTCAC ERIC2:AAGTAAGTGACTGGGGTG AGCG | Variable | Genotyping (ERIC PCR) | [36] |
| Sample Type | No. of Samples | No. (%) of Positive Samples | 95% Cl * |
|---|---|---|---|
| Carcass | 133 | 14 (10.53%) | 6.37–16.89 |
| Hide | 133 | 8 (6.02%) | 3.08–11.42 |
| Rectum | 133 | 9 (6.77%) | 3.60–12.36 |
| Knife | 18 | 1 (5.56%) | 0.99–25.76 |
| Holding pen | 12 | 1 (8.33%) | 1.49–35.39 |
| Workers’ hand | 12 | 3 (25.00%) | 8.89–53.23 |
| Water | 6 | 0 (0.00%) | 0.00–39.03 |
| City | |||
| Diyarbakır | 210 | 17 (8.10%) | 5.12–12.58 |
| Batman | 237 | 19 (8.02%) | 5.19–12.18 |
| Overall prevalence | 447 | 36 (8.00%) | 6.00–11.00 |
| Antibiotic Class | No. (%) Antibiotic-Resistant ExPEC Isolates * | |||||||
|---|---|---|---|---|---|---|---|---|
| Antibiotic Agents | Carcass (n = 14) | Hide (n = 8) | Rectum (n = 9) | Knife (n = 1) | Workers Hand (n = 3) | Holding Pen (n = 1) | Total (n = 36) | |
| Beta-Lactams | ||||||||
| Penicilins | Amoxicillin-Clavulate | 5 (35.7%) | 0 (0%) | 3 (33.3%) | 0 (0%) | 2 (66.7%) | 0 (0%) | 10 (27.8%) |
| Ampicillin | 10 (71.4%) | 3 (37.5%) | 5 (55.6%) | 1 (100%) | 3 (100%) | 0 (0%) | 22 (61.1%) | |
| Ampicilin-Sulbactam | 3 (21.4%) | 0 (0%) | 1 (11.1%) | 0 (0%) | 1 (33.3%) | 0 (0%) | 5 (13.9%) | |
| Cephalosporins | Cefazolin | 4 (28.6%) | 0 (0%) | 1 (11.1%) | 0 (0%) | 1 (33.3%) | 0 (0%) | 6 (16.7%) |
| Cefepime | 3 (21.4%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 3 (8.3%) | |
| Ceftazidime | 3 (21.4%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 3 (8.3%) | |
| Ceftriaxone | 4 (28.6%) | 0 (0%) | 1 (11.1%) | 0 (0%) | 1 (33.3%) | 0 (0%) | 6 (16.7%) | |
| Cefuroxime | 4 (28.6%) | 0 (0%) | 1 (11.1%) | 0 (0%) | 1 (33.3%) | 0 (0%) | 6 (16.7%) | |
| Fluoroquinolones | Ciprofloxacin | 8 (57.1%) | 2 (25%) | 2 (22.2%) | 0 (0%) | 2 (66.7%) | 0 (0%) | 14 (38.9%) |
| Levofloxacin | 8 (57.1%) | 2 (25%) | 2 (22.2%) | 0 (0%) | 2 (66.7%) | 0 (0%) | 14 (38.9%) | |
| Aminoglycosides | Gentamicin | 4 (28.6%) | 0 (0%) | 3 (33.3%) | 0 (0%) | 1 (33.3%) | 0 (0%) | 8 (22.2%) |
| Tetracyclines | Tigecycline | 7 (50%) | 0 (0%) | 2 (22.2%) | 1 (100%) | 1 (33.3%) | 0 (0%) | 11 (30.6%) |
| Sulfonamides | Trimethoprim-Sulfametxazole | 10 (71.4%) | 3 (37.5%) | 0 (0%) | 1 (100%) | 3 (100%) | 0 (0%) | 14 (38.9%) |
| No | Sample Type | City | ExPEC Virulence Gene * | Phylogeny | Phenotypic Antibiotic Resistance Profile | Multidrug Resistance |
|---|---|---|---|---|---|---|
| 1 | Carcass | Diyarbakır | papA, iutA, fimH | D | - | − |
| 2 | Carcass | Diyarbakır | papA, papC, iutA, fimH | D | AMP,CIP,LEV,TGC,TMP-SMX (5) | + |
| 3 | Carcass | Diyarbakır | papA, papC, iutA, fimH | D | AMP,CIP,LEV,TGC,TMP-SMX (5) | + |
| 4 | Carcass | Diyarbakır | papA, papC, iutA, fimH | D | AMP,CIP,LEV,TGC,TMP-SMX (5) | + |
| 5 | Carcass | Diyarbakır | papA, iutA, fimH | D | AMC,AMP,CIP,GEN,LEV,TGC,TMP-SMX (7) | + |
| 6 | Carcass | Diyarbakır | iutA, fimH | B2 | AMP,CFZ,FEP,CAZ,CRO,CXM,CIP,GEN,LEV,TMP-SMX (10) | + |
| 7 | Carcass | Diyarbakır | iutA, fimH | B2 | - | − |
| 8 | Carcass | Diyarbakır | iutA, fimH | D | AMC,AMP,SAM,CFZ,CRO,CXM,CIP,GEN,LEV,TGC,TMP-SMX (11) | + |
| 9 | Carcass | Diyarbakır | iutA, fimH | B2 | AMP,CFZ,FEP,CAZ,CRO,CXM,CIP,GEN,LEV,TMP-SMX (10) | + |
| 10 | Hide | Diyarbakır | iutA, fimH | B1 | AMP,CIP,LEV,TMP-SMX (4) | + |
| 11 | Hide | Diyarbakır | iutA, fimH | B2 | - | − |
| 12 | Rectum | Diyarbakır | iutA, fimH | B2 | AMP,CIP,GEN,LEV,TMP-SMX (5) | + |
| 13 | Rectum | Diyarbakır | iutA, fimH | B1 | - | − |
| 14 | Rectum | Diyarbakır | iutA, fimH | B1 | AMC,AMP,GEN (3) | + |
| 15 | Rectum | Diyarbakır | iutA, fimH | B1 | AMC,AMP,SAM,CFZ,CRO,CXM,CIP,GEN,LEV,TGC (10) | + |
| 16 | Worker hand | Diyarbakır | papC, iutA, fimH | D | AMP,CIP,LEV,TGC,TMP-SMX (5) | + |
| 17 | Worker hand | Diyarbakır | iutA, fimH | D | AMC,AMP,SAM,CFZ,CRO,CXM,CIP,GEN,LEV,TMP-SMX (10) | + |
| 18 | Carcass | Batman | iutA, fimH | B1 | AMC,AMP,SAM,CFZ,FEP,CAZ,CRO,CXM,CIP,LEV,TMP-SMX (11) | + |
| 19 | Carcass | Batman | iutA, fimH | ND | - | − |
| 20 | Carcass | Batman | iutA, fimH | B1 | AMC,AMP,SAM,TGC,TMP-SMX (5) | + |
| 21 | Carcass | Batman | iutA, fimH | D | TGC | + |
| 22 | Carcass | Batman | iutA, fimH | B2 | AMC,AMP,TMP-SMX (3) | + |
| 23 | Hide | Batman | iutA, fimH | D | - | − |
| 24 | Hide | Batman | iutA, fimH | A | - | − |
| 25 | Hide | Batman | papC, fimH | D | - | − |
| 26 | Hide | Batman | iutA, fimH | D | - | − |
| 27 | Hide | Batman | iutA, fimH | B1 | AMP,TMP-SMX (2) | − |
| 28 | Hide | Batman | iutA, fimH | B1 | AMP,CIP,LEV,TMP-SMX (4) | + |
| 29 | Rectum | Batman | iutA, fimH | D | - | − |
| 30 | Rectum | Batman | iutA, fimH | B1 | - | − |
| 31 | Rectum | Batman | papC, iutA, fimH | B2 | - | − |
| 32 | Rectum | Batman | iutA, fimH | B2 | AMP,TMP-SMX (2) | − |
| 33 | Rectum | Batman | iutA, fimH | B2 | AMC,AMP,TGC (3) | + |
| 34 | Knife | Batman | iutA, fimH | D | AMP,TGC,TMP-SMX (3) | + |
| 35 | Worker hand | Batman | iutA, fimH | ND | AMC,AMP,TMP-SMX (3) | + |
| 36 | Holding pen | Batman | iutA, fimH | A | - | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciftci, R.; Guran, H.S. Prevalence, Phylogenetic Distribution, Antimicrobial Resistance, and Genetic Relatedness of Extraintestinal Pathogenic E. coli (ExPEC) Strains Isolated from Beef Cattle and Slaughterhouse Environment. Vet. Sci. 2025, 12, 944. https://doi.org/10.3390/vetsci12100944
Ciftci R, Guran HS. Prevalence, Phylogenetic Distribution, Antimicrobial Resistance, and Genetic Relatedness of Extraintestinal Pathogenic E. coli (ExPEC) Strains Isolated from Beef Cattle and Slaughterhouse Environment. Veterinary Sciences. 2025; 12(10):944. https://doi.org/10.3390/vetsci12100944
Chicago/Turabian StyleCiftci, Resat, and Husnu Sahan Guran. 2025. "Prevalence, Phylogenetic Distribution, Antimicrobial Resistance, and Genetic Relatedness of Extraintestinal Pathogenic E. coli (ExPEC) Strains Isolated from Beef Cattle and Slaughterhouse Environment" Veterinary Sciences 12, no. 10: 944. https://doi.org/10.3390/vetsci12100944
APA StyleCiftci, R., & Guran, H. S. (2025). Prevalence, Phylogenetic Distribution, Antimicrobial Resistance, and Genetic Relatedness of Extraintestinal Pathogenic E. coli (ExPEC) Strains Isolated from Beef Cattle and Slaughterhouse Environment. Veterinary Sciences, 12(10), 944. https://doi.org/10.3390/vetsci12100944






